Abstract
The ATP-binding cassette (ABC) protein superfamily is one of the largest evolutionarily conserved families and is found in all kingdoms of life. The recent completion of the Leishmania genome sequence allowed us to analyze and classify its encoded ABC proteins. The complete sequence predicts a data set of 42 open reading frames (ORFs) coding for proteins belonging to the ABC superfamily, with representative members of every major subfamily (from ABCA to ABCH) commonly found in eukaryotes. Comparative analysis showed that the same ABC data set is found between Leishmania major and Leishmania infantum and that some orthologues are found in the genome of the related parasites Trypanosoma brucei and Trypanosoma cruzi. Customized DNA microarrays were made to assess ABC gene expression profiling throughout the two main Leishmania life stages. Two ABC genes (ABCA3 and ABCG3) are preferentially expressed in the amastigote stage, whereas one ABC gene (ABCF3) is more abundantly expressed in promastigotes. Microarray-based expression profiling experiments also revealed that three ABC genes (ABCA3, ABCC3, and ABCH1) are overexpressed in two independent antimony-resistant strains compared to the parental sensitive strain. All microarray results were confirmed by real-time reverse transcription-PCR assays. The present study provides a thorough phylogenic classification of the Leishmania ABC proteins and sets the basis for further functional studies on this important class of proteins.
ATP-binding cassette (ABC) proteins form the largest family of transmembrane proteins and are found in all living organisms (31). Most of these proteins are involved in the ATP-dependent transport of a variety of molecules across biological membranes, including amino acids, sugars, peptides, lipids, ions, and chemotherapeutic drugs (reviewed in reference 31). The functional significance of this family of proteins is reflected by the diversity of Mendelian and complex disorders associated with human genetic diseases involving ABC genes (reviewed in references 7 and 12). Proteins of the ABC superfamily contain in their sequences a strongly conserved nucleotide-binding domain (NBD) with three major motifs (13, 31). Along with the Walker A and B motifs found in many ATPase families, the NBD is composed of a characteristic ABC signature, the “C” motif, which is located between the two Walker motifs, just upstream of the Walker B site (33). Furthermore, in addition to the nucleotide-binding domains, ABC proteins involved in transport also contain transmembrane domains (TMD) composed of multiple transmembrane α-helices. A functional transporter appears to require a minimum of at least two NBDs coupled with two TMDs. Eukaryotic ABC transporter genes are organized either as full transporters containing two TMDs and two NBDs or as half transporters containing only one of each domain encoded on the same molecule (33). The half transporter molecules need to assemble as homodimers or heterodimers in the membrane to create a functional transporter. Some ABC proteins apparently not involved in transport activities but implicated in other conserved cellular processes do not have any TMDs and are composed of two NBDs fused on the same molecule. On the basis of gene structure similarity and homology in the NBD sequence, eukaryotic ABC proteins can be divided into eight different subfamilies (ABCA to ABCH), seven of which (ABCA to ABCG) are found in the human genome (2). The ABCH subfamily has been identified for the first time in Drosophila melanogaster (14).
In the protozoan parasite Leishmania, several ABC transporters have already been characterized. At least 20 different Leishmania species are responsible for a variety of clinical manifestations ranging from self-healing skin ulcers (e.g., Leishmania major) to life-threatening visceral diseases (e.g., L. donovani and L. infantum) (30). Human leishmaniasis has a prevalence of 12 million cases, an estimated population of 350 million at risk and an incidence of 2 million new cases annually (43). Leishmania has a relatively simple life cycle, with two main stages: the flagellated promastigotes, which are found in the gut of the insect vector, and the intracellular amastigotes, which live inside macrophages of the mammalian host. No effective vaccine is yet available against this parasite, and its control relies primarily on chemotherapy. Antimony containing compounds such as sodium stibogluconate (Pentostam) and N-methylglucamine (Glucantime) remain the mainstay against all forms of Leishmania infections (43), despite the emergence of antimony-resistant parasites now described on a frequent basis in several regions where this organism is endemic (27, 41, 48).
The first ABC protein identified in Leishmania is MRPA (PGPA) (45), a member of the ABCC subfamily able to confer antimony resistance by sequestering thiol-metal conjugates in an intracellular vesicle (40). MRPA was shown to be part of a large gene family with at least four other members (39). More recently, another ABCC member named PRP1 was shown to confer pentamidine resistance and antimony cross-resistance when overexpressed in L. major (11). Among the ABCB subfamily, MDR1 was found to be amplified in L. donovani mutants selected for vinblastine or daunomycin resistance and was shown to confer multidrug resistance by transfection experiments (9, 10, 25, 29, 36). Furthermore, the overexpression of another ABCB protein, named MDR2, was shown to decrease the accumulation of 5-fluorouracil in L. amazonensis (35). Finally, two proteins of the ABCA subfamily termed LtrABC1.1 (46) and LtrABCA2 (5) were recently implicated in phospholipid trafficking and reduced infectivity when overexpressed in L. tropica.
The availability of the genome sequences of L. major (34) and L. infantum (www.genedb.org) now enabled database mining and identification of the complete ABC protein data set in Leishmania. Transcriptional analysis of these family members could provide further insights on the role of these related proteins in the biology of the parasite and in drug resistance. Low-density DNA microarrays are well suited for that purpose given their relative simplicity and reproducibility. As a prelude to and resource for future functional genomic investigations of this relevant group of proteins, we present here a complete inventory and phylogenetic classification of the ABC proteins found in the protozoan parasite Leishmania. Moreover, microarray-based expression analyses were conducted to evaluate the ABC gene expression throughout parasite life stages and in parasites resistant to antimonial drugs.
MATERIALS AND METHODS
Database searches and sequence analyses.
The putative L. major ABC genes were retrieved from the GeneDB database (www.genedb.org) using the Motif search tool with the ABC signature (“C”) motif as query sequence. ORFs identified as encoding proteins containing such motif were then screened for the presence of flanking Walker A and Walker B conserved sequences. Furthermore, a series of BLAST searches were conducted on L. major predicted proteins in GeneDB by using the model ABC domain ABC_tran (accession PF00005) as a query. A series of BLAST searches were done on the assembled contig sequences of L. major Friedlin until no more new ABC coding genes could be identified. Finally, the complete L. major ABC gene data set was used as a query for multiple BLAST searches in the GeneDB database to reveal the complete sequence of the L. infantum orthologues. The amino acid sequence of nucleotide-binding domains of ABC proteins were extracted by performing Pfam searches at the Sanger Institute Web site (www.sanger.ac.uk/Software/Pfam/). Comparisons of the Leishmania ABC genes to homologues present in other sequenced eukaryotic genomes were made using BlastP at the NCBI Web site. Sequences were assigned as orthologues if they showed the highest score in BLAST search analyses and if they clustered on the same tree branch in phylogenetic analyses. The ABC sequences of Trypanosoma brucei and Trypanosoma cruzi were retrieved using the Pfam browse option at GeneDB and using BLAST searches with the PF00005 motif as a query.
Phylogenetic analyses.
Multiple sequence alignments were performed on the complete protein sequences or on the amino acid sequences of the ATP-binding domains by using CLUSTAL W (56) with the default settings. The two NBDs of full-length ABC proteins were treated independently for alignments. If needed, gaps were removed from the alignment by using the BioEdit software (28). The resulting multiple sequence alignments were subjected to analyses using the maximum-parsimony algorithm and the neighbor-joining algorithm (49) with the Poisson correction distance method of the MEGA package version 3.1 (38). The robustness of the neighbor-joining tree was assessed by 1,000 bootstrap resamplings.
Cell lines.
L. infantum amastigotes were kept in culture as axenic amastigotes at 37°C with 5% CO2 in MAA medium as described previously (52). L. infantum promastigotes were derived from the axenic amastigotes by culturing at 25°C in the same medium adjusted to pH 7.0. The L. infantum Sb2000.1 mutant selected for Sb(III) resistance was described previously (20). The L. infantum promastigote Sb4000.4 mutant was selected for Sb(III) resistance using a stepwise selection until it was resistant to 4,000 μM.
Array generation.
The microarrays were generated by arraying 51 70-mer oligonucleotides representing L. infantum ATP-binding cassette transporters and different controls (GAPDH and α-actin as housekeeping control genes). The L. infantum ABC protein coding gene (LinJ12.0790) was not identified at the time the arrays were generated. Furthermore, the BLASTn searches done on the L. infantum assembled contig sequences available at the time failed to distinguish between the ABCG1, ABCG2, and ABCG3 gene sequences. Therefore, an oligonucleotide recognizing these three ABCG genes was designed. Finally, the genes LinJ11.1200 and LinJ11.1230 are 100% identical, and thus one oligonucleotide had been synthesized for the two genes. The gene-specific oligonucleotides were chosen in the last 1,000 nucleotides of each ABC gene and were designed to recognize both L. major and L. infantum genes. BLAST searches against the Leishmania genome were conducted to assess the theoretical specificity of all designed 70-mers. Along with the ABC specific oligonucleotides, 15 genes of the Leishmania trypanothione biosynthesis pathway were represented on the array as PCR fragments generated as described previously (26). The 70-mer oligonucleotides and PCR fragments were dried in a 384-well plate prior of being resuspended in a 50% dimethyl sulfoxide solution. The DNA was denatured at 95°C and printed onto CMT-GAPS II (Corning) or Superchip (ERIE Scientific) slides by using a SDDC-2 Arrayer (Virtek) in a constant 60% humidity atmosphere. Each oligonucleotide and PCR fragment was printed in 24 replicates. Slides were UV cross-linked at 600 mJ (Stratalinker), and the printing quality control of each slide batch was assessed by performing TOTO-3 iodine staining (Molecular Probes).
RNA isolation.
Total RNA was isolated from 107 Leishmania cells during the mid-log phase of growth by using the TRIzol reagent (Invitrogen) as described by the manufacturer. The RNAs were treated with RNase-free DNase I (Ambion) to avoid any genomic contamination and further purified by using RNeasy columns (QIAGEN). The quality and quantity of RNA were assessed by using RNA 6000 Nano assay chips on a Bioanalyzer 2100 (Agilent Technologies). The major criterion for RNA integrity was the presence of three clear ribosomal peaks (18S, 24Sα, and 24Sβ) and the absence of RNA degradation.
RNA labeling and microarray hybridization.
The RNA conversion to cDNAs and the hapten-antibody-based microarray detection was done with the MICROMAX TSA labeling and detection kit (Perkin-Elmer) according to the supplier's recommendations. For each reaction, fluorescein-labeled and biotin-labeled cDNAs were synthesized from 2 μg of purified total RNA previously mixed with two exogenous mRNAs (CAB1 at 2 pg/μl and NAC1 at 2 pg/μl from Arabidopsis thaliana; Stratagene) added as external standards to adjust for variations in the incorporation efficiency of the modified nucleotides and for differences in first-strand cDNA synthesis reaction. The labeled cDNA were precipitated with isopropanol, washed with 70% ethanol, dried, and resuspended each in 15 μl of hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0,1% sodium dodecyl sulfate, 25 μg of salmon sperm DNA/ml, 460 μg of yeast tRNA/ml). Two differentially labeled target cDNAs were pooled for hybridization overnight at 42°C under a coverslip (ERIE Scientific) in a hybridization chamber (Corning). After hybridization, the microarrays were washed once with 2× SSC-0.1% sodium dodecyl sulfate preheated at 42°C and then with 1× SSC, 0.2× SSC, and 0.05× SSC successively at room temperature. Hybridization signals from fluorescein-labeled and biotin-labeled cDNA were sequentially detected on the microarray using Cy3-tyramide and Cy5-tyramide reagents, respectively. Each hybridization experiment was performed in four replicates using four independent RNA extractions and two dye-swapping procedures.
Microarray data processing and analysis.
Detection of the Cy5 and Cy3 signals was sequentially performed on a ScanArray 4000XL scanner (Perkin-Elmer) at a 5-μm resolution. The signal intensity data were extracted from the primary scanned images by using the GenePix Pro 6.0 software (Axon Technologies). The saturated spots, those with an unusual shape, and spots showing a signal-to-noise ratio of <3 were flagged as bad spots and were excluded from further analysis. Signal intensity data were exported into the GeneSpring 7.2 software. The local background value was subtracted from the intensity value of each spot. The Cy3/Cy5 ratio of each spot was normalized with Cy3/Cy5 ratios of the housekeeping genes (GAPDH and α-actin) and of the A. thaliana NAC1 and CAB1 spikes. Measures of significance were assessed by using the Student t test. Genes were considered statistically differentially expressed if they satisfied a P value cutoff of 0.05. (These data can be found in the supplemental material.)
Real-time reverse transcription-PCR (RT-PCR).
Primers were designed based on the ABC sequences of L. infantum using GeneRunner software. cDNA was synthesized from 2 μg of total RNA using Superscript II RNase H− reverse transcriptase (Invitrogen) and Oligo(dT)12-18 primers (Invitrogen) according to manufacturer's instructions. Real-time PCR was performed exactly as described previously (21). The relative amount of PCR products generated from each primer set was determined based on the threshold cycle (CT) value and amplification efficiencies and was normalized by dividing the values by the relative amount of the GAPDH gene used as a control.
RESULTS
The Leishmania ABC protein family.
The combined use of systematic BLAST searches against the L. major genome (www.genedb.org) using the ABC signature sequence as a query and of the analysis of the current annotated database allowed the identification of 42 ORFs that could be assigned to the ABC superfamily. This represents 0.5% of the total number of genes (approximately 8,300) in L. major. The retrieved sequences were designated as ABC proteins based on the presence of the characteristic ABC signature motif flanked by the Walker A and B motifs. Of the 42 ORFs, the L. major genome was found to encode 32 intrinsic membrane ABC proteins and 10 ABC proteins without any TMD. Among the intrinsic membrane proteins, full transporters were more frequent than half transporters (20:12). The number of predicted transmembrane spans varies from 10 to 14 for the full molecules, whereas in half molecules the number of putative transmembrane spans varies from 2 to 6. A detailed inventory of these ORFs, including names, GeneDB database accession numbers for the L. major and L. infantum orthologues, overall structural organization, and most similar ORFs found in the genome of the related parasites T. brucei and T. cruzi is presented in Table 1.
TABLE 1.
Leishmania ABC proteinsa
| Gene nomenclature | L. major | L. infantum | Leishmania alias | Leishmania gene topology | T. brucei | T. cruzi | Orthologous sequences |
|---|---|---|---|---|---|---|---|
| ABCA1 | LmjF2.0300 | LinJ02.0220 | [TM-NBD]2 | Tc00.1047053510045.20* | |||
| Tc00.1047053504881.50* | |||||||
| ABCA2 | LmjF11.1220 | LinJ11.1200 | [TM-NBD]2 | Tc00.1047053507099.80 | |||
| ABCA3 | LmjF11.1240 | LinJ11.1220 | [TM-NBD]2 | Tb11.02.3950 | Tc00.1047053503573.9* | ||
| Tc00.1047053504149.20* | |||||||
| ABCA4 | LmjF11.1250 | LinJ11.1230 | LtrABCA2 | [TM-NBD]2 | Tc00.1047053507099.80 | ||
| ABCA5 | LmjF11.1270 | LinJ11.1250 | [TM-NBD]2 | Tc00.1047053503573.9* | |||
| Tc00.1047053504149.20* | |||||||
| ABCA6 | LmjF11.1290 | LinJ11.1270 | [TM-NBD]2 | ||||
| ABCA7 | LmjF15.0760 | LinJ15.0790 | [TM-NBD]2 | ||||
| ABCA8 | LmjF27.0970 | LinJ27.0550 | LtrABC1.1 | [TM-NBD]2 | |||
| ABCA9 | LmjF27.0980 | LinJ27.0560 | [TM-NBD]2 | ||||
| ABCA10 | LmjF29.0620 | LinJ29.0770 | [TM-NBD]2 | Tb927.3.3730 | Tc00.1047053506989.30* | Yes | |
| Tc00.1047053510149.80* | Yes | ||||||
| ABCA11 | Tc00.1047053811725.80 | ||||||
| ABCB1 | LmjF25.0530 | LinJ25.0540 | TM-NBD | Tb11.03.0540 | Tc00.1047053507093.260 | Yes | |
| ABCB2 | LmjF26.2670 | LinJ26.2650 | LtrMDR2 | [TM-NBD]2 | |||
| ABCB3 | LmjF32.3080 | LinJ32.3640 | TM-NBD | Tb11.01.8700 | Tc00.1047053511021.70* | Yes | |
| Tc00.1047053503749.60* | Yes | ||||||
| Tc00.1047053511537.8* | Yes | ||||||
| ABCB4 | LmjF34.0990 | LinJ34.0950 | MDR1 | [TM-NBD]2 | |||
| ABCC1 | LmjF23.0210 | LinJ23.0230 | PGPC | [TM-NBD]2 | Tb927.8.2160 | Tc00.1047053506417.10 (pseudogene) | |
| ABCC2 | LmjF23.0220 | LinJ23.0240 | PGPB | [TM-NBD]2 | Tb927.8.2160 | Tc00.1047053506417.10 (pseudogene) | |
| ABCC3 | LmjF23.0250 | LinJ23.0290 | MRPA | [TM-NBD]2 | |||
| ABCC4 | LmjF31.1270 | LinJ31.1640 | PGPE | [TM-NBD]2 | |||
| ABCC5 | LmjF31.1280 | LinJ31.1650 | PGPD | [TM-NBD]2 | |||
| ABCC6 | LmjF31.1290 | LinJ31.1660 | [TM-NBD]2 | Tb927.4.4490 | Tc00.1047053508965.14* | ||
| Tc00.1047053457101.30* | |||||||
| Tc00.1047053507079.30* | |||||||
| Tc00.1047053509007.99 (pseudogene) | |||||||
| ABCC7 | LmjF31.1430 | LinJ31.1810 | PRP1 | [TM-NBD]2 | |||
| ABCC8 | LmjF34.0670 | LinJ34.0690 | [TM-NBD]2 | ||||
| ABCC9 | Tb927.4.2510 | Tc00.1047053506559.100* | |||||
| Tc00.1047053447255.29* | |||||||
| Tc00.1047053510231.29* | |||||||
| ABCD1 | LmjF27.0470 | LinJ27.0380 | TM-NBD | Tb11.03.0030 | Tc00.1047053506925.530 | Yes | |
| ABCD2 | LmjF31.0540 | LinJ31.1210 | TM-NBD | Tb927.4.4050 | Tc00.1047053508927.20* | Yes | |
| Tc00.1047053509237.30* | Yes | ||||||
| ABCD3 | LmjF33.1860 | LinJ33.1860 | TM-NBD | Tb11.02.0630 | Tc00.1047053510431.150 | Yes | |
| ABCE1 | LmjF21.0710 | LinJ21.0610 | [NBD]2 | Tb10.70.6290 | Tc00.1047053464879.9* | Yes | |
| Tc00.1047053508637.150* | Yes | ||||||
| Tc00.1047053511913.9 (pseudogene) | |||||||
| ABCF1 | LmjF3.0160 | LinJ03.0010 | [NBD]2 | Tb10.70.4330 | Tc00.1047053504867.20* | Yes | |
| Tc00.1047053510943.80* | Yes | ||||||
| ABCF2 | LmjF19.0800 | LinJ19.0630 | [NBD]2 | Tb10.61.0840 | Tc00.1047053508897.30 | Yes | |
| ABCF3 | LmjF33.0310 | LinJ33.0340 | [NBD]2 | Tb10.329.0040 | Tc00.1047053509105.130 | Yes | |
| ABCG1 | LmjF06.0080 | LinJ06.0080 | NBD-TM | Tb10.6k16.2900 | Tc00.1047053506249.70* | ||
| Tc00.1047053508231.190* | |||||||
| ABCG2 | LmjF06.0090 | LinJ06.0090 | NBD-TM | Tb10.6k16.2900 | Tc00.1047053506249.70* | ||
| Tc00.1047053508231.190* | |||||||
| ABCG3 | LmjF06.0100 | LinJ06.0100 | NBD-TM | Tb10.6k16.2900 | Tc00.1047053505249.70* | ||
| Tc00.1047053505231.190* | |||||||
| ABCG4 | LmjF15.0890 | LinJ15.0940 | NBD-TM | Tb09.160.4600 | Tc00.1047053506579.10* | Yes | |
| Tc00.1047053507241.39* | Yes | ||||||
| ABCG5 | LmjF23.0380 | LinJ23.0420 | NBD-TM | Tb927.8.2380 | Tc00.1047053504425.70* | Yes | |
| Tc00.1047053509331.200* | Yes | ||||||
| ABCG6 | LmjF36.2890 | LinJ36.3580 | NBD-TM | Tb10.8k15.3320 | Tc00.1047053507681.100 | Yes | |
| ABCH1 | LmjF11.0040 | LinJ11.0040 | NBD | Tc00.1047053510381.20* | Yes | ||
| Tc00.1047053506905.40* | Yes | ||||||
| ABCH2 | LmjF29.1640 | LinJ29.1960 | NBD | Tc00.1047053509617.80* | Yes | ||
| Tc00.1047053509669.30* | Yes | ||||||
| ABCH3 | LmjF30.1330 | LinJ30.1700 | NBD | Tb927.6.2810 | Tc00.1047053511501.30* | Yes | |
| Tc00.1047053511753.100* | Yes | ||||||
| Others | LmjF12.1190 | LinJ12.0790 | NBD | Tb927.1.4420 | Tc00.1047053506529.160* | Yes | |
| Tc00.1047053510885.70* | Yes | ||||||
| Others | LmjF32.2060 | LinJ32.2560 | TM-NBD | ||||
| Others | LmjF33.3040 | LinJ33.3040 | NBD | Tb927.2.5410 | Tc00.1047053508809.30* | Yes | |
| Tc00.1047053506619.90* | Yes | ||||||
| Others | LmjF33.3260 | LinJ33.3050 | NBD | Tb927.2.6130 | Tc00.1047053506817.20* | Yes | |
| Tc00.1047053507106.70* | Yes | ||||||
| Total | 42 | 42 | 22 | 28 |
Subfamily names given are according to the HUGO nomenclature (www.gene.ucl.ac.uk/nomenclature/), and gene accession numbers were taken from the GeneDB database (www.genedb.org). In the case of T. cruzi, all alleles were included because of the hybrid nature of this genome (19). An asterisk after the gene name indicates alleles representing the same T. cruzi ORF. The names of genes already studied in Leishmania are included as aliases. Potential orthologous sequences between the Tri-Tryp genomes are represented by a “Yes”in the last column. Tc00.1047053447255.29 and Tc00.1047053510231.29 probably represent two fragments of the same molecule. The same applies to the Tc00.1047053503749.60 and Tc00.1047053511537.8 sequences. TM, transmembrane domain. NBD, nucleotide-binding domain.
Classification of the Leishmania ABC proteins.
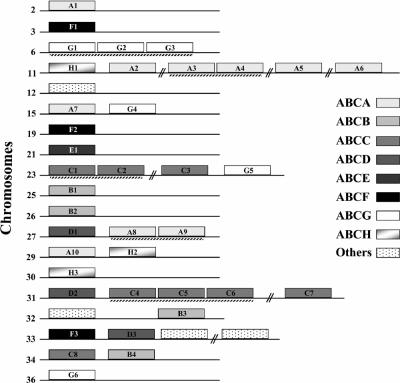
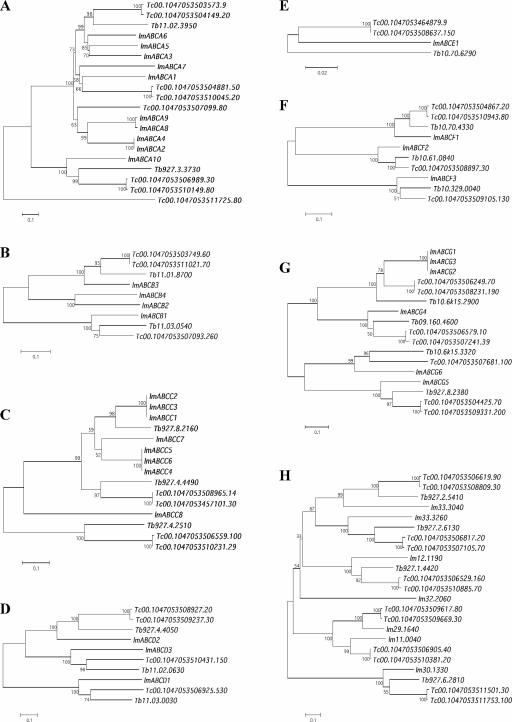
An alignment of the ATP-binding domains was generated and used for phylogenetic analysis to classify the 42 ABC proteins encoded by the Leishmania genome. Figure 1 displays a neighbor-joining tree clustering the 42 ABC proteins into eight subfamilies, seven of which are found in the human genome. As shown in Fig. 1, Leishmania has 10 members of the ABCA family (ABC1), 4 members of the ABCB family (MDR/TAP), 8 members of the ABCC family (MRP/CFTR), 3 members of the ABCD family (ALD), 1 member of the ABCE family (OABP), 3 members of the ABCF family (GCN20), and 6 members of the ABCG family (WHITE). We also found three members belonging to the ABCH family and four proteins not clustering to any of the above-mentioned subfamilies. The same clusters were obtained when a maximum-parsimony phylogenetic analysis was used (data not shown). The Leishmania ABC genes are largely dispersed in the genome and are found on 19 different chromosomes (Fig. 2). Most of the genes are found alone on chromosomes, but some are grouped in a head-to-tail fashion as part of two- or three-gene clusters (Fig. 2). All of these clusters contain members of the same subfamily: ABCA genes (chromosomes 11 and 27), ABCC genes (chromosomes 23 and 31), or ABCG genes (chromosome 6). Interestingly, the noncontiguous ABCA2 and ABCA4 genes of chromosome 11 share a 100% identity at the nucleotide level.
FIG. 1.
Phylogenetic analysis of the L. major ABC proteins. Nucleotide-binding domains were aligned by using CLUSTAL W. The resulting multiple alignment was subjected to phylogenetic analysis by using the neighbor-joining algorithm of the MEGA software. The reliabilities of each branch point were assessed by the analysis of 1,000 bootstrap replicates. A human representative of each mammalian subfamily was incorporated in the analysis to define each subfamily. The subfamily groups are shown as bars on the right of the tree, and I and II represent the N- and C-terminal NBDs of full-length proteins, respectively. lm, Leishmania major; hs, Homo sapiens.
FIG. 2.
Chromosomal location of the ABC protein coding genes in Leishmania. Chromosomes are indicated as horizontal lines, and ABC genes indicated as shaded blocks. A dashed line below genes indicates clusters of tandem genes. A break in the chromosome indicates members of the same subfamily that are not grouped in tandem.
Orthology of the ABC proteins in trypanosomatids.
We compared the ABC data set of L. major to the L. infantum contig sequences and to the related parasite T. brucei (6) and T. cruzi (19) genomes. Table 1 shows that an L. infantum orthologue was found for every L. major ABC gene with a conserved synteny. Furthermore, the L. infantum contig sequences did not reveal any additional ABC protein coding ORF that has not been previously found in the L. major genome. With its 42 ABC genes, Leishmania is the parasite with the largest ABC data set in the Trypanosomatidae family, with the T. cruzi and T. brucei genomes encoding 28 and 22 ABC proteins, respectively (Table 1). This variation in the number of ABC genes between the three organisms seems to be the result of an expansion of the ABCA, ABCC, and ABCG subfamilies that occurred in Leishmania after the split with the Trypanosoma lineage and of the loss of some ABC genes in T. brucei after speciation events within trypanosomes (Fig. 3). The ABCB, ABCD, ABCE, ABCF, and ABCH subfamilies seem to be evolutionary more stable in trypanosomatids, in particular between Leishmania spp. and T. cruzi (Fig. 3). Cross-organism searches revealed multiple homologous ORFs in the genomes of T. brucei and T. cruzi compared to Leishmania, but only some of these could be assigned as orthologous sequences by phylogenetic analyses. These clusters of orthologous genes, where homologues found in the three genomes are grouped on the same cluster in a phylogenetic tree, are observed in the most evolutionary stable subfamilies (Table 1 and Fig. 3). Indeed, only one cluster of orthologous genes was found between Leishmania, T. cruzi, and T. brucei in the ABCA subfamily (Table 1 and Fig. 3A), whereas two have been identified in the ABCB subfamily (Table 1 and Fig. 3B). No ABCC orthologue could be found between Trypanosoma and Leishmania. The ABCD, ABCE, and ABCF subfamilies show the highest level of orthology, with all of the Leishmania genes forming unambiguous pairs with their T. brucei and T. cruzi homologues. Within the ABCG subfamily, three of the six Leishmania genes paired with an orthologue in the Trypanosoma genomes (Table 1 and Fig. 3G). Several Leishmania ABC proteins either included in the ABCH class or still unclassified had orthologues in usually both T. brucei and T. cruzi. One exception is LmjF32.2060, which has no orthologue in the sequenced Trypanosoma genes (Fig. 3H).
FIG. 3.
Phylogenetic analysis of the ABC proteins in the Trypanosomatidae family. Phylogeny derived and displayed according to the procedure outlined in Fig. 1. Subfamilies ABCA to ABCH are shown. For the purpose of the figure, the unclassified proteins were incorporated in the ABCH subfamily tree. The coding sequences Tc00.1047053503749.60 and Tc00.1047053511537.8 in Table 1 probably represent two fragments of the same molecule, so only one sequence was incorporated in panel B. The same applies to the Tc00.1047053447255.29 and Tc00.1047053510231.29 sequences in panel C. lm, L. major; Tc, T. cruzi; Tb, T. brucei.
The Leishmania ABC proteins paralogous relationship.
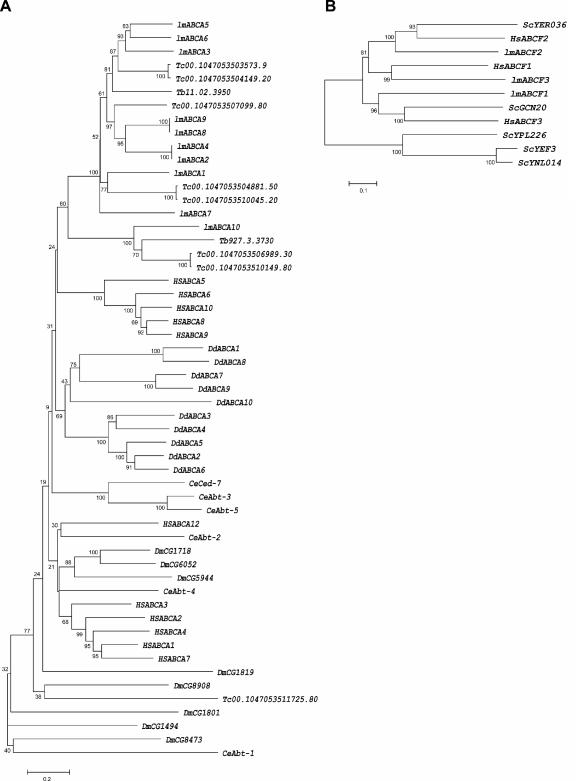
One way to address the putative function of a gene is to identify orthologues of known function. One such approach based on phylogenetic analysis was applied here in order to compare the ABC proteins of Leishmania to those of other eukaryotic genomes whose ABC gene inventory and classification have already been done (Homo sapiens, Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, and Dictyostelium discoideum). As exemplified by the ABCA subfamily in Fig. 4A, the Leishmania proteins cluster in phylogenetic branches along with their trypanosomatid homologues but apart from proteins encoded by the other eukaryotic genomes. This is also the case for the proteins of the ABCC and ABCG subfamilies (results not shown). Among the other subfamilies, very few clear orthologues have been identified by phylogenetic analyses. Among these are the L. major ABCB3 protein, which is found on a strong cluster with the yeast ATM1 protein, and the L. major ABCE1 protein, which is an orthologue of the yeast ABCE1-like YDR091c protein. The three L. major ABCF proteins were also successfully paired; ABCF1 is clustering with the human ABCF3 and the yeast GCN20 proteins, ABCF2 is clustering with the human ABCF2 protein among others, and ABCF3 is found on a cluster along with the human ABCF1 protein (Fig. 4B). In some instances (ABCB1, ABCD3, and ABCG5 proteins), Leishmania ABC proteins are clustering with more than one homologue of another organism (data not shown), rendering the assignment of a putative function more ambiguous.
FIG. 4.
Phylogenetic tree of selected ABC proteins with other eukaryotic ABC proteins. Phylogeny was derived and is displayed according to the procedure outlined in Fig. 1, except that complete protein sequences were used instead of just the NBD. (A) Phylogenetic tree of ABCA proteins in seven eukaryotic genomes. (B) Phylogenetic tree of the ABCF proteins in Leishmania, with human and yeast proteins. lm, L. major; Tc, T. cruzi; Tb, T. brucei; Hs, H. sapiens; Dm, D. melanogaster; Ce, C. elegans; Dd, D. discoideum, Sc, S. cerevisiae.
The Leishmania genome encodes seven ABC proteins not clustering with any of the mammalian ABC subfamilies. These proteins were referred to as members of the ABCH subfamily or as unclassified ABC proteins (“others” in Table 1), whether they clustered or not with representatives of the Drosophila ABCH proteins (Table 1). Most of them do not possess any transmembrane domain, with LmjF32.2060 being the only exception with six predicted amino-terminal membrane-spanning α-helices. Furthermore, with the exception of LmjF30.1330, which possesses a carboxy-terminal NBD and a degenerated amino-terminal NBD, all unclassified proteins possess a single NBD usually located at the carboxy termini of the molecule (Table 1). Phylogenetic analyses revealed that the LmjF11.0040 and LmjF29.1640 proteins are orthologues of the D. discoideum ABCh.2 and ABCh.1 proteins, respectively, and that they are found along with LmjF30.1330 on a cluster containing the D. melanogaster CG9990, CG6162, and CG11147 ABCH proteins (data not shown). All of the other unclassified proteins of Leishmania seem to be more related to bacterial ABC proteins by BLASTp searches.
Generation of DNA microarrays to study ABC gene expression in the two main life stages of Leishmania.
L. major and L. infantum have the same complement of ABC proteins (Table 1), and oligonucleotides were designed to detect the genes of both species. Changes in mRNA abundance were examined by customized DNA microarrays comprising 70-mer oligonucleotides representing the entire ABC data set of Leishmania in multiple replicates in addition to PCR fragments of several genes known to be involved in the trypanothione biosynthetic pathway or in antimony resistance (26). In order to estimate the accuracy of the protocol, arrays were first hybridized to Cy3- and Cy5-labeled cDNA generated from the same RNA preparation, and hybridization was found to be uniform (data not shown). Once the spotting, hybridization, and washing conditions were optimized and hybridization signals could be consistently reproduced, the arrays were used for the parallel analysis of ABC gene expression throughout the life cycle of L. infantum and in antimony-resistant mutants of L. infantum.
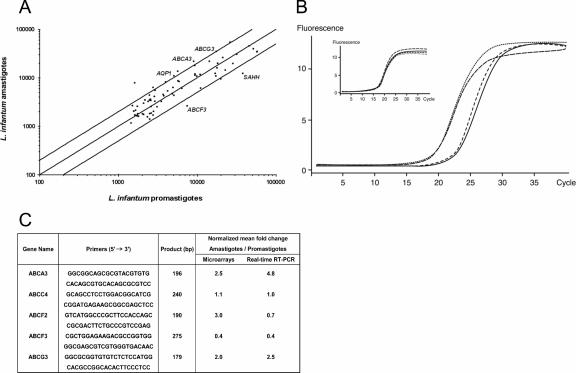
The ABC gene expression profiling between promastigotes and axenic amastigotes of L. infantum is shown in Fig. 5A. From the selected genes printed on the array, one ABC gene, ABCF3, appeared to be consistently upregulated in the promastigote stage (2.5-fold, P < 0.01). The expression of another gene, S-adenosylhomocysteine hydrolase (SAHH), is also increased in promastigotes (2.8-fold, P < 0.01). When we applied a cutoff of at least twofold differential expression, three ABC genes showed a consistent upregulation in the amastigote stage. One corresponds to the ABC transporter ABCA3 (2.5-fold, P < 0.01), the second one corresponds to the ABC transporter ABCG3 (2.0-fold, P < 0.01), and the last one corresponds to ABCF2 (3.0-fold, P < 0.02). The expression of the aquaglyceroporin AQP1 also seems to be increased in axenic amastigotes (2.0-fold, P < 0.02). No other ABC gene showed a significant differential expression in any of the two life stages when using a cutoff between 1.5 and 2. The putative differential expression of ABC genes was further confirmed by real-time RT-PCR, and the increased mRNA levels of ABCF3 in promastigotes and of ABCA3 (Fig. 5B) and ABCG3 in amastigotes were confirmed by this technique (Fig. 5C). The expression of ABCC4, a gene that was not found to be differentially regulated by microarrays, was also found to be similarly expressed between the two stages of Leishmania in real-time RT-PCR experiments (Fig. 5C). Although ABCF2 was found to be significantly overexpressed in the amastigote stage by microarrays, this could not be confirmed by real-time RT-PCR (Fig. 5C).
FIG. 5.
ABC gene expression in life stages of L. infantum. (A) Scatter plot of hybridization intensities representing the average of four independent experiments between L. infantum amastigote and promastigote parasites. The external lines indicate twofold differences, and genes whose expression differs significantly are indicated. The expression of genes represented by dots within the two external lines is considered similar in the two tested conditions, unless indicated otherwise. (B) Real-time PCR fluorescence curves representing duplicates of the ABCA3 gene amplification in promastigotes (dashed and full lines) and amastigotes (dotted and broken lines). The amplification curves of the GAPDH gene used for normalization are shown in the inset. (C) Validation of microarrays data by real-time RT-PCR. The gene ABCC4 was not found to be differentially expressed by microarrays and was used as a negative control. A mean of four independent experiments is shown for microarrays, and a mean of two independent experiments is shown for real-time RT-PCR.
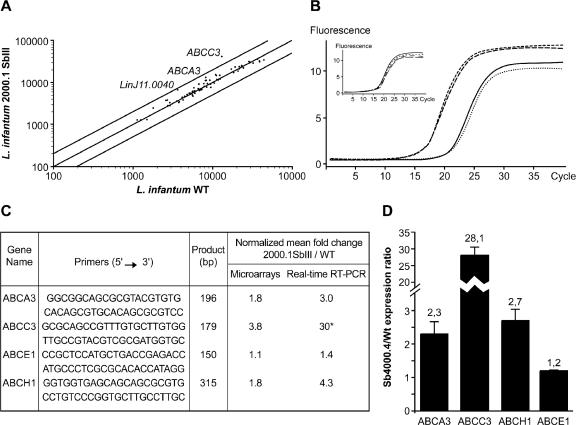
ABC gene expression in antimony-resistant mutants.
The role of the ABC transporter MRPA (ABCC3) in antimonial resistance is well established (20, 40). Since antimony resistance is multifactorial, it is possible that other ABC proteins could also be implicated (44). To test whether antimony resistance is correlated with the differential expression of other ABC genes, we performed microarray analyses using an antimony-sensitive L. infantum promastigote wild-type (WT) strain compared to an antimony-resistant L. infantum promastigote mutant strain (Sb2000.1). The ABC gene expression between the L. infantum-sensitive (WT) and -resistant (Sb2000.1) strains is shown in Fig. 6A. As expected, the majority of the spots aligned along the regression curve, suggesting that the genes represented by these spots are equally expressed in both samples. When applying a cutoff of at least twofold differential expression, the only gene found to be differentially expressed between the sensitive and the resistant strains is the one coding for the ABC transporter MRPA (ABCC3) (3.8-fold, P < 0.01). However, when applying a cutoff of between a 1.5- and a 2-fold difference in mRNA abundance, two more ABC genes, ABCA3 (1.8-fold, P < 0.02) and ABCH1 (1.8-fold, P < 0.01), showed a statistically significant higher expression in the antimony-resistant strain. Interestingly, the same mutant was recently studied with a Leishmania full genome array and, while the expression of several genes was modulated, the expression of the same three ABC protein genes was found to be upregulated (unpublished observations). The differential expression of these three genes in the antimony-resistant mutant was confirmed by real-time RT-PCR experiments (Fig. 6B and C). The overexpression of ABCA3 and ABCH1 is described for the first time in an antimony-resistant Leishmania strain. To test whether this is also the case in other antimony-resistant mutants, we analyzed by real-time RT-PCR the expression of these genes in the L. infantum Sb4000.4 promastigote line selected for resistance to Sb(III). Interestingly, MRPA was grossly overexpressed, but ABCA3 and ABCH1 were also found to be increased in this independent mutant (Fig. 6D). As observed in the Sb2000.1 mutant, the ABCE1 gene was not found to be differentially expressed in the Sb4000.4 line (Fig. 6D). Transfection of MRPA in L. infantum conferred a threefold increase in Sb(III) resistance. Despite the overexpression of ABCA3 and ABCH1 in two independent mutants, attempts to overexpress ABCA3 or ABCH1 in a L. infantum WT background did not result in antimony resistance, however (result not shown). The coexpression of either ABCA3 or ABCH1 with ABCC3 in a WT background did not confer higher resistance than ABCC3 overexpression alone (result not shown).
FIG. 6.
ABC gene expression in L. infantum antimony resistant mutants. (A) Scatter plot of hybridization intensities representing the average of four independent experiments between L. infantum WT and L. infantum Sb2000.1. The external lines indicate twofold differences, and genes whose expression differs significantly are indicated. The expression of genes represented by dots within the two external lines is considered similar in the two tested conditions, unless indicated otherwise. (B) Real-time PCR fluorescence curves representing duplicates of the MRPA gene amplification in the WT strain (dotted and full lines) and the Sb2000.1 strain (dashed and broken lines). The amplification curve of the GAPDH gene used for normalization is shown in the insert. (C) Validation of microarrays data by real-time RT-PCR. The gene ABCE1 was not found to be differentially expressed by microarrays and was used as a negative control. *, The curve for the gene was outside the standard curve, and the fold difference could be underestimated. A mean of four independent experiments is shown for microarrays, and a mean of two independent experiments is shown for real-time RT-PCR. (D) Real-time RT-PCR expression analysis of four ABC genes in L. infantum Sb4000.4 antimony-resistant mutant. The expression ratios of four ABC genes in the Sb4000.4 antimony-resistant mutant relative to the WT antimony-sensitive strain are shown. Results are a mean of three independent experiments performed from three different RNA preparations.
DISCUSSION
The completion of the Leishmania genome sequencing project (34) has allowed the characterization of 42 members of the Leishmania ABC gene family, a number considerably higher in comparison to related trypanosomatids (Table 1) or to the Apicomplexa parasites Plasmodium (fewer than 20 members) (22) and Toxoplasma gondii (20 members) (50). Phylogenetic analysis has allowed the classification of the Leishmania ABC proteins in different subfamilies using a nomenclature ABCA to ABCH adopted by the community of investigators working with eukaryotic ABC proteins (14). Another classification of the L. major ABC genes has also been done using the TCDB system (www.tcdb.org), and 45 Leishmania ABC proteins were classified in 22 different groups belonging to the 3.A.1 TCDB class (6). The discrepancy between the 42 proteins highlighted in the present study and the 45 pinpointed by a preliminary analysis of the L. major genome is explained by the inclusion of four proteins in the latter analysis (6) (LmjF21.0880, LmjF27.1700, LmjF29.0930, and LmjF34.2070) that are, according to all of the search criteria used here, unlikely to be ABC proteins, and by the omission of LmjF33.3040 (see Fig. 3H), which is clearly an ABC protein. For the remainder of this discussion, we shall use the ABCA to ABCH family nomenclature.
Leishmania has representative members of each subfamily, and the proportion of proteins in each of them seems relatively well conserved compared to other eukaryotes, especially for the ABCD, ABCE, and ABCF subfamilies (13). It is interesting that the Leishmania genome encodes for three proteins of the ABCH subfamily originally discovered in D. melanogaster (14). Interestingly, three of the four unclassified ABC proteins are well conserved in trypanosomatids (see Table 1). Homologues of this heterologous group of unclassified proteins have already been identified in other genomes (13, 15, 54) and appear to be more related to bacterial ABC proteins. A comparison of the ABC proteins of Leishmania to those of five other eukaryotic genomes revealed few unambiguous orthologous sequences. This is in agreement with previous studies that evaluated the frequency of orthologous ABC transporter pairs between different eukaryotic genomes (14, 53). Therefore, no detailed predictions of function in Leishmania ABC proteins can be drawn on the basis of phylogeny alone. As previously observed (14, 53), the frequency of orthologous sequences was found to be lower in the transporter subfamilies A, B, C, D, and G compared to those not involved in transport activities (subfamilies E and F).
The difference in ABC gene number in Leishmania compared to other eukaryotic microorganisms is mostly due to an expansion of the ABCA, ABCC, and ABCG genes, several of which have likely resulted from gene duplication events occurring after the split between the Leishmania and Trypanosoma lineages (Fig. 3). Several genes coding for members of those subfamilies have been found grouped in a head-to-tail fashion in the genome of Leishmania (Fig. 2), a finding consistent with gene duplication. The duplication events seem to have occurred recently given the high similarity between the paralogues (Fig. 3A, C, and G). This difference in the ABC complement between Leishmania and Trypanosoma could result from the different environments encountered by the parasitic organisms which translates into distinct needs and maintains a selective pressure on the ABC gene data set. The known function of members of the ABCA, ABCC, and ABCG subfamily in other eukaryotes allows tentative explanations as to why the expansion of these ABC proteins occurred in Leishmania compared to other eukaryotic microorganisms. The lifestyle and life cycle of Leishmania differs considerably from other eukaryotic pathogens. Indeed, it migrates and differentiates in the digestive tract of the sand fly vector and, in contrast to most eukaryotic pathogens, it remains within the phagolysosome of the host macrophages. These inhospitable habitats are likely to produce a number of toxic molecules for which the parasite has to defend itself. The ABCCs are best known to provide protection against a vast repertoire of xeno- and endobiotics and their glutathione, glucuronide, and sulfate conjugates (reviewed in reference 16). Thus, it is possible that the expansion of the ABCC subfamily in Leishmania (Table 1 and Fig. 3) is necessary to protect the cells in both life stages against toxic molecules. Interestingly, two ABCC proteins of Leishmania, MRPA (ABCC3) and PRP1 (ABCC7), have already been shown to have a role in dealing with xenobiotics (11, 40). Some eukaryotic members of the ABCG, most notably ABCG2, are involved in the detoxification of drugs and an expansion of the ABCG proteins in Leishmania (Fig. 3) (the parasite ABCG1, -2, and -3 are distantly related to the human ABCG2) may also help the parasites to cope with the variety of adverse conditions they are encountering.
Leishmania has 10 ABCA proteins, but these are absent in yeast (15) and in Apicomplexa parasites (50), and their number is considerably less in Trypanosoma. The ABCA and the ABCG proteins are best known for the transport of a variety of lipids, including cholesterol, plant sterols, sphingolipids, and phospholipids (37; reviewed in reference 55). At least two Leishmania ABCA transporters have demonstrated an ability to transport phospholipids (5, 46). There is considerable evidence that lipid transport and salvage are implicated in several aspects of Leishmania biology, and this provides a plausible explanation for the expansion of the ABCA and ABCG subfamilies. Indeed, the major sphingolipid of Leishmania, inositol phosphorylceramide, is absolutely required for metacyclogenesis (infectious promastigotes) in L. major (17, 60). Sphingolipid metabolites have been shown to modulate a wide variety of cellular events in a number of cells including Leishmania (18). Surprisingly, Leishmania amastigotes cannot make these sphingolipids and need to salvage them from the host (17, 59, 60) and ABC proteins could possibly be implicated in this phenomenon. Interestingly, and consistent with the proposal described above, our targeted DNA microarrays indicated that the expression of ABCA3 and ABCG3 was consistently upregulated in axenic amastigotes (Fig. 5). A homologue of the L. major ABCA3 in T. cruzi (TcABC1) was shown to be overexpressed in the epimastigote and amastigote stages (57), and the preferential expression in amastigotes of a gene coding for an ABCA protein has recently been reported in Leishmania using random genomic DNA microarrays (1). Glycosylinositol phospholipids are highly abundant and important in host-parasite interactions and were shown to be translocated across membranes (47), and this could also involve ABC proteins. Leishmania has specialized sterols including ergosterol (24) and, if ABCGs were required for their transport, it may also explain partly the expansion of ABCGs in Leishmania.
DNA microarrays are useful in the field of parasitology, as exemplified by numerous studies on stage specific expression and on drug resistance in Leishmania (1, 3, 20, 26, 32, 51). Furthermore, custom-made ABC transporter-targeted microarrays have already been used to study multidrug resistance in cancer cells (4, 23). The only ABC gene found to be preferentially expressed in the promastigote stage of Leishmania is ABCF3, an orthologue of the human ABCF1. The human ABCF1 protein copurifies with the eukaryotic initiation factor 2 and associates with the ribosomes in an ATP-dependent manner (58). Given the orthologous relationship with L. major ABCF3, one can expect a similar role in translation initiation for the Leishmania protein. Stage-specific regulation of gene expression in Leishmania is often controlled at the translation level (8, 42), and thus it is possible that genes expressed in a stage-specific manner such as ABCF3 are involved in stage-specific gene regulation.
The customized DNA microarrays were also used for the analysis of ABC gene expression in antimony-resistant mutants. The gene MRPA was found overexpressed in the Sb2000.1 resistant mutant, in agreement with previously reported results (20). ABCA3 and ABCH1 were also overexpressed in the Sb2000.1 mutant (Fig. 6C). Interestingly, the same three genes—MRPA, ABCA3, and ABCH1—were found to be overexpressed in an independent novel Sb(III) L. infantum resistant mutant (Fig. 6D). Furthermore, given the involvement in vesicular trafficking and exocytosis pathway of an ABCA3 homologue in T. cruzi (57), an attractive scheme to the antimony resistance pathway in the Sb2000.1 or Sb4000.4 mutants would be an increased sequestration of the thiol-Sb(III) complexes in intracellular vesicles by the overexpression of MRPA, followed by an increased exocytosis of those vesicles resulting from ABCA3 overexpression. Preliminary analysis of an antimony-resistant L. donovani field isolate also suggests that ABCA3 expression may be increased in these resistant parasites (unpublished observations). Overexpression of ABCA3 or ABCH1 in a WT background was not sufficient to observe an antimony resistance phenotype (result not shown), so the role, if any, in antimony resistance requires further experimental work. The ABCA3 and/or ABCH1 genes may contribute to resistance, however, in other contexts such as when other mutations are present as in the mutants Sb2000.1 or Sb4000.4.
Our work has highlighted, in contrast to other protozoan parasites, the magnitude of the ABC protein family of Leishmania. Given the multiple proteins found in the transporter subfamilies, Leishmania seems equipped to export a wide variety of compounds. The present study has also illustrated the usefulness of small targeted microarrays of 70-mer oligonucleotides. These ABC arrays will be useful tools for studying the physiological function of ABC proteins and to detect modulation in gene expression in Leishmania parasites resistant to various chemotherapeutic drugs.
Supplementary Material
Acknowledgments
We thank the Sanger Center, D. Smith and J. Mottram for the availability of the L. infantum sequences (www.sanger.ac.uk). We thank Éric Leblanc for help in initial phylogenetic analyses and Jean Morisette for using the GeneSpring 7.2 software.
This study was funded in part by the CIHR group GR14500 and operating grants to M.O., through a FQRNT group grant and through a Wellcome Trust-Burroughs Wellcome Fund new initiative in infectious diseases program grant to M.O., P.L. is a recipient of a CIHR studentship. B.P. and M.O. are Burroughs Wellcome Fund New Investigators and Scholar in Molecular Parasitology, and M.O. holds a Canada Research Chair in Antimicrobial Resistance.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Akopyants, N. S., R. S. Matlib, E. N. Bukanova, M. R. Smeds, B. H. Brownstein, G. D. Stormo, and S. M. Beverley. 2004. Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Mol. Biochem. Parasitol. 136:71-86. [DOI] [PubMed] [Google Scholar]
- 2.Allikmets, R., B. Gerrard, A. Hutchinson, and M. Dean. 1996. Characterization of the human ABC superfamily: isolation and mapping of 21 new genes using the expressed sequence tags database. Hum. Mol. Genet. 5:1649-1655. [DOI] [PubMed] [Google Scholar]
- 3.Almeida, R., B. J. Gilmartin, S. H. McCann, A. Norrish, A. C. Ivens, D. Lawson, M. P. Levick, D. F. Smith, S. D. Dyall, D. Vetrie, T. C. Freeman, R. M. Coulson, I. Sampaio, H. Schneider, and J. M. Blackwell. 2004. Expression profiling of the Leishmania life cycle: cDNA arrays identify developmentally regulated genes present but not annotated in the genome. Mol. Biochem. Parasitol. 136:87-100. [DOI] [PubMed] [Google Scholar]
- 4.Annereau, J. P., G. Szakacs, C. J. Tucker, A. Arciello, C. Cardarelli, J. Collins, S. Grissom, B. R. Zeeberg, W. Reinhold, J. N. Weinstein, Y. Pommier, R. S. Paules, and M. M. Gottesman. 2004. Analysis of ATP-binding cassette transporter expression in drug-selected cell lines by a microarray dedicated to multidrug resistance. Mol. Pharmacol. 66:1397-1405. [DOI] [PubMed] [Google Scholar]
- 5.Araujo-Santos, J. M., A. Parodi-Talice, S. Castanys, and F. Gamarro. 2005. The overexpression of an intracellular ABCA-like transporter alters phospholipid trafficking in Leishmania. Biochem. Biophys. Res. Commun. 330:349-355. [DOI] [PubMed] [Google Scholar]
- 6.Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, D. C. Bartholomeu, N. J. Lennard, E. Caler, N. E. Hamlin, B. Haas, U. Bohme, L. Hannick, M. A. Aslett, J. Shallom, L. Marcello, L. Hou, B. Wickstead, U. C. Alsmark, C. Arrowsmith, R. J. Atkin, A. J. Barron, F. Bringaud, K. Brooks, M. Carrington, I. Cherevach, T. J. Chillingworth, C. Churcher, L. N. Clark, C. H. Corton, A. Cronin, R. M. Davies, J. Doggett, A. Djikeng, T. Feldblyum, M. C. Field, A. Fraser, I. Goodhead, Z. Hance, D. Harper, B. R. Harris, H. Hauser, J. Hostetler, A. Ivens, K. Jagels, D. Johnson, J. Johnson, K. Jones, A. X. Kerhornou, H. Koo, N. Larke, S. Landfear, C. Larkin, V. Leech, A. Line, A. Lord, A. Macleod, P. J. Mooney, S. Moule, D. M. Martin, G. W. Morgan, K. Mungall, H. Norbertczak, D. Ormond, G. Pai, C. S. Peacock, J. Peterson, M. A. Quail, E. Rabbinowitsch, M. A. Rajandream, C. Reitter, S. L. Salzberg, M. Sanders, S. Schobel, S. Sharp, M. Simmonds, A. J. Simpson, L. Tallon, C. M. Turner, A. Tait, A. R. Tivey, S. Van Aken, D. Walker, D. Wanless, S. Wang, B. White, O. White, S. Whitehead, J. Woodward, J. Wortman, M. D. Adams, T. M. Embley, K. Gull, E. Ullu, J. D. Barry, A. H. Fairlamb, F. Opperdoes, B. G. Barrell, J. E. Donelson, N. Hall, C. M. Fraser, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 7.Borst, P., and R. O. Elferink. 2002. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71:537-592. [DOI] [PubMed] [Google Scholar]
- 8.Boucher, N., Y. Wu, C. Dumas, M. Dube, D. Sereno, M. Breton, and B. Papadopoulou. 2002. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 277:19511-19520. [DOI] [PubMed] [Google Scholar]
- 9.Chiquero, M. J., J. M. Perez-Victoria, O. V. F., J. M. Gonzalez-Ros, R. G. del Moral, J. A. Ferragut, S. Castanys, and F. Gamarro. 1998. Altered drug membrane permeability in a multidrug-resistant Leishmania tropica line. Biochem. Pharmacol. 55:131-139. [DOI] [PubMed] [Google Scholar]
- 10.Chow, L. M., A. K. Wong, B. Ullman, and D. F. Wirth. 1993. Cloning and functional analysis of an extrachromosomally amplified multidrug resistance-like gene in Leishmania enriettii. Mol. Biochem. Parasitol. 60:195-208. [DOI] [PubMed] [Google Scholar]
- 11.Coelho, A. C., S. M. Beverley, and P. C. Cotrim. 2003. Functional genetic identification of PRP1, an ABC transporter superfamily member conferring pentamidine resistance in Leishmania major. Mol. Biochem. Parasitol. 130:83-90. [DOI] [PubMed] [Google Scholar]
- 12.Dean, M. 2005. The genetics of ATP-binding cassette transporters. Methods Enzymol. 400:409-429. [DOI] [PubMed] [Google Scholar]
- 13.Dean, M., and T. Annilo. 2005. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6:123-142. [DOI] [PubMed] [Google Scholar]
- 14.Dean, M., A. Rzhetsky, and R. Allikmets. 2001. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11:1156-1166. [DOI] [PubMed] [Google Scholar]
- 15.Decottignies, A., and A. Goffeau. 1997. Complete inventory of the yeast ABC proteins. Nat. Genet. 15:137-145. [DOI] [PubMed] [Google Scholar]
- 16.Deeley, R. G., C. Westlake, and S. P. Cole. 2006. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol. Rev. 86:849-899. [DOI] [PubMed] [Google Scholar]
- 17.Denny, P. W., D. Goulding, M. A. Ferguson, and D. F. Smith. 2004. Sphingolipid-free Leishmania are defective in membrane trafficking, differentiation and infectivity. Mol. Microbiol. 52:313-327. [DOI] [PubMed] [Google Scholar]
- 18.Denny, P. W., and D. F. Smith. 2004. Rafts and sphingolipid biosynthesis in the kinetoplastid parasitic protozoa. Mol. Microbiol. 53:725-733. [DOI] [PubMed] [Google Scholar]
- 19.El-Sayed, N. M., P. J. Myler, D. C. Bartholomeu, D. Nilsson, G. Aggarwal, A. N. Tran, E. Ghedin, E. A. Worthey, A. L. Delcher, G. Blandin, S. J. Westenberger, E. Caler, G. C. Cerqueira, C. Branche, B. Haas, A. Anupama, E. Arner, L. Aslund, P. Attipoe, E. Bontempi, F. Bringaud, P. Burton, E. Cadag, D. A. Campbell, M. Carrington, J. Crabtree, H. Darban, J. F. da Silveira, P. de Jong, K. Edwards, P. T. Englund, G. Fazelina, T. Feldblyum, M. Ferella, A. C. Frasch, K. Gull, D. Horn, L. Hou, Y. Huang, E. Kindlund, M. Klingbeil, S. Kluge, H. Koo, D. Lacerda, M. J. Levin, H. Lorenzi, T. Louie, C. R. Machado, R. McCulloch, A. McKenna, Y. Mizuno, J. C. Mottram, S. Nelson, S. Ochaya, K. Osoegawa, G. Pai, M. Parsons, M. Pentony, U. Pettersson, M. Pop, J. L. Ramirez, J. Rinta, L. Robertson, S. L. Salzberg, D. O. Sanchez, A. Seyler, R. Sharma, J. Shetty, A. J. Simpson, E. Sisk, M. T. Tammi, R. Tarleton, S. Teixeira, S. Van Aken, C. Vogt, P. N. Ward, B. Wickstead, J. Wortman, O. White, C. M. Fraser, K. D. Stuart, and B. Andersson. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409-415. [DOI] [PubMed] [Google Scholar]
- 20.El Fadili, K., N. Messier, P. Leprohon, G. Roy, C. Guimond, N. Trudel, N. G. Saravia, B. Papadopoulou, D. Legare, and M. Ouellette. 2005. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob. Agents Chemother. 49:1988-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagnon, D., A. Foucher, I. Girard, and M. Ouellette. Stage-specific gene expression and cellular localization of two isoforms of the serine hydroxymethyltransferase in the protozoan parasite Leishmania. Mol. Biochem. Parasitol., in press. [DOI] [PubMed]
- 22.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillet, J. P., T. Efferth, D. Steinbach, J. Hamels, F. de Longueville, V. Bertholet, and J. Remacle. 2004. Microarray-based detection of multidrug resistance in human tumor cells by expression profiling of ATP-binding cassette transporter genes. Cancer Res. 64:8987-8993. [DOI] [PubMed] [Google Scholar]
- 24.Goad, L. J., G. G. Holz, and D. H. Beach. 1984. Sterols of Leishmania species: implications for biosynthesis. Mol. Biochem. Parasitol. 10:161-170. [DOI] [PubMed] [Google Scholar]
- 25.Gueiros-Filho, F. J., J. P. Viola, F. C. Gomes, M. Farina, U. Lins, A. L. Bertho, D. F. Wirth, and U. G. Lopes. 1995. Leishmania amazonensis: multidrug resistance in vinblastine-resistant promastigotes is associated with rhodamine 123 efflux, DNA amplification, and RNA overexpression of a Leishmania mdr1 gene. Exp. Parasitol. 81:480-490. [DOI] [PubMed] [Google Scholar]
- 26.Guimond, C., N. Trudel, C. Brochu, N. Marquis, A. E. Fadili, R. Peytavi, G. Briand, D. Richard, N. Messier, B. Papadopoulou, J. Corbeil, M. G. Bergeron, D. Legare, and M. Ouellette. 2003. Modulation of gene expression in Leishmania drug-resistant mutants as determined by targeted DNA microarrays. Nucleic Acids Res. 31:5886-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadighi, R., M. Mohebali, P. Boucher, H. Hajjaran, A. Khamesipour, and M. Ouellette. 2006. Unresponsiveness to glucantime treatment in Iranian cutaneous leishmaniasis due to drug-resistant Leishmania tropica parasites. PLoS Med. 3:e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 29.Henderson, D. M., C. D. Sifri, M. Rodgers, D. F. Wirth, N. Hendrickson, and B. Ullman. 1992. Multidrug resistance in Leishmania donovani is conferred by amplification of a gene homologous to the mammalian mdr1 gene. Mol. Cell. Biol. 12:2855-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herwaldt, B. L. 1999. Leishmaniasis. Lancet 354:1191-1199. [DOI] [PubMed] [Google Scholar]
- 31.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 32.Holzer, T. R., W. R. McMaster, and J. D. Forney. 2006. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol. Biochem. Parasitol. 146:198-218. [DOI] [PubMed] [Google Scholar]
- 33.Hyde, S. C., P. Emsley, M. J. Hartshorn, M. M. Mimmack, U. Gileadi, S. R. Pearce, M. P. Gallagher, D. R. Gill, R. E. Hubbard, and C. F. Higgins. 1990. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance, and bacterial transport. Nature 346:362-365. [DOI] [PubMed] [Google Scholar]
- 34.Ivens, A. C., C. S. Peacock, E. A. Worthey, L. Murphy, G. Aggarwal, M. Berriman, E. Sisk, M. A. Rajandream, E. Adlem, R. Aert, A. Anupama, Z. Apostolou, P. Attipoe, N. Bason, C. Bauser, A. Beck, S. M. Beverley, G. Bianchettin, K. Borzym, G. Bothe, C. V. Bruschi, M. Collins, E. Cadag, L. Ciarloni, C. Clayton, R. M. Coulson, A. Cronin, A. K. Cruz, R. M. Davies, J. De Gaudenzi, D. E. Dobson, A. Duesterhoeft, G. Fazelina, N. Fosker, A. C. Frasch, A. Fraser, M. Fuchs, C. Gabel, A. Goble, A. Goffeau, D. Harris, C. Hertz-Fowler, H. Hilbert, D. Horn, Y. Huang, S. Klages, A. Knights, M. Kube, N. Larke, L. Litvin, A. Lord, T. Louie, M. Marra, D. Masuy, K. Matthews, S. Michaeli, J. C. Mottram, S. Muller-Auer, H. Munden, S. Nelson, H. Norbertczak, K. Oliver, S. O'Neil, M. Pentony, T. M. Pohl, C. Price, B. Purnelle, M. A. Quail, E. Rabbinowitsch, R. Reinhardt, M. Rieger, J. Rinta, J. Robben, L. Robertson, J. C. Ruiz, S. Rutter, D. Saunders, M. Schafer, J. Schein, D. C. Schwartz, K. Seeger, A. Seyler, S. Sharp, H. Shin, D. Sivam, R. Squares, S. Squares, V. Tosato, C. Vogt, G. Volckaert, R. Wambutt, T. Warren, H. Wedler, J. Woodward, S. Zhou, W. Zimmermann, D. F. Smith, J. M. Blackwell, K. D. Stuart, B. Barrell, et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katakura, K., H. Fujise, K. Takeda, O. Kaneko, M. Torii, M. Suzuki, K. P. Chang, and Y. Hashiguchi. 2004. Overexpression of LaMDR2, a novel multidrug resistance ATP-binding cassette transporter, causes 5-fluorouracil resistance in Leishmania amazonensis. FEBS Lett. 561:207-212. [DOI] [PubMed] [Google Scholar]
- 36.Katakura, K., M. Iwanami, H. Ohtomo, H. Fujise, and Y. Hashiguchi. 1999. Structural and functional analysis of the LaMDR1 multidrug resistance gene in Leishmania amazonensis. Biochem. Biophys. Res. Commun. 255:289-294. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi, A., Y. Takanezawa, T. Hirata, Y. Shimizu, K. Misasa, N. Kioka, H. Arai, K. Ueda, and M. Matsuo. Efflux of sphingomyelin, cholesterol and phosphatidylcholine by ABCG1. J. Lipid Res., in press. [DOI] [PubMed]
- 38.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 39.Légaré, D., E. Hettema, and M. Ouellette. 1994. The P-glycoprotein-related gene family in Leishmania. Mol. Biochem. Parasitol. 68:81-91. [DOI] [PubMed] [Google Scholar]
- 40.Légaré, D., D. Richard, R. Mukhopadhyay, Y. D. Stierhof, B. P. Rosen, A. Haimeur, B. Papadopoulou, and M. Ouellette. 2001. The Leishmania ABC protein PGPA is an intracellular metal-thiol transporter ATPase. J. Biol. Chem. 276:26301-26307. [DOI] [PubMed] [Google Scholar]
- 41.Lira, R., S. Sundar, A. Makharia, R. Kenney, A. Gam, E. Saraiva, and D. Sacks. 1999. Evidence that the high incidence of treatment failures in Indian Kala-Azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J. Infect. Dis. 180:564-567. [DOI] [PubMed] [Google Scholar]
- 42.McNicoll, F., M. Muller, S. Cloutier, N. Boilard, A. Rochette, M. Dube, and B. Papadopoulou. 2005. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 280:35238-35246. [DOI] [PubMed] [Google Scholar]
- 43.Murray, H. W., J. Berman, C. R. Davies, and N. G. Saravia. 2005. Advances in leishmaniasis. Lancet 366:1561-1577. [DOI] [PubMed] [Google Scholar]
- 44.Ouellette, M., J. Drummelsmith, and B. Papadopoulou. 2004. Leishmaniasis: drugs in the clinic, resistance, and new developments. Drug Resist. Update 7:257-266. [DOI] [PubMed] [Google Scholar]
- 45.Ouellette, M., F. Fase-Fowler, and P. Borst. 1990. The amplified H circle of methotrexate-resistant Leishmania tarentolae contains a novel P-glycoprotein gene. EMBO J. 9:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parodi-Talice, A., J. M. Araujo, C. Torres, J. M. Perez-Victoria, F. Gamarro, and S. Castanys. 2003. The overexpression of a new ABC transporter in Leishmania is related to phospholipid trafficking and reduced infectivity. Biochim. Biophys. Acta 1612:195-207. [DOI] [PubMed] [Google Scholar]
- 47.Ralton, J. E., K. A. Mullin, and M. J. McConville. 2002. Intracellular trafficking of glycosylphosphatidylinositol (GPI)-anchored proteins and free GPIs in Leishmania mexicana. Biochem. J. 363:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rojas, R., L. Valderrama, M. Valderrama, M. X. Varona, M. Ouellette, and N. G. Saravia. 2006. Resistance to antimony and treatment failure in human Leishmania (viannia) infection. J. Infect. Dis. 193:1375-1383. [DOI] [PubMed] [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 50.Sauvage, V., J. M. Millot, D. Aubert, V. Visneux, M. Marle-Plistat, J. M. Pinon, and I. Villena. 2006. Identification and expression analysis of ABC protein-encoding genes in Toxoplasma gondii: Toxoplasma gondii ATP-binding cassette superfamily. Mol. Biochem. Parasitol. 147:177-192. [DOI] [PubMed] [Google Scholar]
- 51.Saxena, A., E. A. Worthey, S. Yan, A. Leland, K. D. Stuart, and P. J. Myler. 2003. Evaluation of differential gene expression in Leishmania major Friedlin procyclics and metacyclics using DNA microarray analysis. Mol. Biochem. Parasitol. 129:103-114. [DOI] [PubMed] [Google Scholar]
- 52.Sereno, D., G. Roy, J. L. Lemesre, B. Papadopoulou, and M. Ouellette. 2001. DNA Transformation of Leishmania infantum axenic amastigotes and their use in drug screening. Antimicrob. Agents Chemother. 45:1168-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheps, J. A., S. Ralph, Z. Zhao, D. L. Baillie, and V. Ling. 2004. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome. Biol. 5:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szeto, A. C., J. Perez-Rosado, I. Ferrer-Rodriguez, J. Vega, C. Torruella-Thillet, and A. E. Serrano. 2004. Identification and expression analysis of ABC genes in Plasmodium yoelii and P. berghei. Parasitol. Res. 92:1-11. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi, K., Y. Kimura, K. Nagata, A. Yamamoto, M. Matsuo, and K. Ueda. 2005. ABC proteins: key molecules for lipid homeostasis. Med. Mol. Morphol. 38:2-12. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torres, C., F. J. Perez-Victoria, A. Parodi-Talice, S. Castanys, and F. Gamarro. 2004. Characterization of an ABCA-like transporter involved in vesicular trafficking in the protozoan parasite Trypanosoma cruzi. Mol. Microbiol. 54:632-646. [DOI] [PubMed] [Google Scholar]
- 58.Tyzack, J. K., X. Wang, G. J. Belsham, and C. G. Proud. 2000. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 275:34131-34139. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, K., F. F. Hsu, D. A. Scott, R. Docampo, J. Turk, and S. M. Beverley. 2005. Leishmania salvage and remodeling of host sphingolipids in amastigote survival and acidocalcisome biogenesis. Mol. Microbiol. 55:1566-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, K., M. Showalter, J. Revollo, F. F. Hsu, J. Turk, and S. M. Beverley. 2003. Sphingolipids are essential for differentiation but not growth in Leishmania. EMBO J. 22:6016-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.