Abstract
Reverse transcriptase (RT) and integrase (IN) play a central role in the replication and transposition of retroelements. Increasing evidence suggests that the interaction between these two enzymes is functional and plays an important role in replication. In the yeast Saccharomyces cerevisiae retrotransposon Ty1, the interaction of IN with RT is critical for the formation of an active conformation of RT. We show here that the RT associated with VLPs is active only if it is in close interaction with IN. To probe the IN-RT cis-trans relationship, we have used a complementation assay based on coexpressing two transposons. We show that IN acts in cis to activate RT and that a functional integrase provided in trans is not able to complement replication and transposition defects of IN deletion or IN active-site mutant elements. Our data support a model in which IN not only interacts closely with RT during reverse transcription but also remains associated with RT during the formation of the preintegrative complex.
Two key catalytic enzymes, integrase (IN) and reverse transcriptase (RT), play a central role in the replication and transposition of retroviral and retrotransposon cDNA. Increasing evidence suggests that functional and physical interactions between these two enzymes occur during replication (15, 28, 40, 43). In some retroviruses IN is an integral part of the active enzyme; it is an α/β heterodimer composed of a short RT (α) polypeptide and an incompletely processed RT-IN (β) intermediate in the avian leukosis viruses (18, 36) and an oligomer of the α3/β type in human T-cell leukemia virus type 1 (34). It has also been proposed that the active form of the yeast Saccharomyces cerevisiae retrotransposon Ty3 RT is an α/β heterodimer (27). In other retroviruses, RT and IN are fully separated by proteolytic processing during virion maturation, but mutations or deletions in the nonconserved domain of IN affect several stages of viral replication other than integration, including the initiation of reverse transcription, the level of cDNA, 3′-end DNA processing, or nuclear entry (19, 28, 40, 43). Coimmunoprecipitation experiments with antibodies to RT or IN have demonstrated that the human immunodeficiency virus type 1 (HIV-1) or murine leukemia virus proteins interact physically in vitro (16, 17, 32). Recently, Hehl et al. (15) have mapped the domains of interaction of HIV-1 IN and RT in the C-terminal domain of IN and in the finger-palm domain and carboxyl-terminal half of the connection subdomain of RT. Furthermore, Zhu et al. (43) have shown that mutation of an N-terminal Cys residue of HIV-1 IN (C130S) that disrupts the interaction between IN and RT abolishes the ability of the virus to initiate endogenous reverse transcription.
In the yeast Saccharomyces cerevisiae retrotransposon Ty1, interactions between IN and RT are also important for the function of RT (37, 39). In vitro, an active Ty1 recombinant protein can be obtained only if amino acid residues encoded by the C-terminal region of IN are fused to the N-terminal domain of RT (37). In vivo, the IN and RT proteins of Ty1 are expressed and assembled in the virus-like particles (VLPs) as part of a large Gag-Pol-p199 precursor protein (1, 14, 22, 23, 42). After assembly of the VLPs, the Gag-Pol precursor is processed by the pol-encoded protease to liberate the mature Gag-p45, PR-p20, IN-p71, and RT-RH-p63 proteins. The main RT species identified in Ty1 VLPs is a mature p63 protein (63 kDa), but Ty1 RT retains its activity when it is fused to the IN domain in PR-IN-RT and IN-RT fusion proteins (22) or in the Gag-Pol-p199 precursor (42). In a recent study we used IN deletion mutants to investigate the role of IN on RT activity in Ty1 VLPs (39). We identified two domains in the C-terminal region of the Ty1 integrase that affect RT activity in vivo. Deletion of a domain spanning amino acid residues 233 to 520 increases the exogenous specific activity of RT up to 20-fold, whereas removal of a domain rich in acidic amino acid residues between residues 521 and 607 decreases its activity.
We have suggested that interaction between the acidic residues of IN and a basic domain in the N-terminal region of RT could be critical for its correct folding and for the formation of an active conformation of the enzyme. Folding of a protein is a hierarchical process, and the secondary structure of a protein is determined primarily by interaction among amino acid residues that are close in sequence (30). The order of the domains encoded within the pol gene of Ty1 is protease-integrase-RT (PR-IN-RT); thus, it is possible that during synthesis of the Gag-Pol-p199 precursor, the presence of the C-terminal region of IN which is synthesized before RT is necessary in cis to induce proper folding of the RT by facilitating interaction between the acidic domain of IN and the basic domain of RT. To probe the IN-RT cis-trans relationship, we used a complementation assay based on coexpressing two transposons. We show here that the C-terminal domain of IN acts in cis to activate RT. A functional integrase provided in trans is not able to rescue the replication and transposition defects of IN deletion or IN active-site mutant elements. Our data support a model in which IN not only interacts closely with RT during reverse transcription but also remains associated with the RT during formation of the preintegrative complex (PIC).
MATERIALS AND METHODS
Yeast strains and plasmid DNA.
Yeast strain LV47 (MATa ura3Δ851 trp1Δ63 his3Δ200 spt3-10 GAL+) was kindly provided by P. Lesage (33). Yeast strain YH50 (MATα ho ura3-167 trp1Δ1 leu2-3 his3Δ200 spt3-202) and the isogenic rad52::LEU2 strain AGY49 were kindly provided by A. Gabriel. All strains are spt3; they lack a transcription factor required for normal expression of genomic Ty1 elements. This eliminates endogenous Ty1 transcription and thus potential trans-complementation of plasmid-born elements.
pGTy1-H3mHIS3AI and pJEF1105 plasmids were kindly provided by D. Garfinkel and J. Boeke. The pGTy1-H3mHIS3AI plasmid contains the Ty1-H3 element, the expression of which is under the control of the inducible GAL1 promoter. It is marked with a HIS3 reporter gene in the antisense orientation. An artificial intron (AI) in the sense orientation has been inserted in the HIS3 gene. The pJEF1105 plasmid contains the Ty1-H3 element, the expression of which is under the control of the inducible GAL1 promoter. It is marked with a neo gene.
Ty1 VLP purification.
Ty1 VLPs were isolated from yeast strain LV47 cotransformed with a TRP1-based pGPol(−)hisAI (an RT active-site mutant of pGTy1-H3mHIS3AI) and the URA3-based wild-type plasmid pJEF1105 (pJEFWTneo) (8) or IN mutant versions of this plasmid (pJEFΔINneo or pJEFMutINneo) (Fig. 1). The VLPs were purified by using a method described by Eichinger and Boeke (8). RT activity was assayed in the following reaction mix: 4 μl of native or dissociated VLPs was mixed with 16 μl of assay mixture, followed by incubation for 60 min at room temperature (22 to 24°C). The reaction mixture (20 μl) contained final concentrations of 50 mM Tris-HCl (pH 7.8), 40 mM KCl, 20 mM MgCl2, 0.05% NP-40, 1.5 × 10−2 mM dGTP, 8 mM β-mercaptoethanol, 0.01 U of oligo(dG)/poly(rC), and 1 μCi of [α-32P]dGTP. Incorporation of 32P-radiolabeled dGTP into high-molecular-weight poly(dG) was determined by scintillation counting; aliquots of the reaction mixture were spotted onto DE81 filters (Whatman), and the filters were washed three times in 5% Na2HPO4 to remove unincorporated [α-32P]dGTP, washed once in deionized water, and dried after one ethanol wash before counting.
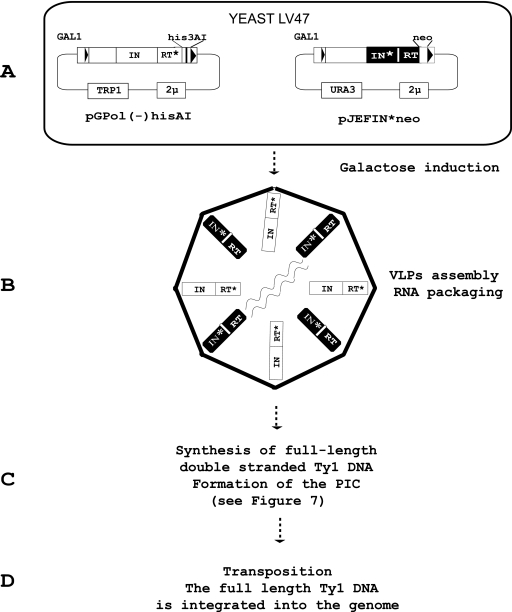
FIG. 1.
Assay for Ty1 activity. (A) The yeast strain LV47 was transformed with two plasmids: pGPol(−)hisAI, a TRP1-based derivative of pGTy1-H3mHIS3AI with an active-site mutation in RT (RT*), and pJEFWTneo, a wild-type URA3-based plasmid or mutant derivatives of this plasmid with deletions or active-site mutations in the IN gene (IN*). (B) On galactose induction, VLPs are formed and assembled in the cytoplasm. Two molecules of genomic RNA are packaged into the VLPs. (C) The genomic Ty1 RNA is reverse transcribed by the IN/RT complex. The full-length double-stranded DNA which is synthesized forms a PIC. (D) The PIC is then transported into the nucleus. IN insert the Ty1 DNA into the host genome (transposition).
VLP dissociation.
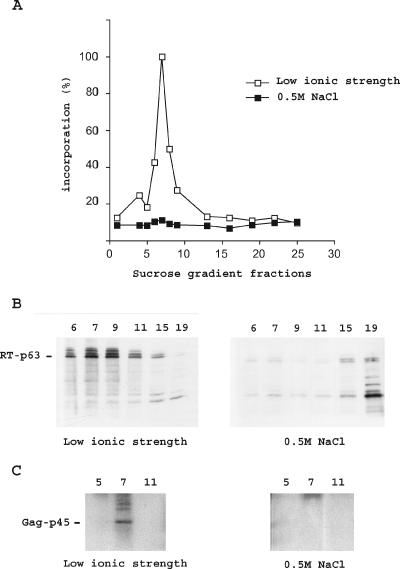
VLPs were dissociated by adding NaCl to a final concentration of 0.5 M. Dissociation in 0.5 M NaCl was checked by fractionating the VLPs on a sucrose step gradient containing the same concentration of NaCl. RT activity assays and immunoblot analyses with RT and Gag (TYA) antibodies were performed on fractions of the density gradient. As shown in Fig. 2, no peak of reverse transcriptase activity was observed with salt-treated VLPs (see Results). Most of the RT proteins remained at the top of the gradient. Because the VLPs are dissociated in high salt concentrations and do not protect RT from protease degradation, a large fraction of RT is degraded during the 3-h centrifugation run. In native VLPs, purified in a low-ionic-strength buffer, the VLP proteins are protected from proteolytic degradation, probably because proteases do not gain access to the interior of the VLPs (5). In this case, the RT and Gag proteins cofractionate with the RT activity.
FIG. 2.
Dissociation of VLPs by salt. (A) VLP-associated RT activity in low-ionic-strength buffer and in a buffer containing 0.5 M NaCl. Wild-type VLPs were prepared as described by Eichinger and Boeke (8). One half of the VLP preparation was fractionated on a step gradient containing a low-ionic-strength buffer. NaCl was added to the other half to a final concentration of 0.5 M. The salt-treated VLPs were fractionated on a sucrose step gradient containing the same concentration of NaCl. RT activity was assayed on fractions of the density gradient. (B and C) Immunoblotting of gradient fractions using anti-RT (B) and anti-Gag (C) antibodies.
DNA oligonucleotides and labeling.
The DNA oligonucleotides were purchased from Thermo Electron (Ulm, Germany) and labeled at their 5′ termini with 32P using [γ-32P]ATP and T4 polynucleotide kinase.
Plasmid construction and mutagenesis.
Deletion mutants of pJEF1105 were made as described previously (39). The QuikChange multi-site-directed mutagenesis kit from Stratagene was used to mutagenize the active site of IN. The oligonucleotides used for generating the mutations were as follows (mutations that also created new restriction sites are indicated in boldface, and new restriction sites are underlined): 5′-CGAACCCTTTCAATACCTACATACTGCAATATTTGGTCCAGTTCACAACCTACC-3′, 5′-CAGGCCAGTGTCTTGGTTATACAAATGGCTCGAGGTTCTGAGTATACTAAC-3′, and 5′-GCGGATTCCCGAGCACATGGAGTCGCTGCGCGCCTAAACCGTACC-3′. Mutants were identified by digestion with SspI, XhoI, or BssHII. The TRP1-marked RT active-site mutant reporter plasmid, pGPol(−), was constructed by subcloning a PvuII-AflII-digested fragment from AGES4 (35) bearing the RT active-site mutation into similarly digested pGTy1-H3mHIS3AI plasmid. Since PvuII-AflII digestion removed a 3.47-kb PvuII-PvuII fragment, the resulting plasmid was digested with PvuII and ligated to the PvuII-PvuII fragment. The resulting plasmid with the correct orientation of the PvuII fragment was digested with EagI-ApaI and ligated to a similarly digested fragment containing the TRP1 gene.
Iterative primer extension.
To detect minus-strand and plus-strand strong-stop DNA replication intermediates, iterative primer extension was carried out with 5′-end-labeled strand-specific oligonucleotide primers (38). Nucleic acids extracted from VLPs were incubated with the labeled primer in a 20-μl volume using a PCR buffer (10 mM Tris-HCl [pH 8.8], 50 mM KCl, 0.1% Triton X-100, 2 mM MgCl2), along with 2 U of Taq polymerase. Primer extension products were generated by 30 cycles of denaturation (30 s at 95°C), annealing (30 s at 46°C), and extension (60 s at 72°C). At the end of the reaction, 10% formamide dye was added to the reaction mixture. Products were denatured by heating at 90°C for 2 min prior to loading them on an 8% polyacrylamide-8 M urea denaturing gel and then analyzed by phosphorimaging. The primers used to detect +sssDNA and −sssDNA were 5′-CAATCCTTGCGTTTCAGCTTCCAC-3′ (complementary to Ty1-H3 positions 5716 to 5697) and 5′-GGAGAACTTCTAGTATATTCTGTATACC-3′ (Ty1-H3 positions 243 to 270 or 5827 to 5854).
Southern blot analysis.
Ty1 VLPs were prepared from 600-ml cultures and purified on sucrose step gradients as described by Eichinger and Boeke (8). The VLPs were concentrated by centrifugation at 40,000 × g for 1 h. The VLP pellet was resuspended in 300 μl of Tris-EDTA (TE), and VLP nucleic acids were prepared by phenol-chloroform extraction, precipitated in ethanol, and resuspended in 75 μl of TE. The nucleic acids were treated with 10 μg of RNase A for 15 min at 37°C, reextracted with phenol and chloroform, precipitated in ethanol, and resuspended in 75 μl of TE. DNA was quantitated by spectrophotometry (NanoDrop ND-1000). A total of 10 μg of VLP DNA was digested with EcoRI or Eco91I (BstEII) and separated on a 1% agarose gel. The nucleic acids were transferred on a Hybond N+ membrane (Amersham) and probed either with a XhoI-HindIII fragment of the Tn903 neo gene or a SacI-NheI HIS fragment of the pGTy1-H3mHIS3AI plasmid. The probes were labeled with [α-32P]dGTP by random priming with the Megaprime DNA labeling system of Amersham. Hybridization of the probe was detected with a phosphorimager screen.
Transposition assay.
Yeast cells were grown at 30°C overnight in yeast nitrogen base medium without amino acids (YNB) supplemented with 2% glucose (wt/vol) and the required amino acids. Cells were washed in water and diluted in YNB medium supplemented with 2% galactose (wt/vol) and the required amino acids and then grown for 2 days at 22°C to induce transposition. Aliquots were plated onto YNB glucose plates without histidine at 30°C to end the transposition induction and to determine the fraction of His+ prototrophs. The cultures were titrated on YPD plates. For the qualitative transposition assay, cells with an optical density at 600 nm of 1 to 1.5 were diluted 10-fold. Then, 10 μl of each dilution was spotted onto YNB glucose plates without histidine and incubated for 2 days at 30°C.
RESULTS
An active Ty1 RT can be obtained from VLPs provided the interaction between RT and IN is maintained.
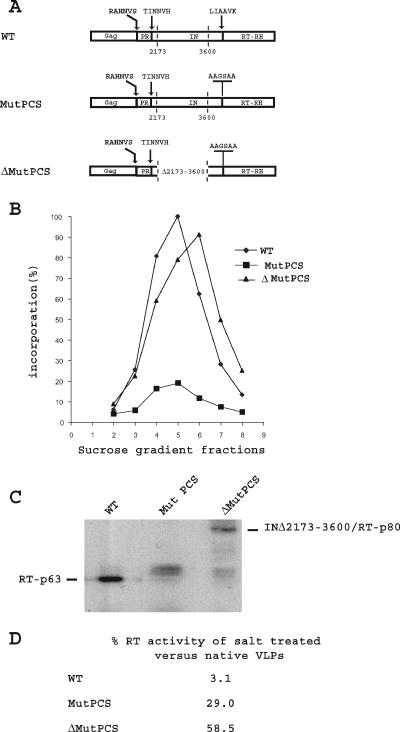
Although RT activity can be detected associated with VLPs in yeast cells overexpressing the Ty1 element, it has not been possible to use VLP preparations as a source of active purified RT. The main RT species identified in Ty1 VLPs is a mature 63-kDa protein generated by proteolytic processing of the Gag-Pol-p199 precursor at the IN/RT cleavage site. The fact that Ty1 RT is active within the VLPs, where it is in close contact with other components of the VLPs, but is inactivated after dissociation of the VLPs suggests that interactions of RT with other VLP components play a role in the activity of RT. As discussed in the introduction, IN is one of the VLP components critical for reverse transcription, and it probably interacts physically with RT (39). To test whether the close association between the two proteins is necessary for the activity of RT, we have blocked the cleavage site between IN and RT by substituting the wild-type IN/RT protease cleavage site (PCS) LIAAVK by AAGSAA (22) so that the two proteins are maintained together after dissociation of the VLPs. It was shown previously by Merkulov et al. (22) that mutations blocking the IN/RT PCS do not affect cleavage at the other sites (Gag/PR or PR/IN). We introduced the IN/RT cleavage site mutation in a wild-type element carrying the full-length IN (pJEFMutPCS) and in an IN deletion mutant pJEFΔ2173-3600 (pJEFΔMutPCS) that retains the 115-amino-acid residue C-terminal domain of IN that is important for RT activity (39) but lacks residues 45 to 520 of IN spanning part of the N-terminal region of IN containing the conserved zinc-binding sequence HHCC and the central core domain with the conserved active-site motif DDE (Fig. 3A). VLPs were purified from cells expressing these elements and fractionated on sucrose step gradients. The exogenous RT activity on fractions from the density gradient was determined in an oligo(dG)/poly(rC) primer-template assay by measuring the incorporation of 32P-radiolabeled dGTP into high-molecular-weight poly(dG) (Fig. 3B). A high level of RT activity was observed with the wild type and the pJEFΔMutPCS element, whereas it was lower for the pJEFMutPCS element containing a full-length IN protein (Fig. 3B). Immunoblotting was performed on the peak fractions of wild type (WT), MutPCS, and ΔMutPCS VLPs using anti-RT antibodies. The volumes of VLPs that were found to incorporate the same amount of dGTP into high-molecular-mass poly(dG) by the oligo(dG)/poly (rC) primer-template assay were loaded onto the gel and probed with Ty1 RT antibodies. The main RT species associated with wild-type VLPs was a fully processed 63-kDa protein (Fig. 3C). In cells transformed with the pJEFΔMutPCS element, an 80-kDa fusion protein (INΔ2173-3600/RT) containing the truncated IN fused to the RT was observed. In agreement with our previous results (39), the activity of RT in the WT and in the Δ2173-3600MutPCS VLPs does not correlate with the amount of RT protein detected by immunoblot analyses and, as previously observed, the specific activity of the Δ2173-3600MutPCS RT was higher than that of the WT RT. In cells containing the pJEFMutPCS element (which carries a full-length IN), a small peak of activity was detected (Fig. 3B), but the specific activity of the 145-kDa IN/RT fusion protein could not be evaluated because the fusion protein was not detected by the RT antibodies (Fig. 3B).
FIG. 3.
(A) Schematic representation of the Gag-Pol polyprotein of the WT element, the IN/RT protease cleavage site (PCS), mutant element and the IN deletion mutant element bearing an IN/RT PCS mutation. The sequences of the Gag/IN, PR/IN, IN/RT, and mutant IN/RT PCS are indicated. The endpoints of the deletion are indicated by vertical dotted lines at positions 2173 and 3600. These numbers are the Ty1-H3 positions (2) and correspond to amino acid residues 45 and 521 of the IN protein, which contains 635 amino acids. (B) VLPs were prepared from cells expressing these elements and purified on a sucrose step gradient. RT activity was assayed using an oligo(dG)/poly(rC) primer template. 100% incorporation is the label incorporated in the peak fraction (fraction 5) of WT VLPs. It is equal to 3 to 7 pmol of dGTP incorporated in 30 min for 4 μl of sucrose gradient-purified VLPs. (C) Immunoblot analysis. The volumes of VLPs that were found to incorporate the same amount of dGTP into high-molecular-mass poly(dG) by the oligo(dG)/poly(rC) primer template assay were loaded onto the gel and probed with Ty1 RT antibodies. (D) The RT activity of the native or salt-treated VLPs was tested on an exogenous primer-template. The percent activities of salt-treated VLPs relative to native VLPs are indicated.
The peak fractions of WT, MutPCS, or Δ2173-3600MutPCS VLPs were then either dissociated by salt by the addition of NaCl to a final concentration of 0.5 M or maintained in a low-salt buffer. The RT activities of VLPs before and after dissociation by salt were compared. As indicated in Fig. 3D, the RT activity of salt-treated wild-type VLPs was only 3% of that of native VLPs. In contrast, in yeast cells expressing the pJEFΔ2173-3600MutPCS plasmid, the activity of salt-treated VLPs was 58.5% of that of native VLPs. The recovery of RT activity in salt-treated VLPs purified from yeast cells containing the pJEFMutPCS element was not as good (29%). Since we have observed (unpublished results) that a recombinant 145-kDa IN/RT fusion protein was not very soluble, it is possible that the low recovery of RT activity of salt-treated MutPCS VLPs is due to a poor solubility of the protein in the experimental conditions used to test the RT activity.
Overall, we can conclude from these results that RT associated with VLPs remains active if it is maintained in close association with IN. This is in line with the observation that an active recombinant protein can be expressed in Escherichia coli only as a fusion protein with amino acid residues encoded by the C-terminal domain of IN fused to the N-terminal domain of RT (37) and confirms that interaction between RT and IN is functional and critical for the polymerase activity of RT (39).
A full-length IN does not trans complement the reverse transcription defect of IN deletion or active site mutant in vivo.
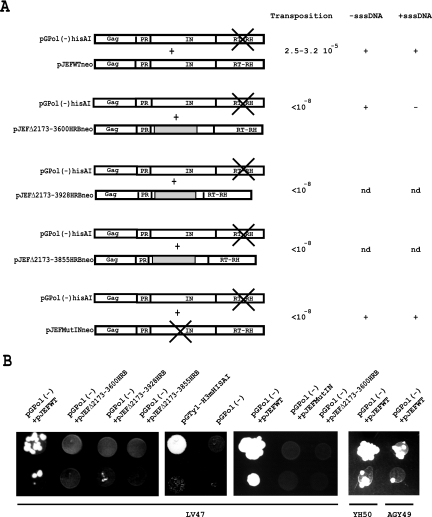
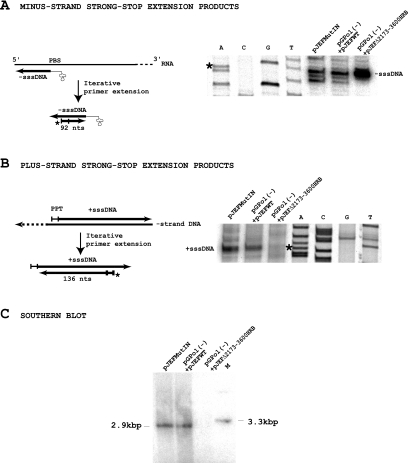
In a previous study we used IN deletion mutants to investigate the role of IN on RT activity in Ty1 VLPs (39). The deletion mutants retained exogenous polymerase activity in a homopolymer primer-template assay and were able to synthesize minus-strand strong-stop DNA (−sssDNA) in vivo, but they failed to synthesize plus-strand strong-stop DNA (+sssDNA). The lack of +sssDNA synthesis was not due to the inhibition of RNase H activity, which generates the polypurine tract primer for plus-strand synthesis, but was attributed to a defect in minus-strand strong-stop DNA transfer from the 5′ end to the 3′ end of the Ty1 RNA (39). To test whether a full-length IN provided in trans can rescue the strand transfer defect of the IN deletion mutants, we used a complementation assay based on coexpressing two transposons (Fig. 1). A TRP1-based reporter plasmid, pGPol(−)hisAI (a derivative of pGTy1-H3mHIS3AI [7]; see Materials and Methods) bearing a Ty1 element with the D211E active-site mutant of RT (35), was introduced into the spt3 yeast cells LV47, together with a URA3-based plasmid carrying different kinds of IN-deletion Ty1 elements (pJEFΔneo). The transformants were analyzed for Ty1 transposition. In pGPol(−)hisAI and pJEFΔneo plasmids, Ty1 is placed under the transcriptional control of the inducible GAL-1-10 promoter which is SPT3 independent; growth of yeast cells in a medium supplemented with galactose induces the transcription of both GAL-Ty1 elements and, as a result, Gag-Pol precursors of both elements are synthesized and incorporated in the VLPs (Fig. 1). Therefore, after processing of the precursors, VLPs assembled in the cotransformed cells will contain full-length IN and inactive RT molecules derived from the pGPol(−)hisAI plasmid and full-length RT and truncated IN molecules derived from the pJEFΔneo plasmid. mRNA transcribed from both plasmids is also packaged within the particles. Since the Ty1 mRNA in the VLPs is dimeric (12), VLPs with two RNA transcripts from pGPol(−)hisAI, two RNA transcripts from pJEFΔneo or one of each transcript coexist in the yeast cells. If the full-length IN provided in trans by the pGPol(−)hisAI plasmid is able to complement the strand transfer defect of the IN deletion mutant, double-stranded cDNA bearing HIS3 should be synthesized in VLPs containing one or two molecules of pGPol(−) RNA. A His+ phenotype should be observed if this cDNA is integrated in the host DNA. As a control, yeast cells were transformed with a wild-type pJEF1105 (pJEFWTneo) plasmid, together with the pGPol(−)hisAI plasmid, to check that VLPs containing full-length active IN and RT from the pJEFWTneo plasmid were able to induce transposition. As shown in Fig. 4, His+ prototrophs were generated in this control experiment. To control that the His+ prototrophs were generated by integrase activity independently of homologous recombination (31), we have repeated the transposition assay in a rad52 strain (AGY49) incapable of homologous recombination. His+ prototrophs were also generated in this strain (Fig. 4B), indicating that transposition in the cells transformed by the pJEFWTneo and the pGPol(−)hisAI plasmids indeed occurred by integrase activity independently of homologous recombination. Furthermore, this result indicates that the active-site mutation of RT is recessive and does not inhibit reverse transcription of the Ty1 element by the active RT encoded by the pJEFWTneo element. This is in agreement with previous studies showing that GAL-Ty1 elements with mutations in IN or RT are complemented at a low level by genomic Ty1 elements in SPT3+ strains (6). The production of His+ cells was decreased 30- to 40-fold in cells cotransformed with pGPol(−)hisAI and pJEFWTneo plasmids compared to a yeast strain expressing a wild-type Ty1-HIS3 element that has a transposition frequency of 1.2 × 10−3 (Fig. 4A). This is in part due to the fact that HIS3 cDNA can be synthesized only in VLPs containing one or two molecules of the Ty1-HIS3 transcripts and also because the VLPs contain an inactive RT derived from the pGPol(−)hisAI plasmid which probably competes with the active enzyme for binding to the primer that initiates reverse transcription. We next checked whether yeast cells transformed with the reporter pGPol(−)hisAI plasmid and pJEFΔneo plasmids containing different kinds of IN deletions were able to generate His+ prototrophs (Fig. 4). In contrast to the control experiment, none of these combinations were able to generate His+ prototrophs. Since recombination between plasmids is a concern in these experiments, this result shows that wild-type Ty1 elements were not generated by recombination between the two mutant plasmids. To determine whether the mutant VLPs containing the IN deletions were able to synthesize Ty1 replication intermediates, an iterative primer extension assay (Materials and Methods) was carried out to detect minus- and plus-strand strong-stop DNA. As shown in Fig. 5A and B, the two strong-stop DNAs were detected in control VLPs. In VLPs containing the Δ2173-3600 deletion, only minus-strand strong-stop DNA was detected. A cDNA synthesis assay developed for the fission yeast Schizosaccharomyces pombe (21) and adapted for Ty1 by Merkulov et al. (22) confirmed that double-stranded cDNA was synthesized in the control VLPs and that VLPs containing the IN deletion mutant pJEFΔ2173-3600 failed to make cDNA (Fig. 5C). Therefore, the full-length IN protein supplied in trans was unable to complement the strand transfer defect of IN deletion mutants. This suggests that the N-terminal domain and/or the central domain containing the active site of IN are involved in strand transfer.
FIG. 4.
(A) Schematic representation of the Gag-Pol polyproteins expressed in yeast cells cotransformed with the pGPol(−) plasmid and the WT pJEF1105 plasmid (pJEF WTneo), the IN deletion mutant derivatives of pJEF1105 (pJEFΔ2173-3600HRB, pJEFΔ2173-3928HRB, and pJEFΔ2173-3855HRB), or the IN active-site mutant of pJEF1105 (pJEFMutIN). The deletions were filled in by inserting the yeast gene Hrb1 (gray rectangle) in order to avoid steric hindrance between the PR/IN and IN/RT cleavage sites and allow complete processing of the Gag-Pol polyprotein (37). In a previous study we showed that the deletion mutants Δ2173-3600 and Δ2173-3855 retained RT activity. The level of RT activity in Δ2173-3855 VLPs was low compared to that of WT or Δ2173-3600 VLPs. Transposition was induced at 22°C in a medium supplemented with 2% galactose and ended at 30°C in a medium containing 2% glucose. The fraction of His+ prototrophs was determined to calculate the frequency of transposition. The transposition frequency of a positive control with yeast cells containing the WT pGTy1-H3mHIS3AI plasmid was 1.2 10−3. Transposition of yeast cells cotransformed with pGPol(−) and WT pJEF1105 plasmids is decreased 30- to 40-fold (see text). Synthesis of -sssDNA and +sssDNA was determined by iterative primer extension. (B) Qualitative transposition assay in yeast strains LV47, YH50, or AGY49. A total of 10 μl of induced cells with an optical density at 600 nm of 1 to 1.5 and 10 μl of a 10-fold dilution were spotted onto glucose plates without histidine and incubated for 2 days at 30°C. A positive control with the WT pGTy1-H3mHIS3AI plasmid and a negative control with the pGPol(−)hisAI plasmid are shown.
FIG. 5.
(A and B) Iterative primer extension analysis of Ty1 replication intermediates in the VLPs. 5′-end-labeled primers complementary to -sssDNA (A) or +sssDNA (B) were annealed to VLP-derived nucleic acids and repeatedly extended with Taq polymerase. The extension products were loaded on an 8% denaturing polyacrylamide gel and mapped with sequence ladders as size markers. The asteriks mark the positions where the -sssDNA and +sssDNA should map. PBS, primer binding site; PPT, polypurine tract. (C) Southern blot analysis of cDNA produced in yeast cells transformed with mutant elements. VLP-extracted nucleic acids were digested with EcoRI and RNase A, separated on a 1% agarose gel, and transferred onto a Hybond N+ membrane (Amersham). The membrane was probed with a XhoI-HindIII fragment of the Tn903 neo gene 32P labeled by random priming. Equal amounts of DNA were loaded in all lanes. The EcoRI VLP cDNA fragment containing the neo gene migrates at 2.9 kbp. M, marker obtained by digesting the pJEF1105 plasmid with SmaI. The SmaI fragment containing the neo gene migrates at 3.3 kbp.
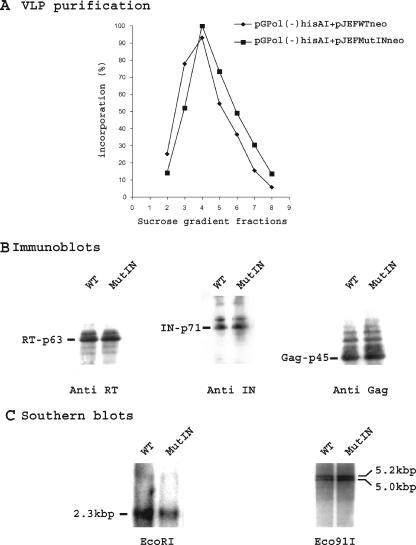
We next checked whether the active site of IN plays a role in strand transfer. The conserved aspartate and glutamate residues of the active site (DD35E) of IN were replaced with alanine, and yeast cells were cotransformed with this IN active-site mutant element (pJEFMutINneo) and with the reporter pGPol(−)hisAI plasmid. VLPs purified from these cells had the same RT activity (Fig. 6A) and contained the same amount of RT, IN, and Gag as the VLPs purified from control cells containing the reporter plasmid pGPol(−)hisAI and the pJEFWTneo plasmid (Fig. 6B). Southern blot analyses showed that equivalent amounts of double-stranded cDNA were synthesized in these cells (Fig. 6C). Therefore, mutation of the active site of IN does not impair DNA synthesis. The surprising observation was that transposition was inhibited in spite of cDNA synthesis. This cannot be attributed to a dominant effect of the active-site mutation in IN since Moore et al. (26), using a similar intragenic complementation test, showed that IN active-site mutations were recessive.
FIG. 6.
Characterization of VLPs isolated from yeast cells containing the reporter plasmid pGPol(−)hisAI and the pJEFWTneo plasmid or the pJEFMutINneo plasmid. (A) VLP-associated RT activity. (B) Immunoblotting with anti-RT, anti-IN, and anti-Gag antibodies. The volumes of VLPs that were found to incorporate the same amount of dGTP into high-molecular-mass poly(dG) by the oligo(dG)/poly(rC) primer template assay were loaded onto the gel and probed with the antibodies. (C) Production of double-stranded cDNA in VLPs. The VLP DNA was digested with EcoRI or Eco91I (BstEII), separated on a 1% agarose gel, transferred to a hybond N+ membrane, and probed with 32P-labeled DNA. The EcoRI-digested DNA was probed with a HIS-labeled fragment. The 2.3-kbp band is the Ty1-HIS digested DNA. The Eco91I-digested DNA was probed with a neo-labeled fragment which hybridizes with a 5.0-kbp Ty1-neo digested cDNA and cross-hybridizes with the 5.2-kbp Ty1-HIS digested cDNA.
DISCUSSION
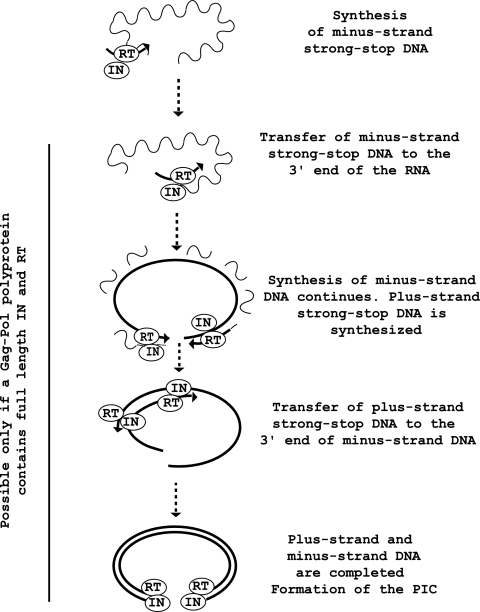
Experiments in this report suggest that IN and RT form a stable complex and do not dissociate during the entire Ty1 reverse transcription cycle. To explain our result, we propose a model of Ty1 replication (Fig. 7) in which the IN and RT domains of the Gag-Pol precursor remain tightly associated after maturation of the precursor and during the whole replication cycle. All of the steps of reverse transcription are then performed by an IN/RT complex containing an active RT. In the last step of replication, plus-strand and minus-strand DNA syntheses are completed simultaneously by two IN/RT complexes. After completion of DNA synthesis, the two IN/RT complexes are poised for the formation of the PIC. If the IN/RT complexes that have performed replication carry an inactive IN, as in yeast cells cotransformed with the pJEFMutINneo and the pGPol(−)hisAI plasmid, the PIC that is transported to the nucleus would be incapable of integrating the Ty1 cDNA into the host cell DNA, and this would explain the lack of transposition. Our model is not in contradiction with the intragenic complementation observed by Moore et al. (26) using catalytic and NLS defective IN mutants because in cells where these mutants are coexpressed, the IN/RT complexes both carry an active RT that can participate in replication. Therefore, PICs containing a dimer (or multimer) with NLS(−) and IN(−) proteins can be formed and translocated to the nucleus by the NLS function of the catalytically inactive IN molecule, while the catalytic activity of the NLS mutant molecule performs the integration reaction.
FIG. 7.
Model of Ty1 reverse transcription outlining the role of the IN/RT complex. For clarity, only one molecule of genomic RNA is shown, and five important steps of reverse transcription are schematized. After completion of cDNA synthesis, the PIC is formed. We suggest that the IN/RT complex which has carried out reverse transcription remains bound to the cDNA in the PIC, which is transported to the nucleus.
In the scheme presented in Fig. 7, one IN/RT complex is postulated to bind stably to each end of the cDNA to form the core of the PIC. The intragenic complementation experiments of Moore et al. (26) also suggested that IN proteins could dimerize (or multimerize) and form a PIC on a given cDNA molecule. Tight binding of Ty1 RT to the ends of the primer-template was recently inferred by Pandey et al. (29) from in vitro studies with a recombinant enzyme. These authors have shown that Ty1 RT binds its primer-template much more strongly than either avian myeloblastosis virus or Moloney murine leukemia virus RTs and have speculated that stable binding of Ty1 RT to blunt double-stranded DNA ends could help recruit IN to the ends of the cDNA in the PIC. In the model presented in Fig. 7, we propose that IN is recruited by RT from the beginning of reverse transcription and that the two proteins remain together during formation of the PIC. The composition and structure of the Ty1 PIC have not been investigated, but it would be interesting to know whether, as in some retroviruses (4, 9-11, 13), RT remains associated with the Ty1 PIC after entry in the nucleus and whether RT is involved in the integration process in vivo. The observation that cytoplasmic viral DNA of some retroviruses (Rous sarcoma virus, avian sarcoma and leukosis virus, or HIV) is not completely double stranded suggests that RT may complete DNA synthesis after entry of the PIC in the nucleus (20, 24). RT could also be used to fill in the four- to six-nucleotide single-stranded gaps created by the IN-catalyzed staggered cleavage of the target DNA (3, 41). A better characterization of the composition of the nuclear PIC of Ty1 would permit to determine whether RT remains closely associated with IN after entry of the PIC into the nucleus or whether, given that IN does not require other proteins to catalyze correct two-ended integration (25), RT and IN are dissociated at some stage before entry of the PIC in the nucleus.
Acknowledgments
We thank our anonymous reviewers for their thoughtful comments that greatly improved this article.
We are grateful to P. Lesage for providing the yeast strain LV47, to D. Garfinkel for the pGTy-H3mHIS3AI plasmid, to J. Boeke for the pJEF1105 plasmid, and to A. Gabriel for the D211E polymerase active-site mutant of Ty1 (AGES4) and the yeast strains YH50 and AGY49. We thank T. Menees for the anti-IN and anti-RT antibodies.
REFERENCES
- 1.Adams, S. E., J. Mellor, K. Gull, R. B. Sim, M. F. Tuite, S. M. Kingsman, and A. J. Kingsman. 1987. The functions and relationships of Ty-VLP proteins in yeast reflects those of mammalian retroviral proteins. Cell 49:111-119. [DOI] [PubMed] [Google Scholar]
- 2.Boeke, J. D., D. Eichinger, D. Castrillon, and G. R. Fink. 1988. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol. Cell. Biol. 8:1432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brin, E., J. Yi, A. M. Skalka, and J. Leis. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287-39295. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns, N. R., H. R. Saibil, N. S. White, J. F. Pardon, P. A. Timmins, S. M. H. Richardson, B. M. Richards, S. E. Adams, S. M. Kingsman, and A. J. Kingsman. 1992. Symmetry, flexibility, and permeability in the structure of yeast retrotransposon virus-like particles. EMBO J. 11:1155-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curcio, M. J., and D. J. Garfinkel. 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 12:2813-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichinger, D. J., and J. D. Boeke. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54:955-966. [DOI] [PubMed] [Google Scholar]
- 9.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, Y. X., S. P. Moore, D. J. Garfinkel, and A. Rein. 2000. The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 74:10819-10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 14.Garfinkel, D. J., A. M. Hedge, S. D. Youngren, and T. D. Copeland. 1991. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J. Virol. 65:4573-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hehl, E. A., P. Joshi, G. V. Kalpana, and V. R. Prasad. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 78:5056-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, S. C., D. L. Court, M. Zweig, and J. G. Levin. 1986. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J. Virol. 60:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa, T., N. Okui, N. Kobayashi, R. Sakuma, T. Kitamura, and Y. Kitamura. 1999. Monoclonal antibodies against the minimal DNA-binding domain in the carboxyl-terminal region of human immunodeficiency virus type 1 integrase. J. Virol. 73:4475-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 19.Lai, L., H. Liu, X. Wu, and J. C. Kappes. 2001. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J. Virol. 75:11365-11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, Y. M. H., and J. M. Coffin. 1991. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol. Cell. Biol. 11:1419-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin, H. L. 1995. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol. Cell. Biol. 15:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkulov, G. V., J. F. Lawler, Y. Eby, and J. D. Boeke. 2001. Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J. Virol. 75:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merkulov, G. V., K. M. Swiderek, C. B. Brachmann, and J. D. Boeke. 1996. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J. Virol. 70:5548-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, M. D., B. Wang, and F. D. Bushman. 1995. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J. Virol. 69:3938-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, S. P., and D. J. Garfinkel. 1994. Expression and partial purification of enzymatically active recombinant Ty1 integrase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 91:1843-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, S. P., L. A. Rinckel, and D. J. Garfinkel. 1998. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell. Biol. 18:1105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nymark-McMahon, M. H., N. S. Beliakova-Bethell, J. L. Darlix, S. F. Le Grice, and S. B. Sandmeyer. 2002. Ty3 integrase is required for initiation of reverse transcription. J. Virol. 76:2804-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padow, M., L. Lai, C. Deivanayagam, L. J. DeLucas, R. B. Weiss, D. M. Dunn, X. Wu, and J. C. Kappes. 2003. Replication of chimeric human immunodeficiency virus type 1 (HIV-1) containing HIV-2 integrase (IN): naturally selected mutations in IN augment DNA synthesis. J. Virol. 77:11050-11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandey, M., S. Patel, and A. Gabriel. 2004. Insights into the role of an active site aspartate in ty1 reverse transcriptase polymerization. J. Biol. Chem. 279:47840-47848. [DOI] [PubMed] [Google Scholar]
- 30.Roder, H. 2004. Stepwise helix formation and chain compaction during protein folding. Proc. Natl. Acad. Sci. USA 101:1793-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharon, G., T. J. Burkett, and D. Garfinkel. 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 14:6540-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasara, T., G. Maga, M. O. Hottiger, and U. Hubscher. 2000. HIV-1 reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett. 507:39-44. [DOI] [PubMed] [Google Scholar]
- 33.Todeschini, A. L., A. Morillon, M. Springer, and P. Lesage. 2005. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:7459-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trentin, B., N. Rebeyrotte, and R. Z. Mamoun. 1998. Human T-cell leukemia virus type 1 reverse transcriptase (RT) originates from the pro and pol open reading frames and requires the presence of RT-RNase H (RH) and RT-RH-Integrase proteins for its activity. J. Virol. 72:6504-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzun, O., and A. Gabriel. 2001. A Ty1 reverse transcriptase active-site aspartate mutation blocks transposition but not polymerization. J. Virol. 75:6337-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner, S., P. Hindmarsh, M. Napirei, K. Vogel-Bachmayr, and B. M. Wohrl. 2002. Subcellular localization and integration activities of rous sarcoma virus reverse transcriptase. J. Virol. 76:6205-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm, M., M. Boutabout, and F. X. Wilhelm. 2000. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem. J. 348:337-342. [PMC free article] [PubMed] [Google Scholar]
- 38.Wilhelm, M., T. Heyman, M. Boutabout, and F. X. Wilhelm. 1999. A sequence immediately upstream of the plus-strand primer is essential for plus-strand DNA synthesis of the Saccharomyces cerevisiae Ty1 retrotransposon. Nucleic Acids Res. 27:4547-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilhelm, M., and F. X. Wilhelm. 2005. Role of Integrase in Reverse Transcription of the Saccharomyces cerevisiae Retrotransposon Ty1. Eukaryot. Cell 4:1057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youngren, S. D., J. D. Boeke, N. J. Sanders, and D. J. Garfinkel. 1988. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol. Cell. Biol. 8:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu, K., C. Dobard, and S. A. Chow. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]