Abstract
SWI/SNF is a well-characterized chromatin remodeling complex that remodels chromatin by sliding nucleosomes in cis and/or displacing nucleosomes in trans. The latter mechanism has the potential to remove promoter nucleosomes, allowing access to transcription factors and RNA polymerase. In vivo, histone acetylation often precedes apparent nucleosome loss; therefore, we sought to determine whether nucleosomes containing acetylated histones could be displaced by the SWI/SNF chromatin remodeling complex. We found that SAGA-acetylated histones were lost from an immobilized nucleosome array when treated with the SWI/SNF complex. When the nucleosome array was acetylated by SAGA in the presence of bound transcription activators, it generated a peak of acetylation surrounding the activator binding sites. Subsequent SWI/SNF treatment suppressed this acetylation peak. Immunoblots indicated that SWI/SNF preferentially displaced acetylated histones from the array relative to total histones. Moreover, the Swi2/Snf2 bromodomain, an acetyl-lysine binding domain, played a role in the displacement of acetylated histones. These data indicate that targeted histone acetylation by the SAGA complex predisposes promoter nucleosomes for displacement by the SWI/SNF complex.
SAGA (Spt-Ada-Gcn5-acetyltransferase) is a 1.8-MDa multi-subunit complex that is an important coactivator for many yeast genes (13, 18). SAGA is recruited to promoters of genes by sequence-specific DNA-binding transcription activators that interact with its Tra1 subunit (5, 12, 52). Once recruited to target promoters Gcn5, the catalytic subunit of SAGA, acetylates a patch of nucleosomes surrounding the promoter (25, 57).
SWI/SNF is a 1.15-MDa nucleosome remodeling complex composed of 12 subunits (27, 54). It also serves as an important coactivator for a subset of yeast genes (16a, 55). SWI/SNF is recruited to gene promoters by sequence-specific DNA-binding transcription activators that interact with the Snf5 and Swi1 subunits (14, 38, 39, 47). The SWI/SNF complex utilizes the ATPase activity of the Swi2/Snf2 subunit to disrupt and/or mobilize nucleosomes (6, 10, 15, 19, 26, 43, 45, 48). SWI/SNF activity can result in histones being removed from a segment of DNA either by nucleosome sliding in cis or by nucleosome displacement (octamer transfer) in trans (46, 58).
There are clear instances in vivo where SAGA and SWI/SNF work in concert during the process of gene activation. For example, at the cell-cycle-regulated HO endonuclease gene both the SAGA and the SWI/SNF complexes play an important role in providing an epigenetic memory of the action of the Swi5 transcription activator until later in the cell cycle when HO transcription is induced (9). SAGA and SWI/SNF remain stably associated with the HO promoter after the loss of Swi5, which was required to recruit them. A more detailed analysis of the HO promoter has revealed that SWI/SNF binding requires histone acetylation by Gcn5 to overcome repression by Ash1 and Sin3/Rpd3 (35). Stable association of SAGA and SWI/SNF with promoter nucleosomes can be achieved through the bromodomains found in both the Swi/Snf2 subunit of SWI/SNF and the Gcn5 subunit of SAGA (14, 15, 39, 47). These bromodomains have been found to recognize acetylated lysine residues (11, 17, 21, 34, 41, 42).
Other examples of genes activated by SAGA and SWI/SNF are the PHO5 and PHO8 genes. Induction of these genes involves transient acetylation of promoter nucleosomes by SAGA. Loss of the acetylated histones is dependent on the presence of SWI/SNF (1, 49, 50). This observation is most consistent with SWI/SNF playing a role in the displacement of acetylated nucleosomes at these promoters. Throughout the genome, activated promoters are depleted of nucleosomes (28), and indeed, the promoters of these genes seem to have lost contact with histones during activation (2, 3, 24).
In light of these in vivo observations we sought to determine whether the SWI/SNF complex was capable of displacing nucleosomes that contained SAGA-acetylated histones. We report here that SWI/SNF is able to remove SAGA-acetylated histones from nucleosomal arrays in vitro and that this activity is partly dependent on the bromodomain of Swi2/Snf2. Moreover, acetylated histones were displaced more readily than bulk histones, indicating that SAGA-acetylated promoter nucleosomes were marked for displacement by SWI/SNF.
MATERIALS AND METHODS
Purification of SWI/SNF, SAGA, and mutant complexes.
All protein complexes were purified by using the tandem affinity purification (TAP) protocol (51). For SWI/SNF purification, Snf6 was TAP tagged at the C terminus by using a TRP1 marker and transformed into W303 by Mike Carrozza. The endogenous Swi2/Snf2 bromodomain deletion Snf6-TAP strain was generated by using the Cre-Lox recombination system. Briefly, a kanamycin resistance cassette flanked by LoxP recombination sites was integrated into Swi2/Snf2 after amino acid 1552, effectively deleting the bromodomain. Positive clones were selected by plating on kanamycin medium. By transforming positive clones with a galactose-inducible plasmid expressing Cre recombinase, the LoxP sites excised the kanamycin marker. The Swi2/Snf2 bromodomain mutants were replica plated and negatively selected for sensitivity to kanamycin. The Swi2/Snf2 subunit was also hemagglutinin tagged at the C terminus by transformation with a kanamycin marker. The strain used to purify the SAGA complex was a gift from F. Winston (FY2021) (59).
Reconstitution of immobilized arrays.
Immobilized template was made by digesting the pG5E4T array with NgoMIV (NEB) overnight at 37°C. The fragment was biotinylated by Klenow fill-in using biotin dCTP for 30 min at room temperature. After heat inactivation, the template was ethanol precipitated and digested with NheI overnight at 37°C, so the template was biotinylated at only one end. The template was reconstituted by step dilution with HeLa core histones into a nucleosomal array (44). The array was bound to streptavidin-magnetic beads at 30°C for 3 h on an inline rotator. The immobilized array was washed extensively and stored at 4°C with 100 ng of HeLa oligonucleosomes/μl. The array was quantified by titration with known quantities of HeLa core histone by silver staining (45).
Immunoblots and fluorography.
For immunoblots, approximately 100 ng of yeast or HeLa nucleosome was incubated with acetyl coenzyme A (acetyl-CoA) and/or SAGA for 30 min in 1× HAT buffer (50 mM Tris-HCl [pH 8], 25% glycerol, 0.5 mM EDTA pH 8, 50 mM KCl, 5 mM dithiothreitol [DTT], 5 μl, 5 mM phenylmethylsulfonyl fluoride [PMSF]). One-half of the reaction was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver stained, while the other half was electrophoresed and immunoblotted with either an anti-acetyl H3 antibody or an anti-acetyl H4 antibody. For fluorography, approximately 2 μg of yeast or HeLa nucleosome were incubated with H3-acetyl-CoA and/or SAGA for 30 min in 1× HAT buffer. One-half of the reaction was subjected to SDS-PAGE and Coomassie blue stained, while the other half was subjected to SDS-PAGE and incubated in ENHANCE, and the gel was subjected to fluorography.
Radiolabeled histone eviction assay.
An approximately 5 nM concentration of immobilized array was washed and resuspended in 1× binding buffer (10 mM HEPES [pH 7.8], 50 mM KCl, 2 mM MgCl2, 5 mM DTT, 0.5 mM PMSF, 10 mM sodium butyrate, 0.25 μg bovine serum albumin/μl, 5% glycerol). The template was incubated with 100 nM recombinant Gal4-VP16 for 15 min at room temperature (56). After washing to remove unbound Gal4-VP16, the array was acetylated by 2 nM SAGA for 1 h at 30°C with H3-acetyl-CoA and 5 μg of competitor chromatin. After radiolabeling, the array was washed to remove free H3-acetyl-CoA. For nucleosome displacement, 2.5 nM SWI/SNF was incubated with 0.7 nM array for 10 min before the addition of 0.3 nM linear acceptor DNA and 3 mM ATP. After 90 min, the supernatant was removed, the immobilized array was washed twice, and both supernatant and washes were retained for scintillation counting. The beads were resuspended in the same volume of buffer as the combined volume of the supernatant and washes and were counted in 5 ml of Scintisafe Econo 2 scintillation cocktail (Fisher Scientific). Nucleosome displacement was shown to be ATP dependent by inhibiting the reaction with 6 mM ATP-γ-S [adenosine-5′-O-(3-thiotriphosphate)].
Scanning in vitro ChIP assay on immobilized array.
Approximately 250 fmol of immobilized array was washed and resuspended in 1× binding buffer (10 mM HEPES [pH 7.8], 50 mM KCl, 2 mM MgCl2, 5 mM DTT, 0.5 mM PMSF, 10 mM sodium butyrate, 0.25 μg of bovine serum albumin/μl, 5% glycerol) to a final concentration of 5 nM. The array was incubated with 100 nM recombinant Gal4-VP16 for 15 min at room temperature (56). After a washing step, the activator-bound template was then acetylated with 0.2 nM SAGA for 30 min at 30°C in the presence of 5 μg of competitor chromatin. If the activator was removed, the template was washed twice and resuspended in 1× binding buffer with 5 μg of competitor chromatin and 200 nM Gal4 oligonucleotide. After acetylation and oligonucleotide competition, the template was either micrococcal nuclease (MNase) digested or subjected to SWI/SNF nucleosome displacement. For the nucleosomes displacement reaction, 2.5 nM SWI/SNF or 5 nM RSC (remodels the structure of chromatin) was incubated with 0.7 nM array for 10 min before the addition of 0.3 nM linear acceptor DNA and 3 mM ATP. After 1.5 h, the supernatant was removed, and the array was washed twice with 1× binding buffer. The template was resuspended in 1× binding buffer with 5 μg of competitor chromatin and digested with 10 U of MNase (Worthington LS004797) for 10 min at room temperature. The mononucleosomes and dinucleosomes were precleared with salmon sperm DNA and protein A-agarose beads (Upstate catalog no. 16-157). The supernatant was mixed with 1 μl of anti-acetyl K9 H3 antibody (Upstate catalog no. 06-599) or anti-histone H3 antibody (Abcam) overnight. The mixture was bound to protein A-agarose beads for 2 h and washed extensively. The supernatant was retained, and the immunoprecipitate was washed extensively prior to elution. Both the supernatant and the immunoprecipitate eluate were subjected to proteinase K digestion at 55°C for 1 h, phenol-chloroform extracted, and ethanol precipitated. The resulting DNA fragments were slot blotted onto a positively charged nylon membrane, Zeta-Probe GT (Bio-Rad catalog no. 162-0196). The membrane was probed with a series of radiolabeled DNA fragments that span the length of the pG5E4T array (57). The membrane was analyzed on Typhoon (Amersham) and quantified by using ImageQuant (Amersham). The level of acetylation on each segment of the array was quantified by using the percent IP (%IP), which is the immunoprecipitated fraction divided by the sum of the immunoprecipitated fraction and the supernatant. The relative acetylation on the template was compared by using the relative %IP, which normalizes the %IP to promoter proximal acetylation.
Immunoblots.
Using conditions identical to the ChIP assay, 5 nM array was acetylated with 0.2 nM SAGA, after binding to activator Gal4VP16. The array could then be subjected to nucleosome displacement by SWI/SNF as described above. After gel electrophoresis, transfer, and blocking, the polyvinylidene difluoride membrane was immunoblotted with anti-acetyl K9 H3 antibody (Upstate catalog no. 06-599) and anti-histone H4 (Upstate catalog no. 05-858). The immunoblots were developed with ECL Plus and detected by scanning on Typhoon (Amersham GE). The data was quantified on an ImageQuant TL (Amersham GE).
RESULTS
SWI/SNF removes histone acetylated by SAGA from immobilized nucleosome arrays.
Our studies utilized immobilized nucleosome arrays. Once bound to magnetic beads, these nucleosomal arrays could be sequentially modified by chromatin modifying and/or chromatin remodeling complexes, which were added and removed, allowing for detailed mechanistic studies of chromatin dynamics. In this particular study, we used the pG5E4T template (31), which contains five Gal4 binding sites and an E4 promoter. The pG5E4T template was linearized by restriction enzyme digestion and subsequently biotinylated at one end. Upon reconstitution with HeLa core histones, the nucleosome array was bound to streptavidin magnetic beads, and excess core histones were removed by washing. The divalent cation concentration never exceeded 2 mM, and no linker histone was present, so the arrays did not aggregate into higher-order chromatin structures (8). The quality of the nucleosome array was confirmed by MNase digestion, and we observed that the array could be digested into mononucleosomes and dinucleosomes (57). The amount of recovered array was quantified by silver staining (45).
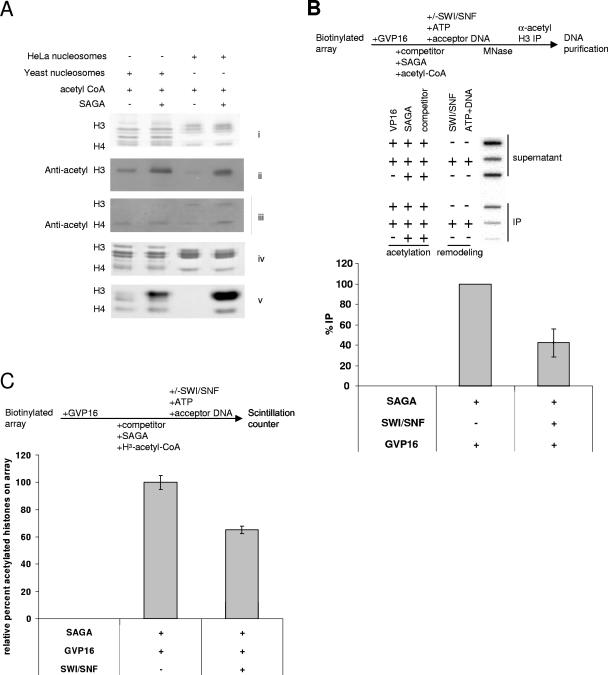
Since our system uses heterologous components, we tested whether HeLa nucleosomes and yeast nucleosomes were acetylated equivalently by yeast SAGA. As a loading control, we normalized for histone content by silver staining (Fig. 1A, i). While in parallel, immunoblots detected equivalent levels of H3 and H4 acetylation on yeast and HeLa nucleosomes, after incubation with SAGA and acetyl-CoA (Fig. 1A, ii and iii). The immunoblots also revealed a low level of acetylation with the yeast nucleosomes when SAGA was not present, which was due to either preexisting acetylation or acetylation by substoichiometric amounts of nucleosomal HATs in the yeast nucleosome preparation. To distinguish between these two causes, we used fluorography and Coomassie blue staining as a loading control for histone content (Fig. 1A, iv). After incubation with H3-acetyl-CoA and SAGA, a strong acetylation signal was detected on H3, H2B, and H4 for both yeast and HeLa nucleosomes (Fig. 1A, v). However, the fluorography also revealed that yeast nucleosomes were acetylated in the absence of SAGA, implying that the yeast nucleosomes contained HATs that modestly acetylated the nucleosomes.
FIG. 1.
SWI/SNF reduces the amount of acetylation detected on immobilized nucleosomal arrays. (A) Yeast and HeLa nucleosomes are equivalent substrates for yeast SAGA acetylation. Yeast or HeLa nucleosomes were incubated with acetyl-CoA and/or yeast SAGA. (i) As a loading control, half the reaction was silver stained. (ii and iii) The nucleosomes were immunoblotted with antibodies directed against anti-acetyl H3 and anti-acetyl H4, respectively. For fluorography, yeast or HeLa nucleosomes were incubated with H3-acetyl-CoA and/or yeast SAGA. (iv and v) As a loading control, half the reaction was Coomassie blue stained (iv) prior to electrophoresis, ENHANCE treatment, and fluorography (v). (B) SWI/SNF treatment causes a decrease in acetylation on the nucleosome array. Using competitor chromatin, the artificial activator Gal4-VP16 targets SAGA acetylation, and the array is subjected to SWI/SNF treatment, with acceptor DNA and ATP. After nucleosome displacement, the supernatant is removed, and the array is MNase digested and immunoprecipitated with an anti-acetyl H3 antibody. The immunoprecipitated nucleosomes are subjected to DNA purification and slot blotted onto nylon membrane, along with the supernatant. The membrane is probed with end-labeled full-length pG5E4T template. The %IP is the immunoprecipitated fraction divided by the sum of the immunoprecipitated fraction and the supernatant. In this case, the %IP was normalized to the %IP without SWI/SNF treatment. (C) SWI/SNF targets and displaces activator-targeted SAGA acetylation. The artificial activator Gal4-VP16 is bound to the immobilized nucleosomal array. After the addition of competitor, the array is acetylated with SAGA. The array is subjected to SWI/SNF nucleosome displacement with ATP and acceptor DNA, and the beads are counted on a scintillation counter. The loss of acetylation on the beads corresponds to nucleosomes displaced by SWI/SNF from the array. The change in acetylation on the array is expressed as the relative percent acetylated histones on the beads.
Initially, we investigated whether SWI/SNF could affect the levels of SAGA acetylated histones on an immobilized nucleosome array. Interaction of SWI/SNF with an activator is important for the recruitment of this complex to promoters and subsequent transcription stimulation (37, 38, 60). Therefore, we included the artificial activator, Gal4-VP16 in our assays. We previously demonstrated that Gal4-VP16 could target SAGA acetylation to promoter nucleosomes (57). To track acetylated histones, the nucleosomal array was acetylated with H3-acetyl-CoA by SAGA. After a washing step to remove the excess H3-acetyl-CoA, the array was incubated with or without SWI/SNF and ATP. DNA was included in all reactions, which accepts transferred histones and simultaneously competes for SWI/SNF binding. Incubation of the array with SAGA and H3-acetyl-CoA resulted in the incorporation of H3-acetate into the nucleosome array, which could be measured by using a scintillation counter (Fig. 1C). After incubation with SWI/SNF and ATP, we observed that 40% of [H3]acetate on the array was displaced into the supernatant. The loss of acetylation required the presence of both SWI/SNF and ATP, implying that SWI/SNF displaced SAGA-acetylated histones (Fig. 1 and data not shown).
We confirmed the role of SWI/SNF in the loss of acetylated histones by using an in vitro chromatin immunoprecipitation (ChIP) assay (Fig. 1B). Immobilized arrays were bound by Gal4-VP16, and competitor chromatin was included prior to the addition of SAGA so acetylation would be targeted to the promoter region. The acetylated array was then incubated with or without SWI/SNF, ATP, and acceptor DNA. After being washed, the array was digested into mononucleosomes and dinucleosomes with MNase and then immunoprecipitated with an antibody against acetylated K9 on histone H3. Nucleosomes that bound the antibody were separated with protein A-agarose beads from the supernatant, and the beads were washed extensively. DNA was purified from the immunoprecipitated material, as well as the supernatant, and slot blotted onto a nylon membrane. After hybridization to a probe that spanned the length of the pG5E4T template, we observed a decrease in total signal (supernatant and beads) after SWI/SNF treatment, representing an overall loss of histones prior to MNase digestion. Immunoprecipitation experiments (i.e., %IP) revealed that a smaller fraction of nucleosomes remained on the array in the sample treated with SWI/SNF compared to that not treated with SWI/SNF (Fig. 1B). These data indicated that SAGA-acetylated histones were lost during SWI/SNF treatment. In fact, SWI/SNF treatment resulted in the loss of over half of the SAGA-acetylated histones (Fig. 1B).
SWI/SNF suppresses the histone acetylation peak generated by activator targeting of SAGA.
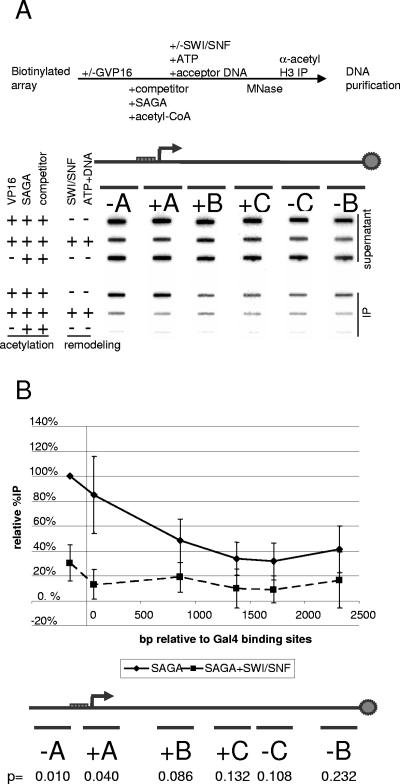
In previous studies, GCN5 was shown to produce a peak of acetylation in vivo at the HIS3 promoter. These results were recapitulated in vitro on a nucleosomal array, where the SAGA complex produced a peak of acetylation that surrounded activator binding sites (25, 57). We hypothesized that displacement of SAGA acetylated histones might suppress this activator-dependent SAGA acetylation peak on nucleosomal arrays.
We performed a scanning in vitro ChIP assay to detect the effects of SWI/SNF nucleosome displacement on the SAGA acetylation peak. The immobilized array was bound by Gal4-VP16. Competitor chromatin was added to the arrays, prior to the addition of acetyl-CoA and SAGA. The acetylation should be preferentially targeted at the promoter by Gal4-VP16 recruitment of SAGA in the presence of competitor chromatin, as previously described (57). The acetylated array was then treated with SWI/SNF in the presence of acceptor DNA and ATP. After MNase digestion, immunoprecipitation (IP) with the acetyl H3 antibody, and DNA purification, the template was slot blotted onto a nylon membrane. The membrane was sequentially hybridized with a series of probes that spanned the length of the template (Fig. 2A). The IP efficiency, or %IP of each segment was normalized to the −A segment of the promoter region, where the SAGA acetylation peaked near the activator binding sites as expected (57). After treatment with SWI/SNF, the overall levels of acetylation were decreased. Moreover, the loss of acetylation was most pronounced at the promoter where the SAGA acetylation peak was suppressed (Fig. 2B).
FIG. 2.
Nucleosome displacement suppresses the SAGA acetylation peak. (A) Scanning in vitro ChIP assay, with the relative positions of the probes for the pG5E4T array indicated. The array was subjected to nucleosome displacement and ChIP as described in Fig. 1B. The membrane was hybridized to a series of labeled probes shown in the diagram. The positions of the probes relative to the HindIII restriction enzyme site are also indicated on the diagram. The blots are shown below each corresponding probe. (B) The SAGA acetylation peak is suppressed by SWI/SNF. The %IP was normalized to the %IP at the −A probe, which is upstream of the Gal4 binding sites and the E4 promoter. The solid line depicts the H3 acetylation profile along the template, while the dashed line corresponds to the effect of SWI/SNF nucleosome displacement on the H3 acetylation profile. A Student t test was used to determine the statistical significance of the difference in the acetylation profile before and after SWI/SNF treatment. The P values for the t test are given below each segment of the array.
SWI/SNF preferentially displaces nucleosomes acetylated by SAGA.
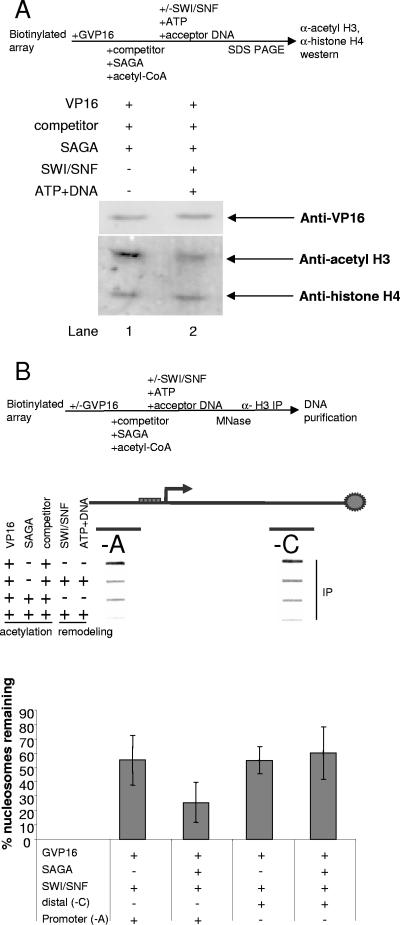
While the in vitro ChIP illustrated that SWI/SNF displaced acetylated promoter nucleosomes, we wanted to directly test whether SWI/SNF preferentially displaced acetylated nucleosomes. This was measured by immunoblotting the acetylated nucleosome arrays after SWI/SNF treatment with an antibody against acetylated H3 and an antibody against an unmodified patch of histone H4. The anti-acetyl H3 antibody recognized nucleosomes that are acetylated by SAGA at lysine 9 on histone H3, while the anti-histone H4 antibody recognized both acetylated and unmodified nucleosomes. At 50 mM KCl, the H3/H4 tetramer is stable and does not dissociate, so the anti-histone H4 antibody measured total nucleosome content on the array, including the acetylated nucleosomes that are also detected by the anti-acetyl H3 antibody. A comparison of acetylated H3 signal to histone H4 signal should indicate whether acetylated nucleosomes are lost to a greater extent than unmodified nucleosomes. SAGA acetylation was targeted to the promoter region of nucleosome arrays using Gal4-VP16 and competitor chromatin. After removal of the free acetyl-CoA, the arrays were then treated with SWI/SNF. The nucleosome arrays were Western blotted with the anti-acetyl H3 antibody and the anti-histone H4 antibody, which could detect all modified and unmodified nucleosomes. After SWI/SNF treatment, we observed a larger decrease of acetyl H3 signal relative to H4 signal (Fig. 3A, compare lanes 1 and 2). The peaked acetylation profile from Fig. 2A indicated that most SAGA acetylation occurred on the two promoter nucleosomes flanking the Gal4 binding sites. SWI/SNF specifically displaced the acetylated promoter nucleosomes, but the majority of nucleosomes remained on the array. The Western blot was not sensitive enough to detect the loss of a small fraction of the total nucleosome population. However, if SWI/SNF indiscriminately displaced nucleosomes from the array, we would expect to see a greater loss of total nucleosome content on the array. Using Western blot analysis, we detected a significant decrease in acetylation, and the Western blot did not show an appreciable decrease in total nucleosome content, as measured by the anti-histone H4 antibody.
FIG. 3.
Acetylated histones are preferentially lost after SWI/SNF nucleosome displacement. (A) The array is bound by activator Gal4-VP16 prior to the addition of competitor chromatin, followed by acetylation under competitive conditions with SAGA. After SWI/SNF nucleosome displacement, free histones are washed away, and the array is subjected to immunoblotting with an anti-VP16 antibody, an anti-acetyl H3 antibody, and an anti-histone H4 antibody. (B) Scanning in vitro ChIP assay with histone H3 antibody. The array is subjected to nucleosome displacement and ChIP as described in Fig. 1B. The membrane is hybridized to a promoter specific probe (−A) and a distal probe (−C) as shown in the diagram. The blots are shown below each corresponding probe, along with a bar graph for nucleosomes remaining on the array. The %IP was normalized to the %IP before nucleosome displacement.
To further examine the preferential displacement of acetylated nucleosomes by SWI/SNF, a ChIP was performed with a C-terminal H3 antibody that detects all modified an unmodified forms of histone H3 to measure of histone density on the array (28). We sought to determine whether, compared to unacetylated arrays, SAGA acetylation stimulated the displacement of nucleosomes, specifically in the promoter region. After Gal4-VP16 binding and the addition of competitor chromatin, the immobilized array was either acetylated with SAGA or subjected to a mock acetylation reaction. SWI/SNF treatment was carried out in parallel on the acetylated array and the unacetylated array. After a washing step, the template was digested into mononucleosomes and dinucleosomes using MNase, followed by IP with the anti-histone H3 antibody. After SWI/SNF treatment, we observed a decrease in the %IP (Fig. 3B). After the probes were scanned at the promoter (−A) and distal segments (−C), we found that SWI/SNF nucleosome displacement was enhanced by SAGA acetylation at the promoter (Fig. 3B). When the array was not acetylated prior to SWI/SNF treatment, we observed that 55 and 60% of the nucleosomes remained on the array at the promoter and distal regions, respectively. However, if the array was acetylated prior to SWI/SNF treatment, 25% of the nucleosomes remained at the promoter, whereas 55% remained at the distal regions. Therefore, while SWI/SNF was able to target the activator-bound array in the absence of SAGA, acetylation significantly enhanced nucleosome displacement by SWI/SNF. Thus, SWI/SNF preferentially displaced SAGA-acetylated nucleosomes.
The Swi2/Snf2 bromodomain contributes to acetylated nucleosome displacement by SWI/SNF.
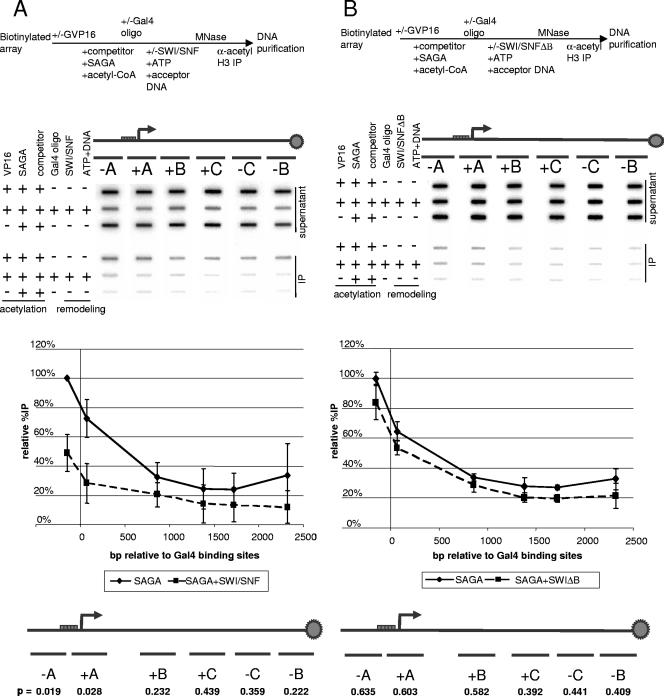
SWI/SNF is targeted to promoters by transcription activators in a manner similar to that of the SAGA complex (14, 38, 39, 47). Thus, SWI/SNF might preferentially remove histones acetylated by SAGA because it is targeted to the same location on the nucleosome array by Gal4-VP16. To test the importance of the activator in targeting SWI/SNF to the SAGA acetylation peak, we sought to determine whether SWI/SNF targeted acetylated histones, independent of activator. After Gal4-VP16 binding, the immobilized array was acetylated with SAGA, and then Gal4-VP16 was removed by using Gal4 oligonucleotide competition. SWI/SNF treatment was then carried out in the absence of activator. After IP with the anti-acetyl H3 antibody, we observed a decrease in the relative %IP after SWI/SNF treatment (Fig. 4A). After scanning with probes spanning the entire template, we found that SWI/SNF treatment did decrease the SAGA acetylation peak (Fig. 4A), but to a lesser extent than when activator was present (compare with Fig. 2B). When Gal4-VP16 was present during SWI/SNF treatment, 70 and 72% of the acetylated histones were displaced at positions −A and +A, respectively, whereas 51 and 44% of the acetylated histones were displaced at positions −A and +A, respectively, when GAL4-VP16 was removed prior to SWI/SNF treatment. With or without activator, SWI/SNF displacement of acetylated histones in these regions was significant, with P values of <0.025, when analyzed by the Student t test. Therefore, the presence of activator enhances SWI/SNF-mediated nucleosome displacement but is not required for this activity.
FIG. 4.
The Swi2/Snf2 bromodomain is required for the transfer of acetylated histones. (A) SWI/SNF reduction of acetylated histone peak can occur without Gal4-VP16 targeting of this complex. Scanning in vitro ChIP analysis after SWI/SNF nucleosome displacement in the absence of activator Gal4-VP16. The experiment was previously described in Fig. 2. The template is bound to activator Gal4-VP16, acetylated by SAGA with competitor chromatin, and activator is removed by Gal4 oligonucleotide competition. The array was then subjected to SWI/SNF nucleosome displacement and immunoprecipitated with an anti-acetyl H3 antibody. The graph depicts the loss of promoter-acetylated nucleosomes after SWI/SNF nucleosome displacement without activator. The %IP was normalized to the %IP at the −A probe. The solid line depicts the H3 acetylation profile along the template, while the dashed line corresponds to the effect of SWI/SNF nucleosome displacement on the H3 acetylation profile. (B) Scanning in vitro ChIP analysis after activator is removed and SWI/SNF bromodomain mutant treatment. The experiment was as described in Fig. 2. Gal4-VP16 targeted SAGA acetylation at the promoter nucleosomes using competitor chromatin. The activator is removed by Gal4 oligonucleotide competition, and the array is subjected to nucleosome displacement by the bromodomain mutant complex, followed by IP with an anti-acetyl H3 antibody. The graph depicts the loss of promoter-acetylated nucleosomes after nucleosome displacement by the Swi2/Snf2 bromodomain mutant complex. The %IP was normalized to the %IP at the −A probe. The solid line depicts the H3 acetylation profile along the template, and the dashed line corresponds to the effect of Swi2/Snf2 bromodomain mutant on the H3 acetylation profile. A Student t test was used to determine the statistical significance of the difference in the acetylation profile before and after SWI/SNF treatment. The P values for the t test are given below each segment of the array.
Preferential displacement of acetylated nucleosomes by SWI/SNF could result from acetylation enhancing nucleosome displacement and/or increased recognition of acetylated nucleosomes by SWI/SNF. The latter possibility was consistent with two previous observations. First, the Swi2/Snf2 bromodomain was shown to recognize and bind to acetylated nucleosomes (15). Second, it was demonstrated that preferential displacement of acetylated nucleosomes did not require Gal4-VP16 targeting and that the bromodomain could provide acetylated nucleosome recognition. Since acetylated nucleosomes are the preferred substrate for SWI/SNF displacement, the Swi2/Snf2 bromodomain may play a role in the preferential action of SWI/SNF on these nucleosomes. To test this possibility, we used an in vitro ChIP assay to compare the acetylated nucleosome displacement activity of wild-type SWI/SNF complex to that of a complex lacking the Swi2/Snf2 bromodomain.
SAGA acetylation was targeted to the promoter by the artificial activator Gal4-VP16 and competitor chromatin. After the activator was removed with Gal4 oligonucleotide competition, we incubated the array with the Swi2 bromodomain mutant complex. When assayed with the scanning in vitro ChIP, the Swi2 bromodomain mutant complex showed reduced nucleosome displacement, compared to wild type as indicated by the lack of suppression of the SAGA nucleosome acetylation peak (compare Fig. 4A and B). The Swi2/Snf2 bromodomain mutant was not able to specifically target acetylation in the promoter region. Rather, hyperacetylated and hypoacetylated nucleosomes were equally displaced along the length of the array by the mutant complex. The wild-type SWI/SNF complex displaced 51% of acetylated nucleosomes at the −A probe and 11% at the −C probe, whereas the bromodomain mutant displaced only 16% at the −A probe, and 6% at the −C probe (Fig. 4B). Although some displacement occurred at the promoter, the mutant complex did not target acetylated nucleosomes as well as the wild type. Therefore, the Swi2/Snf2 bromodomain is important for SWI/SNF-mediated displacement of SAGA-acetylated histones.
RSC suppresses the histone acetylation peak generated by activator targeting of SAGA.
The Swi/Snf-related RSC complex also displaces nucleosomes in trans (7, 32). Indeed, Reinke et al. propose that other chromatin remodelers displace nucleosomes in the absence of SWI/SNF (50). Although ChIP and microarray analysis localized RSC at the promoters of RNA polymerase III-transcribed genes, those authors acknowledge that RSC is difficult to immunoprecipitate and may bind at other promoters (40). Moreover, RSC interacts genetically with SAGA and binds H3 peptides acetylated at lysine 14 (22), while in higher eukaryotes, PBAP, the Drosophila RSC homolog, localizes at hyperacetylated nucleosomes in polytene stains (36). Thus, RSC may be functionally redundant with SWI/SNF and displaces promoter-acetylated nucleosomes in the absence of the latter remodeling complex.
We tested whether RSC displaced SAGA-acetylated nucleosomes by using the scanning in vitro ChIP experiment. The nucleosomal array was bound by the artificial activator Gal4-VP16 and acetylated by SAGA in the presence of competitor chromatin. After targeting acetylation at the promoter, the array was washed, and the activator was removed by Gal4 oligonucleotide competition. With the activator removed, the array was incubated with RSC, ATP, and acceptor DNA. After MNase digestion, IP with the acetyl H3 antibody, and DNA purification, the template was slot blotted onto a nylon membrane. The membrane was sequentially hybridized with a series of probes that spanned the length of the template (Fig. 5A). RSC decreased the level of acetylation at the promoter (Fig. 5B). Interestingly, we observed a slight increase in the level of acetylation at segments distal to the Gal4 binding sites, suggesting that acetylated nucleosomes were transferred from the promoter to the distal region. Thus, RSC nucleosome displacement suppressed the acetylation profile of SAGA.
FIG. 5.
RSC nucleosome displacement suppresses the SAGA acetylation peak. (A) RSC reduction of acetylated histone peak can occur without Gal4-VP16 targeting of this complex. Scanning in vitro ChIP analysis was performed after RSC nucleosome displacement in the absence of activator Gal4-VP16. The experiment was as described in Fig. 2. The template is bound to activator Gal4-VP16, acetylated by SAGA with competitor chromatin, and the activator is removed by Gal4 oligonucleotide competition. The array was then subjected to RSC nucleosome displacement and immunoprecipitated with an anti-acetyl H3 antibody. (B) The graph depicts the loss of promoter-acetylated nucleosomes after RSC nucleosome displacement without activator. The %IP was normalized to the %IP at the −A probe. The solid line depicts the H3 acetylation profile along the template, while the dashed line corresponds to the effect of RSC nucleosome displacement on the H3 acetylation profile. The graph shows the average of two independent experiments.
DISCUSSION
In this study, we have shown that the SWI/SNF complex can displace SAGA-acetylated nucleosomes from nucleosome arrays. SAGA acetylated a pronounced peak of approximately two promoter-proximal nucleosomes. Thus, the bulk of acetylation occurred in a peak surrounding the Gal4 binding sites of the nucleosomal array. SAGA-acetylated nucleosomes were predisposed to displacement by SWI/SNF through a reaction that required ATP hydrolysis and was facilitated by the Swi2/Snf2 bromodomain. Displacement of acetylated nucleosomes by SWI/SNF resulted in the generation of nucleosome-free DNA surrounding activator binding sites. These data are consistent with a model in which activator recruitment of the SAGA complex results in a patch of acetylation of promoter and surrounding nucleosomes. These acetylated nucleosomes are then targeted for displacement by the SWI/SNF complex, resulting in a nucleosome-free gap to accommodate the formation of preinitiation transcription complexes.
For several years the SWI/SNF complex has been known to be capable of displacing nucleosomes in trans through a process termed octamer transfer (32, 58). Our biochemical data and the in vivo observations of Reinke and Horz (50) suggest it may also play a more direct functional role in promoter activation. Histone acetylation may predispose nucleosomes for displacement in at least two non-mutually exclusive ways. First, histone acetylation may facilitate octamer transfer and/or nucleosome disassembly. Although evidence that histone acetylation may destabilize nucleosomes in some way has been elusive, several reports are consistent with such a possibility. It has been reported that acetylation and ubiquitination of histones H2A and H2B increase the lability of H2A-H2B dimers in chicken erythrocyte nucleosomes (30). Histone H4 tetra-acetylation has been found to reduce the thermal stability of nucleosome cores (53), and H3 and H4 acetylation reduces some of the strong interactions of the histone octamers with DNA (4), a finding consistent with the fact that acetylation can in some instances increase the binding of transcription factors to nucleosomes (29). Moreover, acetylation by p300 has been shown to increase the transfer of H2A/H2B dimers onto histone chaperones during action of the ATP-dependent chromatin remodeling complex ACF (20).
Another possibility is that histone acetylation marks nucleosomes for displacement by bromodomain-containing chromatin remodeling complexes such as SWI/SNF. This possibility is consistent with our observation that displacement of acetylated histones by SWI/SNF is at least partly dependent on the Swi2/Snf2 bromodomain. Bromodomains are acetyl-lysine binding domains (11, 61), and the SWI/SNF bromodomain has been shown to anchor the complex onto acetylated nucleosomes (14, 16). Interactions of the Swi2/Snf2 bromodomain with acetylated nucleosomes might participate in the process of histone displacement directly or act by concentrating SWI/SNF on those nucleosomes. Numerous transcription- or chromatin-related proteins contain bromodomains and may have reasons for interacting with acetylated histones (23). However, the large number of bromodomains found in the RSC chromatin remodeling complex in yeast, a complex similar to SWI/SNF (22), is consistent with the possibility that bromodomain acetylated histone interactions are widely used in nucleosome remodeling and displacement.
Acknowledgments
We are grateful to Ahmed Hassan, Kiranmai Kocherlakota, Bing Li, Daeyoup Lee, and Samantha Pattenden for many helpful comments during this study.
P.P. is a Senior Research Fellow with the Leukemia and Lymphoma Society. J.L.G. was supported by the Pew Latin American Fellows Program in the Biomedical Sciences. This study was supported by NIGMS Grant R37 GM047867 to J.L.W.
REFERENCES
- 1.Barbaric, S., H. Reinke, and W. Horz. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23:3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 3.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 4.Brower-Toland, B., D. A. Wacker, R. M. Fulbright, J. T. Lis, W. L. Kraus, and M. D. Wang. 2005. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 346:135-146. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, B. R., Y. J. Kim, M. H. Sayre, B. C. Laurent, and R. D. Kornberg. 1994. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 91:1950-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 8.Carruthers, L. M., J. Bednar, C. L. Woodcock, and J. C. Hansen. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37:14776-14787. [DOI] [PubMed] [Google Scholar]
- 9.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 10.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 11.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 12.Fishburn, J., N. Mohibullah, and S. Hahn. 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18:369-378. [DOI] [PubMed] [Google Scholar]
- 13.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 14.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-1064. [DOI] [PubMed] [Google Scholar]
- 15.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 16.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 16a.Holstege, F. C., E. G. Jennings, J. J. Wyricks, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 17.Hudson, B. P., M. A. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 304:355-370. [DOI] [PubMed] [Google Scholar]
- 18.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 19.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 20.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899-1907. [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 22.Kasten, M., H. Szerlong, H. Erdjument-Bromage, P. Tempst, M. Werner, and B. R. Cairns. 2004. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23:1348-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura, A., K. Matsubara, and M. Horikoshi. 2005. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J. Biochem. 138:647-662. [DOI] [PubMed] [Google Scholar]
- 24.Korber, P., T. Luckenbach, D. Blaschke, and W. Horz. 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 24:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, H., A. N. Imbalzano, P. A. Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370:477-481. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K. K., P. Prochasson, L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2004. Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem. Soc. Trans. 32:899-903. [DOI] [PubMed] [Google Scholar]
- 28.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 29.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 30.Li, W., S. Nagaraja, G. P. Delcuve, M. J. Hendzel, and J. R. Davie. 1993. Effects of histone acetylation, ubiquitination and variants on nucleosome stability. Biochem. J. 296(Pt. 3):737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, Y. S., M. F. Carey, M. Ptashne, and M. R. Green. 1988. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell 54:659-664. [DOI] [PubMed] [Google Scholar]
- 32.Lorch, Y., M. Zhang, and R. D. Kornberg. 1999. Histone octamer transfer by a chromatin-remodeling complex. Cell 96:389-392. [DOI] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Matangkasombut, O., and S. Buratowski. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11:353-363. [DOI] [PubMed] [Google Scholar]
- 35.Mitra, D., E. J. Parnell, J. W. Landon, Y. Yu, and D. J. Stillman. 2006. SWI/SNF binding to the HO Promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol. Cell. Biol. 26:4095-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohrmann, L., K. Langenberg, J. Krijgsveld, A. J. Kal, A. J. Heck, and C. P. Verrijzer. 2004. Differential targeting of two distinct SWI/SNF-relatedDrosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24:3077-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 38.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 39.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ornaghi, P., P. Ballario, A. M. Lena, A. Gonzalez, and P. Filetici. 1999. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol. 287:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen-Hughes, T., R. T. Utley, J. Cote, C. L. Peterson, and J. L. Workman. 1996. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273:513-516. [DOI] [PubMed] [Google Scholar]
- 44.Owen-Hughes, T., R. T. Utley, D. J. Steger, J. M. West, S. John, J. Cote, K. M. Havas, and J. L. Workman. 1999. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol. Biol. 119:319-331. [DOI] [PubMed] [Google Scholar]
- 45.Owen-Hughes, T., and J. L. Workman. 1996. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 15:4702-4712. [PMC free article] [PubMed] [Google Scholar]
- 46.Phelan, M. L., G. R. Schnitzler, and R. E. Kingston. 2000. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol. 20:6380-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prochasson, P., K. E. Neely, A. H. Hassan, B. Li, and J. L. Workman. 2003. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell 12:983-990. [DOI] [PubMed] [Google Scholar]
- 48.Quinn, J., A. M. Fyrberg, R. W. Ganster, M. C. Schmidt, and C. L. Peterson. 1996. DNA-binding properties of the yeast SWI/SNF complex. Nature 379:844-847. [DOI] [PubMed] [Google Scholar]
- 49.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 50.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 51.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 52.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siino, J. S., P. M. Yau, B. S. Imai, J. M. Gatewood, and E. M. Bradbury. 2003. Effect of DNA length and H4 acetylation on the thermal stability of reconstituted nucleosome particles. Biochem. Biophys. Res. Commun. 302:885-891. [DOI] [PubMed] [Google Scholar]
- 54.Smith, C. L., R. Horowitz-Scherer, J. F. Flanagan, C. L. Woodcock, and C. L. Peterson. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10:141-145. [DOI] [PubMed] [Google Scholar]
- 55.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 57.Vignali, M., D. J. Steger, K. E. Neely, and J. L. Workman. 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalyzed by the yeast SWI/SNF complex. Nature 400:784-787. [DOI] [PubMed] [Google Scholar]
- 59.Wu, P. Y., and F. Winston. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22:5367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]