Abstract
The opportunistic human pathogen Aspergillus fumigatus produces a large quantity of asexual spores (conidia), which are the primary agent causing invasive aspergillosis in immunocompromised patients. We investigated the mechanisms controlling asexual sporulation (conidiation) in A. fumigatus via examining functions of four key regulators, GpaA (Gα), AfFlbA (RGS), AfFluG, and AfBrlA, previously studied in Aspergillus nidulans. Expression analyses of gpaA, AfflbA, AffluG, AfbrlA, and AfwetA throughout the life cycle of A. fumigatus revealed that, while transcripts of AfflbA and AffluG accumulate constantly, the latter two downstream developmental regulators are specifically expressed during conidiation. Both loss-of-function AfflbA and dominant activating GpaAQ204L mutations resulted in reduced conidiation with increased hyphal proliferation, indicating that GpaA signaling activates vegetative growth while inhibiting conidiation. As GpaA is the primary target of AfFlbA, the dominant interfering GpaAG203R mutation suppressed reduced conidiation caused by loss of AfflbA function. These results corroborate the hypothesis that functions of G proteins and RGSs are conserved in aspergilli. We then examined functions of the two major developmental activators AfFluG and AfBrlA. While deletion of AfbrlA eliminated conidiation completely, null mutation of AffluG did not cause severe alterations in A. fumigatus sporulation in air-exposed culture, implying that, whereas the two aspergilli may have a common key downstream developmental activator, upstream mechanisms activating brlA may be distinct. Finally, both AffluG and AfflbA mutants showed reduced conidiation and delayed expression of AfbrlA in synchronized developmental induction, indicating that these upstream regulators contribute to the proper progression of conidiation.
The genus Aspergillus represents the most widespread fungi in the environment and includes industrially, agriculturally, and medically important species. All aspergilli reproduce in asexual mode, which involves the formation of multicellular organs termed conidiophores bearing thousands of mitotically derived asexual spores (conidia). The study of asexual development (conidiation) in the model fungus Aspergillus nidulans has provided important information on the mechanisms controlling growth and development (reviewed in references 3 and 4).
Conidiation in A. nidulans is a continual sequence from vegetative growth to asexual development. It is a precisely timed and genetically programmed event responding to internal and external cues. Previous studies demonstrated that vegetative growth signaling is primarily mediated by a heterotrimeric G protein system composed of FadA, SfaD, and GpgA (Gα, Gβ, and Gγ subunits, respectively); PhnA (a Gβγ activator); and the cyclic AMP (cAMP)-dependent protein kinases PkaA and PkaB (23, 25, 28, 29, 32, 39; reviewed in reference 43). Activation of this G protein signaling stimulates hyphal proliferation, which in turn represses conidiation and production of the mycotoxin sterigmatocystin (ST; Fig. 1A) (10, 25, 39). Constitutive activation of FadA signaling causes uncontrolled accumulation of hyphal mass and the absence of sporulation, resulting in the fluffy autolytic phenotype (39). Initiation of conidiation requires both inhibition of this G protein signaling and activation of development-specific functions. FlbA is an RGS (regulator of G protein signaling) domain protein, which plays a crucial role in antagonizing vegetative growth signaling, likely by facilitating the intrinsic GTPase activity of FadA (15, 39).
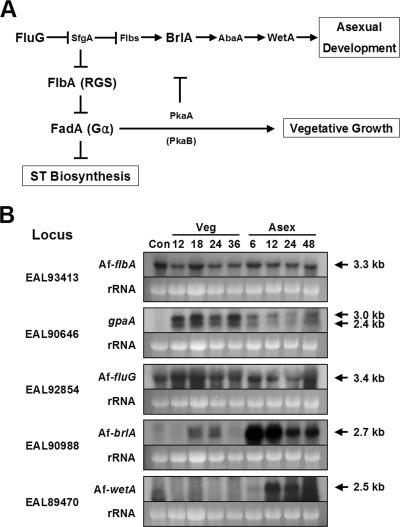
FIG. 1.
Model for growth and developmental control in A. nidulans and expression of major regulators in A. fumigatus. (A) Activation of FadA-mediated vegetative signaling represses asexual development and ST production (10, 39). FlbA is an RGS protein that down-regulates FadA signaling, which in turn allows asexual development to proceed. Commencement of conidiation requires the activities of FluG and other developmental genes (reviewed in reference 4; see also reference 44). Key regulators examined in this study are shown in a larger font. (B) Northern blots showing mRNA levels of AfflbA, gpaA, AffluG, AfbrlA, and AfwetA throughout the life cycle including conidia (Con) of the WT strain AF293. Numbers indicate time (hours) of incubation in liquid-submerged culture (Veg) and synchronized asexual developmental induction conditions (Asex). Equal loading of total RNA was evaluated by ethidium bromide staining of rRNA.
FluG is a key upstream activator of conidiation and is associated with the production of a small diffusible molecule (16). Loss-of-function fluG mutants form colonies exhibiting the nonsporulating fluffy phenotype (16). Our recent study showed that this FluG-dependent commencement of development in A. nidulans occurs via removal of the negative regulation imposed by the novel Zn(II)2Cys6 domain protein SfgA (30; reviewed in reference 44). Derepression of conidiation caused by FluG activity leads to the activation of the key downstream developmental activator brlA encoding a C2H2 zinc finger transcription factor, which activates expression of other genes required for asexual development (Fig. 1A) (1, 8; reviewed in references 4 and 44). Further genetic and biochemical studies identified two additional regulators of conidiation, abaA and wetA, that function downstream of brlA. The abaA gene encodes another developmental regulator that is activated by brlA during the middle stages of conidiophore development (5). The wetA gene functions in the late phase of conidiation for the synthesis of crucial cell wall components (19, 31). These three genes act in concert with other genes to control conidiation-specific gene expression and determine the order of gene activation during conidiophore development and spore maturation (Fig. 1A) (22, reviewed in reference 4).
The opportunistic human pathogen Aspergillus fumigatus is the most prevalent airborne fungal pathogen, and it causes severe and usually fatal invasive aspergillosis in immunocompromised patients (reviewed in reference 14). Moreover, the airborne A. fumigatus conidia are the primary agent for allergic bronchopulmonary aspergillosis, a severe pulmonary complication resulting from hypersensitivity to A. fumigatus proteins. Probably, among many characteristics, the ability to produce a massive number of small (2- to 3-μm-diameter) hydrophobic conidia and the thermotolerant nature of this fungus have contributed significantly to its ubiquitous presence and fitness in the environment (14). However, the mechanisms controlling asexual sporulation in A. fumigatus are largely unknown.
Based on the useful framework obtained from the study of A. nidulans development, we attempted to dissect the upstream and downstream regulatory mechanisms of conidiation in A. fumigatus. The facts that these two fungi are distantly related (9) and are different in the reproductive processes, i.e., A. fumigatus lacks a sexual cycle and produces a structurally different conidiophore (lacking metulae), led us to the hypothesis that the two aspergilli may have both conserved and distinct mechanisms controlling the conidiation process. Via comparative genome analyses, we have identified the homologues of FadA, FlbA, FluG, BrlA, and WetA in A. fumigatus, which are designated GpaA (17), AfFlbA, AfFluG, AfBrlA, and AfWetA, respectively. Deletion and additional genetic analyses in conjunction with expression and phenotypic studies revealed that AfFlbA and GpaA constitute the crucial G protein signaling components that coordinate vegetative growth and asexual development, implying that functions of these signaling elements are conserved in both species. Moreover, as found in A. nidulans, AfBrlA is essential for conidiophore formation, and AfFlbA and AfFluG are necessary for proper conidiation and AfbrlA expression in A. fumigatus. However, somewhat distinct from A. nidulans, AfFlbA or AfFluG is not absolutely required for conidiation or activation of AfbrlA in A. fumigatus. Taken together, we propose that, whereas a G protein (GpaA) signaling pathway and its regulation (AfFlbA) as well as downstream activation of conidiation are conserved in these distantly related model and pathogenic aspergilli, the imperfect fungus A. fumigatus has distinct and persistent mechanisms activating conidiation through AfBrlA.
MATERIALS AND METHODS
Aspergillus strains, growth conditions, and transformation.
A. fumigatus strains used in this study are listed in Table 1. Both A. fumigatus AF293 (wild type [WT] [7]) and AF293.1 (AfpyrG1 [36]) strains were used as WT. Standard culture and genetic techniques for A. nidulans were used (12, 24). The composition of minimal medium was as follows (per liter): 10 g glucose, 6 g NaNO3, 0.52 g MgSO4 · 7H2O, 0.52 g KCl, 1.52 g KH2PO4, and 1 ml of the 1,000× trace element solution [22 g/liter ZnSO4 · 7H2O, 11 g/liter H3BO3, 5 g/liter MnCl2 · 4H2O, 5 g/liter FeSO4 · 7H2O, 1.6 g/liter CoCl2 · 5H2O, 1.6 g/liter CuSO4 · 5H2O, 1.1 g/liter (NH4)6Mo7O24 · 4H2O, 50 g/liter Na2EDTA]. The mixture was then pH adjusted to 6.5 with 1.0 N NaOH. All strains were inoculated on solid (or liquid) minimal medium with appropriate supplements (5 mM uridine and 10 mM uracil; simplified as MM) and incubated at 37°C. If needed, yeast extract (YE) was added (0.1% or 0.5% final concentration). To observe development in liquid-submerged culture, all strains were inoculated with 5 × 105 conidia/ml in 100 ml liquid MM and incubated at 250 rpm at 37°C. The mycelial aggregates of each strain were observed microscopically every 3 h starting at 18 h of growth in liquid culture. Under these experimental conditions AF293 and AF293.1 elaborated conidiophores consistently. Standard A. nidulans transformation techniques (21, 37) were used.
TABLE 1.
Aspergillus strains used in this study
| Strain name | Relevant genotype | Source or reference |
|---|---|---|
| A. fumigatus | ||
| AF293 | Wild type | 7 |
| AF293.1 | AfpyrG1 | 36 |
| ΔAffluG1 | AfpyrG1 ΔAffluG::AfpyrG+ | This study |
| ΔAfflbA4 | AfpyrG1 ΔAfflbA::AfpyrG+ | This study |
| ΔAfbrlA7 | AfpyrG1 ΔAfbrlA::AfpyrG+ | This study |
| mGF01 | AfpyrG1 AfflbA1 white1 | This study |
| mGF24a | AfpyrG1 AfflbA24 | This study |
| tJH5.01 | AfpyrG1 AfflbA88 AfflbA+ AfpyrG+ | This study |
| tJH4.02 | AfpyrG1 AfpyrG+ | This study |
| tJH4.04 | AfpyrG1 gpaAQ204L AfpyrG+ | This study |
| tJH3.06 | AfpyrG1 AfflbA88 AfpyrG+ | This study |
| tJH3.09 | AfpyrG1 AfflbA88 gpaAG203R AfpyrG+ | This study |
| tJH6.05 | AfpyrG1 AfflbA88 gpaAG203R AfpyrG+ | This study |
| A. nidulans | ||
| FGSC26 | biA1 veA1 (wild type) | FGSCb |
| RJA4.4 | pyrG89 yA2 ΔfluG::trpC veA1 | 26 |
| RJA5.9 | pyrG89 ΔflbA::argB+pyroA4 veA1 | 27 |
| AJC11.32 | biA1 trpC801 brlA42 veA1 | 11 |
mGF24, -49, -50, -80, -88, -104, -112, -114, -115, -129, -130, -131, and -132 are likely isogenic except for the AfflbA mutant allele (Fig. 2C).
FGSC, Fungal Genetic Stock Center.
For synchronized asexual developmental induction, about 1 × 108 conidia of WT and relevant mutant strains were inoculated in 100 ml liquid MM with 0.1% YE and incubated at 37°C and 250 rpm for 18 h (0 h for developmental induction). Then, mycelia were harvested by being filtered through Miracloth (CalBiochem, California), transferred to solid MM with 0.1% YE, and further incubated at 37°C. Samples for RNA isolation were collected (conidia, 12, 18, 24, and 36 h of liquid culture and 6, 12, 24, and 48 h post-asexual developmental induction; Fig. 1B), squeeze dried, stored at −80°C, and subjected to total RNA isolation.
The AffluG, AfflbA, and AfbrlA null (deletion) mutants were generated by transforming AF293.1 with the individual PCR-generated deletion constructs (see below). The gpaAQ204L mutants were generated by transformation of AF293.1 with the PCR-generated gpaAQ204L construct along with the wild-type AfpyrG gene. The gpaAG203R mutants were generated by introducing the gpaAG203R construct and AfpyrG+ together into AF293.1 or mGF88 (AfflbA88; Table 1). The relevant genotype of each mutant was confirmed by PCR amplification of the coding regions followed by restriction enzyme digestion of the amplicons.
Mutagenesis and isolation of AfflbA loss-of-function mutants.
About 105 conidia of AF293.1 were inoculated on solid MM with 0.5% YE and supplements (5 mM uridine and 10 mM uracil) and incubated at 37°C for 4 days, and the conidia were collected for mutagenesis. Approximately 108 conidia of AF293.1 were treated with 1 μg/ml or 10 μg/ml (final concentration) of 4-nitroquinoline-1-oxide (6) for 0, 30, and 60 min, respectively, as previously described (26). The mean survival rate of a treatment with 1 μg/ml 4-nitroquinoline-1-oxide for 30 min was ∼70%, and more than 110,000 survivors of this condition were screened for morphological abnormalities.
Nucleic acid isolation and manipulation.
Genomic DNA isolation was carried out as previously described (41). Briefly, about 105 conidia of individual strains were inoculated in 2 ml liquid MM with 0.5% YE in 10-ml test tubes and incubated at 37°C for 18 h (stationary culture), and the mycelial mats were harvested and squeeze dried. Samples (0.2 to 0.5 g) were transferred to microcentrifuge tubes containing 400 μl of 0.5-mm zirconia/silica beads (BioSpec Products, Oklahoma), 500 μl of breaking buffer (2% Triton X-100, 1% sodium dodecyl sulfate, 0.1 M NaCl, 10 mM Tris-Cl [pH 8.0], 1 mM EDTA), and 500 μl of phenol-chloroform-isoamyl alcohol (25:24:1) and ground with a Mini Bead-Beater (BioSpec Products) for 2 min. The aqueous phase was separated by centrifugation, and genomic DNA was isolated as described previously (41).
Total RNA isolation and Northern blot analyses were carried out as described previously (26, 41). Total RNA was isolated from individual samples (about 0.2 g) by adding 400 μl of 0.5-mm zirconia/silica beads and 1 ml of Trizol reagent (Invitrogen, California) and grinding the mixture in a Mini Bead-Beater for 2.5 min. Subsequent RNA isolation was performed following the manufacturer's instructions. Total RNA (6 μg/lane) was separated by electrophoresis using a 1.1% agarose gel containing 6% formaldehyde. The nucleic acids were transferred to the MagnaProbe nylon membrane (0.45 μm; Osmonics, Minnesota). Probes were prepared by amplifying the coding regions of the individual genes from WT (AF293) genomic DNA. Primers are listed in Table S1 in the supplemental material. Each amplicon (1.43-kb AfflbA, 1.65-kb gpaA, 1.45-kb AffluG, 1.51-kb AfbrlA, and 1.34-kb AfwetA) was labeled with [32P]dCTP using the Prime-a-Gene system (Promega, Madison, WI) and used as a probe for Northern blot analyses. Hybridization was carried out using modified Church buffer (1 mM EDTA, 0.25 M Na2HPO4 · 7H2O, 1% hydrolyzed casein, 7% sodium dodecyl sulfate; adjusted to pH 7.4 with 85% H3PO4) as previously described (38).
Deletion constructs of AffluG, AfflbA, and AfbrlA were generated employing the double-joint PCR method (41). OKH237 and OKH238, OKH243 and OKH244, and OJH75 and OJH88 primer pairs (see Table S1 in the supplemental material) were used to amplify the 5′-flanking regions (∼1 kb) of AffluG, AfflbA, and AfbrlA, respectively. The 3′-flanking regions (∼1 kb) of the individual genes were amplified with OKH239 and OKH240 (AffluG), OKH245 and OKH246 (AfflbA), and OJH89 and OJH78 (AfbrlA). Primers away from the open reading frames (ORFs) contained 22 bases of homologous sequences overlapping with the ends of AfpyrG+. The selective marker AfpyrG+ was amplified with OKH235 and OKH236 (for ΔAffluG and ΔAfflbA) or OJH84 and OJH85 (for ΔAfbrlA). Three amplicons were mixed in a 1:2:1 ratio, and the second round of PCR was carried out (41). Using the second-round PCR products as templates, the final deletion constructs were generated with nested primer pairs OKH241 and OKH242 (ΔAffluG), OKH247 and OKH248 (ΔAfflbA), and OJH79 and OJH80 (ΔAfbrlA) and used for transformation of AF293.1.
For sequencing analyses of the AfflbA mutant alleles, the AfflbA coding regions from 14 AfflbA− mutants were amplified by PCR using OJH68 and OJH42 (for the N-terminal region) and OJH43 and OKH246 (for the C-terminal region). The gpaA coding region was amplified with OJH12 and OJH17. The resulting amplicons were sequenced directly.
The gpaAQ204L dominant activating mutant allele was generated by site-directed mutagenesis with oligonucleotides OJH69 (paired with OJH12 for 5′-flanking region) and OJH70 (paired with OJH17 for 3′-flanking region). These oligonucleotides introduced a BglII site for screening convenience. The gpaAG203R dominant interfering mutant allele was generated with OJH71 (paired with OJH12 for 5′-flanking region) and OJH72 (paired with OJH17 for 3′-flanking region), which introduced a NaeI site. The resulting amplicons were joined as described previously (41), and each construct was confirmed by digestion with BglII and NaeI. The final gpaAQ204L and gpaAG203R amplicons were cointroduced with AfpyrG+ to AF293.1 (and AfflbA88 for gpaAG203R). Transformants were screened for the presence of the mutant (gpaAQ204L or gpaAG203R) allele by PCR followed by restriction enzyme digestion.
Quantitative analyses of conidiation levels.
The numbers of conidia in various strains were determined in two ways: incubation time and the age of the colony section (distance from the center of the colony). In both cases, WT and relevant mutant strains were point inoculated and grown on solid MM with 0.5% YE at 37°C. The conidia were collected in 0.01% Tween 20 at 2, 3, 4, and 5 days of incubation from the entire plate (colony) and counted. To measure spores in the different regions of the colony grown for 5 days, conidia were collected from the center, middle, and edge of the colonies and counted using a hemocytometer.
Microscopy.
The colony photographs were taken using a Sony DSC-F828 digital camera. Photomicrographs were taken using an Olympus BH2 compound microscope installed with an Olympus DP-70 digital imaging system.
RESULTS
Identification and expression of key developmental regulators in A. fumigatus.
To begin to understand the regulatory mechanisms of growth and development in A. fumigatus, we first identified the A. fumigatus homologues of the five key A. nidulans regulators (Fig. 1A). Blastp analyses of the A. fumigatus genome (TIGR [http://www.tigr.org/tdb/e2k1/afu1/]) using the A. nidulans FadA, FlbA, FluG, BrlA, and WetA proteins as queries have identified GpaA (97% identity, 98% similarity with FadA in A. nidulans [17]), AfFlbA (79% identity, 85% similarity), AfFluG (69% identity, 83% similarity), AfBrlA (68% identity, 77% similarity), and AfWetA (56% identity, 66% similarity). The locus numbers are presented in Fig. 1B.
To check whether these genes are expressed, we examined the mRNA levels of individual genes throughout the life cycle of an A. fumigatus WT strain (AF293) by Northern blot analyses. As shown in Fig. 1B, the AfflbA and AffluG genes were found to encode 3.3-kb and 3.4-kb transcripts, respectively, which were present at relatively constant levels throughout the life cycle. Hybridization with the gpaA probe resulted in the detection of two (∼3.0-kb and 2.4-kb) transcripts, whose levels are high in vegetative growth, low in conidiation, and absent in conidia. While it can be speculated that the gpaA gene may encode two transcripts, a potential cross-hybridization cannot be excluded due to the high nucleotide level identity between gpaA and gpaB (EAL90625; 63% identity) or AfganA (EAL92343; 61% identity). The AfbrlA and AfwetA genes were specifically expressed during conidiation. The AfbrlA transcript reached the highest level at 6 h and then decreased, whereas the AfwetA mRNA began to accumulate at 12 h post-developmental induction and continued to accumulate. In accordance with the occurrence of conidiophore formation in WT, AfbrlA transcripts (reviewed in reference 4) were clearly visible at 18 and 24 h of liquid MM-submerged culture conditions (see below).
Deletion and 14 additional loss-of-function AfflbA mutations cause reduced conidiation.
We first attempted to dissect the role of AfFlbA in developmental regulation by generating the ΔAfflbA mutant. Multiple ΔAfflbA mutants were isolated and examined for phenotypic changes. Similar to the A. nidulans ΔflbA mutant, the ΔAfflbA mutant exhibited the fluffy phenotype during the first 2 days of growth. In contrast, while the A. nidulans ΔflbA mutant continues to accumulate hyphal mass without development, resulting in autolysis of the colony (15), the ΔAfflbA mutant started to produce conidiophores from the center of the colony and did not undergo hyphal disintegration (Fig. 2A). However, the levels of conidiation (and spore pigmentation) in the ΔAfflbA mutant were dramatically reduced (∼30% of WT), indicating that AfFlbA is necessary for the normal levels of conidiation in A. fumigatus, but the fungus can overcome the developmental defect caused by the lack of a key RGS protein.
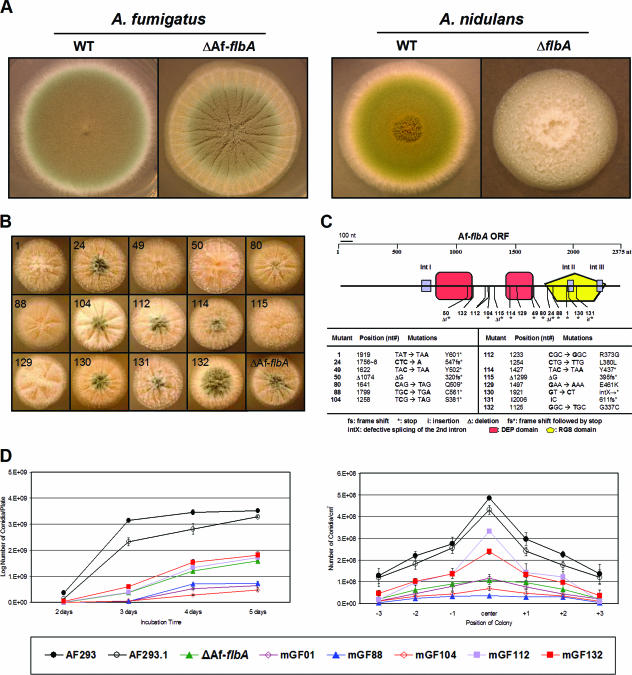
FIG. 2.
Phenotypes of various AfflbA mutants. (A) WT (AF293 and FGSC26) and ΔflbA (ΔAfflbA4 and RJA5.9) strains of A. fumigatus and A. nidulans were point inoculated on solid MM with 0.5% YE and incubated for 3 days at 37°C. Note the differences between the A. nidulans and A. fumigatus flbA mutant phenotypes. (B) Fourteen AfflbA mutant strains and the ΔflbA mutant were point inoculated on solid MM with 0.5% YE and incubated for 2 days at 37°C. Numbers indicate mutant strains and alleles. (C) The AfflbA ORF composed of 2,375 nucleotides (nt) (including three introns, IntI, IntII, and IntIII) and the approximate position of each AfflbA mutation are schematically presented. In the table, the allele number, position, and nature of the nucleotide and resulting amino acid change(s) for each AfflbA mutant allele are presented. (D) Two graphs present the results of quantitative analyses of conidiation in WT (AF293 and AF293.1) and selected AfflbA mutant strains inoculated on solid MM with 0.5% YE. Error bars indicate standard deviations calculated from biological triplicates. Note that mutations in AfflbA result in a reduced number of conidia in both cases.
To further confirm that reduced conidiation is due to loss of AfflbA function, we isolated 14 mutants exhibiting phenotypes similar to those of the ΔAfflbA mutant via random chemical mutagenesis (see Materials and Methods). These mutants showed various levels of enhanced vegetative growth (at the early phase of growth) with reduced sporulation (Fig. 2B). To test whether such phenotypic changes were related to loss of AfflbA function, the AfflbA coding regions from all 14 mutants were PCR amplified and the individual amplicons were directly sequenced. As presented in Fig. 2C, sequence analyses revealed that all 14 mutants had mutations within the AfflbA ORF.
Among 14 mutants, mGF104, mGF114, mGF49, mGF80, mGF88, and mGF1 are derived from nonsense mutations and exhibit phenotypes similar to those of the AfflbA deletion mutant. Three mutants that have missense mutations prior to the RGS domain show various degrees of sporulation. mGF129 has a G-to-A transition, causing a mutation of the 461st amino acid Glu (acidic) to Lys (basic), which likely abolishes AfFlbA function. mGF112 has both missense (Arg373Gly) and silent (Leu380Leu) mutations, where the R373G substitution is likely responsible for the mutant phenotype. Sporulation of mGF132 that had the G337C substitution was affected less severely than that of other mutants. mGF24, mGF50, and mGF115 are all derived from deletion followed by frameshift and early termination within or before the RGS domain. The mGF131 mutant has a C insertion followed by frameshift and early termination. mGF130 is derived from a GT-to-CT transversion, which likely blocks the splicing of the second introns, resulting in frameshift and early termination.
In order to confirm whether the mutations in AfflbA are solely responsible for the phenotype, mGF01, mGF88, and mGF104 were transformed with the wild-type AfflbA gene and AfpyrG+, and 40%, 54%, and 22% of the transformants in each case restored the WT phenotype (Fig. 3 shows results for AfflbA88 [mGF88] and AfflbA88 with AfflbA+). Interestingly, introduction of AfflbA+ into mGF01, which produces white conidia, restored conidiation, but not spore pigmentation, to the WT level, indicating that mGF01 may have two mutations (AfflbA1 and white1; Table 1).
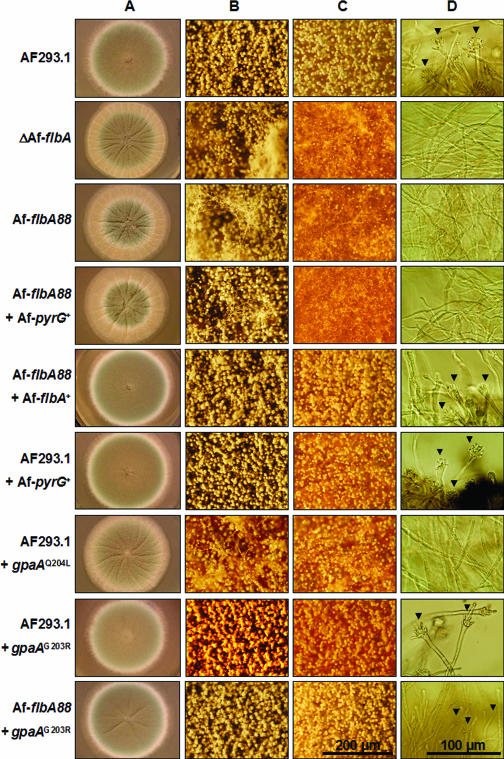
FIG. 3.
GpaA is the cognate Gα for AfFlbA. WT (AF293.1 and AF293.1 with AfpyrG+), ΔAfflbA, AfflbA88, AfflbA88 with AfpyrG+ alone, AfflbA88 with AfflbA+, AF293.1 with gpaAQ204L, AF293.1 with gpaAG203R, and AfflbA88 with gpaAG203R strains were point inoculated on solid MM with 0.5% YE (A, B, and C) and incubated for 3 days at 37°C. (A to C) Entire colonies (A) and close-up views of the center (B) and the edges (C) of individual colonies (bar, 200 μm). (D) Developmental status of tested strains in liquid-submerged culture (MM) was photographed at 24 h of incubation. Note that, while WT (AF293.1 and AF293.1 with AfpyrG+), AfflbA88 with AfflbA+, AF293.1 with gpaAG203R, and AfflbA88 with gpaAG203R strains form conidiophores (marked by arrowheads), ΔAfflbA, AfflbA88, AfflbA88 with AfpyrG+ alone, and AF293.1 with gpaAQ204L strains do not sporulate (bar, 100 μm).
Due to the evident reduction in conidiation levels of various AfflbA− mutants, quantitative analyses of spore formation were carried out by measuring the number of conidia produced by WT, ΔAfflbA, and five selected AfflbA− strains. This was accomplished in two ways: (i) counting conidia from the entire point-inoculated colony grown for 2 to 5 days and (ii) counting conidia from the center (old region), middle, and edge (actively growing region) of the 5-day-old colony. In both cases, all AfflbA− mutants tested exhibited reduced (10 to 70% of WT) levels of conidiation (Fig. 2D). Moreover, unlike WT, AfflbA− mutants did not produce conidiophores in liquid-submerged culture (Fig. 3D). These results indicate that, while it is not absolutely required for conidiation, AfFlbA is needed for proper asexual sporulation in A. fumigatus.
GpaA is the primary target of AfFlbA.
High (97%) identity between GpaA (17) and FadA led us to hypothesize that GpgA is the primary target of AfFlbA and that uncontrolled activation of GpaA-mediated signaling causes reduced conidiation. We tested this hypothesis by generating the constitutively active (Q204L) and dominant interfering (G203R) mutant GpaA alleles (references 39 and 40 and references therein) and examining the phenotypic changes caused by these mutations. If AfFlbA regulates GpaA negatively, GpaAQ204L should cause the phenotypic alterations similar to those resulting from loss of AfflbA function and GpaAG203R should suppress the altered sporulation caused by AfflbA mutations in a dominant manner. Keeping this in mind, we first generated gpaAQ204L and gpaAG203R mutant strains that are heterozygous for gpaA by cointroducing each construct with AfpyrG+ into a WT strain (AF293.1, pyrG1). To generate the AfflbA− gpaAG203R double mutant, mGF88 (AfflbA88; pyrG1) was transformed with the gpaAG203R construct and AfpyrG+ or with AfpyrG+ alone. As shown in Fig. 3, introduction of the gpaAQ204L allele into AF293.1 yielded the colonies exhibiting reduced conidiation and the absence of conidiophore formation in liquid-submerged culture as observed in AfflbA− mutants. Moreover, somewhat similarly to those found in A. nidulans (39), the gpaAG203R mutants showed reduced radial growth with normal conidiation levels. Importantly, the introduction of the gpaAG203R mutant allele into the AfflbA88 mutant restored conidiation in both air-exposed and liquid-submerged culture conditions (Fig. 3, bottommost panel). Collectively, these results corroborate the hypothesis that GpaA is the cognate Gα for AfFlbA and that GpaA signaling stimulates hyphal growth while inhibiting asexual sporulation in A. fumigatus.
A potential role of AffluG in sporulation.
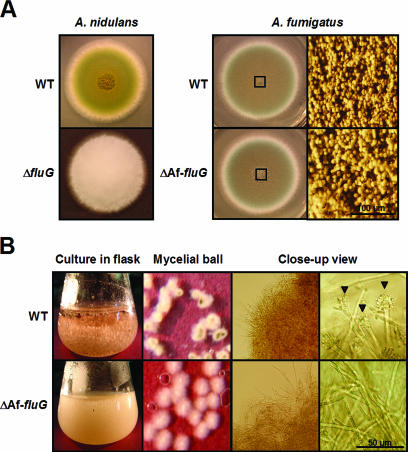
As the upstream developmental activator FluG is required for the commencement of conidiation in A. nidulans, the ΔfluG mutant exhibits the nonconidial fluffy (but not autolytic) phenotype (Fig. 4A) (16). To test whether the A. fumigatus FluG homologue is needed for conidiation in A. fumigatus, the AffluG deletion mutant was generated. Somewhat unexpectedly, the AffluG deletion mutant could sporulate normally like WT but formed slightly increased levels of aerial hyphae in air-exposed culture (solid medium) conditions (Fig. 4A), indicating that activation of A. fumigatus conidiation in the presence of air does not require the activity of AffluG. However, while A. fumigatus WT strains sporulated within 24 h in liquid-submerged culture, the AffluG deletion mutant never produced conidiophores up to 32 h. Moreover, the AffluG deletion mutant formed less compact mycelial aggregates (Fig. 4B). Taken together, it can be speculated that, while AfFluG may play a certain role in conidiation, the presence of air can bypass the need for AfFluG in conidiophore development in A. fumigatus. A potential role of AfFluG in conidiation was further tested by examining the expression of AfbrlA (see below).
FIG. 4.
Phenotypes of the ΔAffluG mutant. (A) WT (FGSC26 and AF293) and ΔfluG (RJA4.4 and ΔAffluG1) strains of A. nidulans and A. fumigatus were point inoculated on solid MM with 0.5% YE and incubated at 37°C for 3 days. Note that deletion of fluG in A. nidulans caused the fluffy nonconidial phenotype, whereas deletion of AffluG in A. fumigatus did not affect conidiation severely (bar, 100 μm). The rightmost panel shows the close-up views of the centers of the colonies (squares). (B) Morphology of the mycelial aggregates and developmental status in liquid MM-submerged culture were photographed at 24 h of incubation. Note that the ΔAffluG mutant forms less compact mycelial aggregates and does not sporulate (bar, 50 μm). The arrowheads indicate conidiophores elaborated by the wild type.
AfBrlA is required for conidiophore formation.
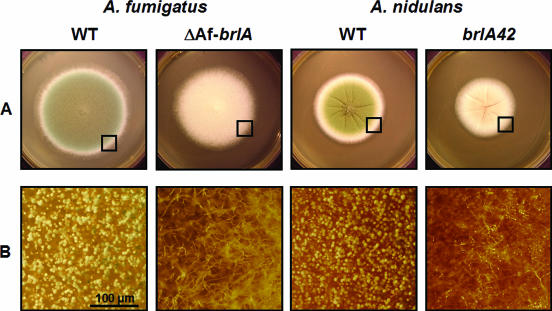
The result suggesting that A. fumigatus may have a distinct upstream regulatory mechanism(s) for the activation of sporulation led us to test whether downstream regulation of conidiation by BrlA is divergent in the two aspergilli. To test this, multiple AfbrlA deletion mutant strains were generated. As shown in Fig. 5, deletion of AfbrlA completely eliminated asexual development in A. fumigatus, resulting in the colonies displaying elongated aerial hyphae and increased hyphal mass. These characteristics of the ΔAfbrlA mutant are more similar to those of the A. nidulans fluffy mutants than the A. nidulans brlA mutants, which form flat colonies. In any case, it is clear that AfbrlA is essential for the formation of conidiophores in A. fumigatus and that the role of the key downstream developmental activator BrlA in asexual development is conserved in the two aspergilli.
FIG. 5.
Requirement of AfbrlA in A. fumigatus conidiation. (A) WT (AF293 and FGSC26) and brlA mutant (ΔAfbrlA7 and AJC11.32) strains of A. fumigatus and A. nidulans were point inoculated on solid MM with 0.5% YE and incubated at 37°C for 3 days. (B) Close-up views of the edges of the colonies (marked by squares; bar, 100 μm). Note that the AfbrlA deletion mutant fails to produce conidiophores.
AfFluG and AfFlbA are required for proper expression of AfbrlA.
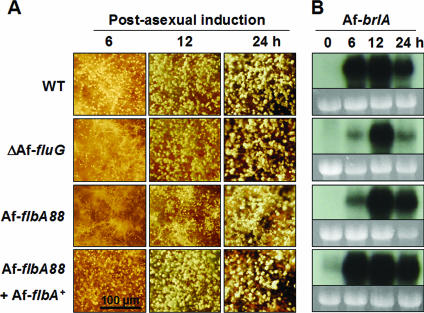
In A. nidulans, both fluG and flbA are required for the expression of brlA (15, 16). The facts that AfbrlA is necessary for conidiophore formation and that accumulation of its transcript(s) is specifically coupled with conidiation (Fig. 1B) led us to test whether AfFlbA and AfFluG affect AfbrlA expression. Synchronous developmental induction of WT (AF293), ΔAffluG, AfflbA88, and AfflbA88 with AfflbA+ strains was carried out, and changes in the development and brlA expression patterns were examined. We found that the ΔAffluG and AfflbA88 mutants, but not WT or AfflbA88 with AfflbA+, exhibited delayed conidiation with increased aerial hyphae during the early phase (6 to 12 h) of post-asexual developmental induction (Fig. 6A). Furthermore, while WT and AfflbA88 with AfflbA+ strains accumulated high levels of AfbrlA transcript at 6 h post-developmental induction, the ΔAffluG and AfflbA88 mutants showed the highest level of AfbrlA accumulation at 12 h (Fig. 6B). These results indicate that AfFluG and AfFlbA (at least partially) function in conidiation by influencing expression of AfbrlA. However, in contrast to A. nidulans, these two upstream regulators are not absolutely required for activation of AfbrlA expression or conidiation in A. fumigatus.
FIG. 6.
AffluG and AfflbA are required for proper activation of AfbrlA. (A) Progression of conidiation during synchronous asexual developmental induction is shown (bar, 100 μm). Numbers indicate time (hours) of post-asexual developmental induction. Note that AffluG and AfflbA mutant strains exhibit enhanced formation of aerial hyphae and reduced (and delayed) sporulation. (B) Corresponding Northern blots of the samples shown in panel A (0 h = 18 h in culture submerged in liquid MM with 0.1% YE). Equal loading of total RNA was evaluated by ethidium bromide staining of rRNA. Note that mutational inactivation of AffluG or AfflbA causes delayed and reduced AfbrlA mRNA accumulation. The AfflbA88 with AfflbA+ strain shows slightly elevated AfbrlA expression levels, which is likely due to the presence of two copies of AfflbA+ (data not shown).
DISCUSSION
The ease of genetic analyses and the availability of various experimental tools have made A. nidulans an excellent model system for studying signal transduction, multicellular development, and secondary metabolism (20, 34, 42, 43). While the study of developmental regulation in A. nidulans has provided valuable information, the potential use of such knowledge in dissecting the mechanisms controlling growth and development in other aspergilli remained to be tested. In this study, we examined the roles of the four key A. nidulans regulators in controlling development of the pathogenic fungus A. fumigatus and demonstrated that these two Aspergillus species have conserved Gα-RGS signaling components and a core downstream activator of sporulation, but A. fumigatus may have distinct upstream mechanisms activating AfbrlA.
We previously showed that the A. nidulans RGS protein FlbA (fluffy low brlA locus A) has a major role in determining the balance between vegetative growth and development through its ability to down-regulate FadA (39). When FadA-dependent signaling is activated in response to some unknown factor, it stimulates growth and blocks both asexual and sexual development. Attenuation of FadA-mediated vegetative growth signaling by FlbA allows development to occur. Inactivation of FlbA or constitutive activation of FadA, e.g., by the G42R, R178L, G183S, R178C, or Q204L mutation predicted to cause reduced (or lack of) intrinsic GTPase activity of FadA, results in uncontrolled FadA signaling and leads to proliferation of undifferentiated aerial hyphae that autolyze as colonies mature (35, 39, 40). In contrast, overexpression of flbA or a dominant interfering mutation in FadA (G203R) results in inhibited hyphal growth coupled with hyperactive conidiation (15, 39). The flbA loss-of-function or dominant activating fadA mutations result in the fluffy-autolytic phenotype regardless of the veA1 or veA+ alleles (our unpublished data). VeA is a novel multifunctional protein balancing sexual and asexual development in A. nidulans and influencing production of pigments and secondary metabolite in other aspergilli (see reference 42 and references therein).
Due to such a critical function of FlbA in A. nidulans, we first investigated the role of the FlbA homologue AfFlbA in A. fumigatus. Deletion and 14 other loss-of-function AfflbA mutations resulted in reduced levels of conidiation and conidial pigmentation. Furthermore, it also caused increased hyphal proliferation during the early period of growth (up to 2 days), and the mutant colonies exhibited an expanded growing edge with delayed conidiation, while WT colonies showed vigorous production of conidiophores (Fig. 3A and C). In addition, the ΔAfflbA or AfflbA88 mutants did not produce conidiophores in liquid-submerged culture conditions, whereas WT and AfflbA complemented (AfflbA88 with AfflbA+) strains elaborated conidiophores abundantly. Collectively, these findings suggest that AfFlbA functions in down-regulation of hyphal proliferation and (indirect) activation of development in A. fumigatus, too. However, there is a noticeable difference between the phenotypes of the A. nidulans ΔflbA and ΔAfflbA mutants, where the latter never undergoes autolysis. Such a difference can be explained by a speculation that A. fumigatus has multiple mechanisms activating development, which bypass the requirement of AfFlbA in sporulation and allow the AfflbA mutants to escape hyphal disintegration. It is important to note that the A. nidulans ΔflbA mutant cannot proceed to development. This speculation is further studied by examining the role of AfFluG (see below).
As the FadA homologue GpaA is the primary target of AfFlbA, the constitutively active GpaAQ204L mutation caused increased hyphal proliferation and reduced sporulation in a dominant manner. Moreover, the dominant interfering GpaAG203R mutation restored conidiation in the AfflbA88 mutant to the WT level in both air-exposed and liquid-submerged culture conditions (Fig. 3). The G203R mutation is predicted to block the conformational change in the switch II region of GpaA and thereby prevent dissociation of GTP-Gα from Gβγ (39, 40). These results indicate that inactivation of GpaA signaling circumvents the need for AfFlbA in proper progression of conidiation and corroborate the idea that GpaA and AfFlbA constitute a Gα-RGS pair, which functions as a major coordinator of growth and development in A. fumigatus. Interestingly, the levels of gpaA mRNA(s) appeared to be low during asexual development and absent in conidia in comparison to those observed during vegetative growth. If this is a part of the means by which the level of GpaA signaling is controlled, it indicates that the two aspergilli may have different regulatory mechanisms, because both the mRNA and the protein levels of fadA were relatively constant throughout the life cycle of A. nidulans (39).
In A. nidulans, both the FadA and GanB (another Gα) signaling pathways are involved in activation of cAMP-dependent protein kinase A (PKA [13, 32]). Two PKA catalytic subunits, PkaA and PkaB, have been shown to stimulate vegetative growth, where PkaA plays a primary role (23, 32). Deletion of pkaA resulted in hyperactive conidiation with restricted vegetative growth and suppressed developmental defects caused by ΔflbA as well as the dominant activating fadAG42R mutation. Furthermore, overexpression of pkaA led to reduced sporulation with elevated hyphal proliferation (32). Later, PkaA was also shown to function downstream of GanB for conidial germination (13). Similarly, in A. fumigatus, GpaB (GanB homologue)-mediated signaling is associated with activation of the predominant PKA catalytic subunit PkaC1 (18). However, one critical difference between two fungi is that, while deletion of pkaC1 also resulted in restricted hyphal growth, it caused drastically reduced sporulation (18). These findings indicate that, whereas the Gα-RGS level regulatory mechanism is conserved, asexual development is regulated differently at the level of PKA in the two aspergilli. The potential involvement of PkaC1 in the GpaA signaling pathway remains to be investigated.
The study of asexual development in A. nidulans has identified a number of genes required for the activation of conidiation, where FluG functions most upstream (reviewed in reference 4; see also reference 44). The FluG-dependent sporulation requires the key downstream transcription factor BrlA, a C2H2 zinc finger DNA-binding protein, which activates development-specific gene expression beginning at the time of conidiophore vesicle formation (1, 2). Since FluG and BrlA represent key upstream and downstream activators of conidiation, we attempted to characterize the homologues of these regulators in A. fumigatus. Expression patterns of AffluG and AfbrlA were almost identical to those found in A. nidulans (reviewed in reference 4). However, mRNA of AfbrlA accumulates highly at 6 h post-developmental induction, whereas it takes about 12 h for the A. nidulans brlA mRNA to reach the same level (Fig. 1B) (30). Moreover, AfbrlA mRNA(s) started to accumulate as early as 18 h of vegetative growth in liquid-submerged culture conditions, at which time no A. nidulans brlA mRNA is clearly detectable. These results are in agreement with our observations that A. fumigatus WT strains sporulate in liquid-submerged culture and produce conidiophores much faster than A. nidulans does under synchronous developmental induction conditions (not shown).
Deletion of AfbrlA completely eliminated conidiation in all conditions tested, indicating that the activation of AfbrlA expression early in conidiophore development also represents a foremost and essential control point for initiating the conidiation pathway in A. fumigatus and that the two aspergilli have a common core downstream activator for conidiophore development. However, somewhat unexpectedly, AfFluG is found to be dispensable for conidiation in the presence of air, which is in contrast to the necessity for FluG in A. nidulans conidiation. On the other hand, we also found that AfFluG (at least partially) contributes to the commencement of development under different culture conditions, i.e., liquid MM-submerged culture and synchronous developmental induction, through affecting expression of AfbrlA (Fig. 4 and 6). We also demonstrated that AfFlbA is necessary for the proper expression of AfbrlA and thereby progression of conidiation. Collectively, our phenotypic, expression, and genetic studies all suggest that the pathogenic fungus A. fumigatus may have more than one mechanism activating AfbrlA and unique and powerful strategies for its asexual reproduction. In A. nidulans, both inhibition of G protein-mediated growth signaling by FlbA and activation of developmental functions by FluG must occur in order for the development to proceed (reviewed in reference 4). Thus, together with the fact that both AfFluG and AfFlbA are required for proper expression of AfbrlA, it will be interesting to test whether removal of both AfflbA and AffluG functions would cause additive detrimental effects on development of A. fumigatus.
Regarding a possible FluG-independent developmental activation branch, it is noteworthy that the newly identified A. nidulans tmpA gene regulates conidiation independently of the FluG pathway (33). The TmpA protein belongs to a novel family of putative membrane flavoproteins that may be involved in the synthesis of a (different) developmental signal. The absence of tmpA resulted in decreased brlA expression and conidiation on solid medium, and overexpression of tmpA tagged alleles caused conidiation in liquid-submerged culture. Three lines of evidence indicate that TmpA and FluG regulate conidiation through independent pathways: (i) conidiation of the ΔtmpA mutant could be restored by juxtaposed growth with WT or the ΔfluG mutant, (ii) overexpression of fluG induced conidiation independently of tmpA, and (iii) the ΔtmpA ΔfluG double mutants exhibited an additive fluffy phenotype (33). If the homologue of TmpA plays a similar role in A. fumigatus conidiation, it can be speculated that the presence of either AfFluG or AfTmpA function alone may be sufficient to confer the progression of conidiation in A. fumigatus.
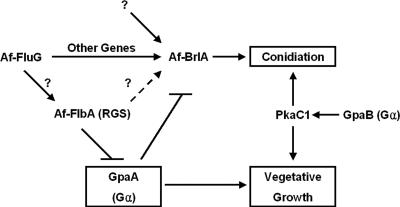
Finally, based on our findings, we present a genetic model for regulation of asexual development and vegetative growth in A. fumigatus (Fig. 7). In this model, similar to the one proposed for A. nidulans, AfFlbA functions as the major negative regulator of GpaA-mediated signaling that stimulates vegetative growth, which in turn inhibits sporulation. The GpaB-PkaC signaling pathway has been proposed to induce both hyphal growth and conidiation (18). AfBrlA is essential for the activation of conidiophore formation, and its expression is influenced in part by AfFluG and AfFlbA. The potential presence of an upstream mechanism(s) activating AfbrlA that is independent of AfFluG is indicated. The roles of other G protein components and FLB genes in conidiation as well as the involvement of negative regulators of conidiation including SfgA in A. fumigatus remain to be studied. Experiments testing the roles of these A. fumigatus developmental regulators in gliotoxin production (reviewed in reference 14) and virulence establishment are currently being carried out.
FIG. 7.
Model for growth and developmental control in A. fumigatus. Activation of GpaA stimulates hyphal growth and represses conidiation. This GpaA-dependent signaling pathway is attenuated by AfFlbA. A potential role of AfFluG in activating AfFlbA is indicated by a question mark. Activation of conidiation does not absolutely require the activity of AfFluG or AfFlbA. The potential existence of an AfFluG-independent pathway(s) activating AfbrlA is indicated. The GpaB-PkaC1 pathway has been proposed to be responsible for both conidiation and vegetative growth (18).
Supplementary Material
Acknowledgments
We thank Kap-Hoon Han and Greg Flygt for experimental support and Ellin Doyle for critically reviewing the manuscript.
This work was supported by University of Wisconsin Graduate School and National Science Foundation grants (MCB-0421863) to J.-H.Y. J.-H.M. was in part supported by the Korea Research Foundation (KRF-2004-214-F00041).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 2.Adams, T. H., H. Deising, and W. E. Timberlake. 1990. brlA requires both zinc fingers to induce development. Mol. Cell. Biol. 10:1815-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams, T. H. 1994. Asexual sporulation in higher fungi, p. 367-382. In N. A. R. Gow and G. M. Gadd (ed.), The growing fungus. Chapman & Hall, London, United Kingdom.
- 4.Adams, T. H., J. K. Wieser, and J.-H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrianopoulos, A., and W. E. Timberlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bal, J., E. M. Kajtaniak, and N. J. Pieniazek. 1977. 4-Nitroquinoline-1-oxide: a good mutagen for Aspergillus nidulans. Mutat. Res. 56:153-156. [Google Scholar]
- 7.Brookman, J. L., and D. W. Denning. 2000. Molecular genetics in Aspergillus fumigatus. Curr. Opin. Microbiol. 3:468-474. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and W. E. Timberlake. 1992. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galagan, J. E., S. E. Calvo, C. Cuomo, L.-J. Ma, J. R. Wortman, S. Batzoglou, S.-I. Lee, M. Bastürkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. d'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. Á. Peñalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 10.Hicks, J. K., J.-H. Yu, N. P. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnstone, I. L., S. G. Hughes, and A. J. Clutterbuck. 1985. Cloning an Aspergillus nidulans developmental gene by transformation. EMBO J. 4:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Käfer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 13.Lafon, A., J.-A. Seo, K.-H. Han, J.-H. Yu, and C. d'Enfert. 2005. The heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latgé, J.-P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, B. N., and T. H. Adams. 1994. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14:323-334. [DOI] [PubMed] [Google Scholar]
- 16.Lee, B. N., and T. H. Adams. 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 8:641-651. [DOI] [PubMed] [Google Scholar]
- 17.Liebmann, B., S. Gattung, B. Jahn, and A. A. Brakhage. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269:420-435. [DOI] [PubMed] [Google Scholar]
- 18.Liebmann, B., M. Müller, A. Braun, and A. A. Brakhage. 2004. The cyclic AMP-dependent protein kinase A network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72:5193-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall, M. A., and W. E. Timberlake. 1991. Aspergillus nidulans wetA activates spore-specific gene expression. Mol. Cell. Biol. 11:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinelli, S. D. 1994. Aspergillus nidulans as an experimental organism. Prog. Ind. Microbiol. 29:33-58. [PubMed] [Google Scholar]
- 21.Miller, B. L., K. Y. Miller, and W. E. Timberlake. 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 5:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirabito, P. M., T. H. Adams, and W. E. Timberlake. 1989. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57:859-868. [DOI] [PubMed] [Google Scholar]
- 23.Ni, M., S. Rierson, J.-A. Seo, and J.-H. Yu. 2005. The pkaB gene encoding the secondary protein kinase A catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans. Eukaryot. Cell 4:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. A. Macdonald, and W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 25.Rosén, S., J.-H. Yu, and T. H. Adams. 1999. The Aspergillus nidulans sfaD gene encodes a G protein β subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo, J.-A., Y. Guan, and J.-H. Yu. 2003. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans. Genetics 165:1083-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo, J.-A., K.-H. Han, and J.-H. Yu. 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53:1611-1623. [DOI] [PubMed] [Google Scholar]
- 28.Seo, J.-A., K.-H. Han, and J.-H. Yu. 2005. Multiple roles of a heterotrimeric G-protein γ-subunit in governing growth and development of Aspergillus nidulans. Genetics 171:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo, J.-A., and J.-H. Yu. 2006. The phosducin-like protein PhnA is required for Gβγ-mediated signaling for vegetative growth, developmental control, and toxin biosynthesis in Aspergillus nidulans. Eukaryot. Cell 5:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo, J.-A., Y. Guan, and J.-H. Yu. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172:1535-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sewall, T. C., C. W. Mims, and W. E. Timberlake. 1990. Conidium differentiation in Aspergillus nidulans wild-type and wet-white (wetA) mutant strains. Dev. Biol. 138:499-508. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soid-Raggi, G., O. Sánchez, and J. Aguirre. 2006. TmpA, a member of a novel family of putative membrane flavoproteins, regulates asexual development in Aspergillus nidulans. Mol. Microbiol. 59:854-869. [DOI] [PubMed] [Google Scholar]
- 34.Timberlake, W. E. 1990. Molecular genetics of Aspergillus development. Annu. Rev. Genet. 24:5-36. [DOI] [PubMed] [Google Scholar]
- 35.Wieser, J., J.-H. Yu, and T. H. Adams. 1997. Dominant mutations affecting both sporulation and sterigmatocystin biosynthesis in Aspergillus nidulans. Curr. Genet. 32:218-224. [DOI] [PubMed] [Google Scholar]
- 36.Xue, T., C. K. Nguyen, A. Romans, D. P. Kontoyiannis, and G. S. May. 2004. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch. Microbiol. 182:346-353. [DOI] [PubMed] [Google Scholar]
- 37.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, J.-H., and T. J. Leonard. 1995. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J. Bacteriol. 177:4792-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, J.-H., J. Wieser, and T. H. Adams. 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15:5184-5190. [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, J.-H., S. Rosén, and T. H. Adams. 1999. Extragenic suppressors of loss-of-function mutations in the Aspergillus FlbA regulator of G-protein signaling domain protein. Genetics 151:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, J.-H., Z. Hamari, K.-H. Han, J.-A. Seo, Y. Reyes-Domínguez, and C. Scazzocchio. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 42.Yu, J.-H., and N. P. Keller. 2005. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43:437-458. [DOI] [PubMed] [Google Scholar]
- 43.Yu, J.-H. 2006. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44:145-154. [PubMed] [Google Scholar]
- 44.Yu, J.-H., J.-H. Mah, and J.-A. Seo. 2006. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot. Cell 5:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.