Abstract
The pellicle of the protozoan parasite Toxoplasma gondii is a unique triple bilayer structure, consisting of the plasma membrane and two tightly apposed membranes of the underlying inner membrane complex. Integral membrane proteins of the pellicle are likely to play critical roles in host cell recognition, attachment, and invasion, but few such proteins have been identified. This is in large part because the parasite surface is dominated by a family of abundant and highly immunogenic glycosylphosphatidylinositol (GPI)-anchored proteins, which has made the identification of non-GPI-linked proteins difficult. To identify such proteins, we have developed a radiolabeling approach using the hydrophobic, photoactivatable compound 5-[125I]iodonaphthalene-1-azide (INA). INA can be activated by photosensitizing fluorochromes; by restricting these fluorochromes to the pellicle, [125I]INA labeling will selectively target non-GPI-anchored membrane-embedded proteins of the pellicle. We demonstrate here that three known membrane proteins of the pellicle can indeed be labeled by photosensitization with INA. In addition, this approach has identified a novel 22-kDa protein, named PhIL1 (photosensitized INA-labeled protein 1), with unexpected properties. While the INA labeling of PhIL1 is consistent with an integral membrane protein, the protein has neither a transmembrane domain nor predicted sites of lipid modification. PhIL1 is conserved in apicomplexan parasites and localizes to the parasite periphery, concentrated at the apical end just basal to the conoid. Detergent extraction and immunolocalization data suggest that PhIL1 associates with the parasite cytoskeleton.
Toxoplasma gondii is a ubiquitous protozoan pathogen capable of infecting warm-blooded mammals worldwide and an opportunistic human pathogen in immunocompromised individuals and the fetus. Like other parasites of the phylum Apicomplexa, T. gondii is an obligate intracellular parasite; repeated cycles of invasion, replication, and lysis of host cells by the tachyzoite stage are both a critical part of the parasite's life cycle and a cause of tissue damage during acute infection.
The shape and structural integrity of the tachyzoite are maintained by several unique structural components. Together, the plasma membrane and flattened vesicles of the inner membrane complex (IMC) form the pellicle, a triple bilayer characteristic of apicomplexan parasites. Underlying the pellicle is a filamentous cytoskeletal structure known as the subpellicular network. Two characterized proteins of this network, IMC1 and IMC2, are homologous to the articulins of ciliate protozoa and have structural similarity to mammalian intermediate filament proteins (22). The subpellicular network associates on its outer face with the IMC and on its inner face with 22 subpellicular microtubules. The subpellicular microtubules, which extend in a spiral fashion two-thirds the length of the parasite, are organized at the apical end of the parasite by the basal polar ring, just posterior to the tubulin-based conoid. The subpellicular network, the subpellicular microtubules, and the pellicle are all exceptionally stable structures.
Integral membrane proteins of the pellicle are likely to function in signal transduction, interaction with extracellular substrates (such as host cell ligands), force transduction for the actin-myosin motor during invasion, and maintenance of interactions among the pellicle, subpellicular network, and microtubules. However, very few such proteins have been identified. Parasite attachment is mediated in part by proteins secreted onto the surface from the micronemes, specialized apical organelles involved in host cell invasion (reviewed in references 39 and 42). The characterization of a large family of glycosylphosphatidylinositol (GPI)-anchored proteins known as the SAGs (surface antigens) has suggested that a subset of these proteins may also play a role in host cell adhesion (10, 13, 20, 21). Attempts to identify other non-GPI-anchored pellicle proteins by conventional surface labeling methods have been severely hindered by the abundance and immunodominance of the SAG proteins (21).
In order to identify novel, non-GPI-anchored integral membrane proteins of the parasite pellicle, we utilized the photoactivatable compound 5-[125I]iodonaphthalene-1-azide (INA). INA is highly hydrophobic and partitions into the lipid bilayer of cellular membranes (1, 2). Upon activation, the azide converts to a reactive nitrene, which covalently binds to adjacent protein domains within the lipid bilayer, tagging the membrane-embedded portions with 125I (1, 2, 12). Activation of INA can occur through two processes (36). Because INA partitions into all membranes of the parasite, direct activation, which occurs when INA is exposed to UV light (314 nm), identifies membrane-embedded proteins in multiple membrane compartments. While direct activation of INA at wavelengths greater than 370 nm is negligible, INA can be indirectly activated at these wavelengths through a process called photosensitization, which involves energy transfer from an excited fluorochrome to any INA molecule within collisional distance (26, 29, 30, 34-36). We therefore reasoned that, by confining photosensitizing fluorochromes to the parasite pellicle, membrane-embedded proteins of the pellicle could be selectively labeled with INA.
We demonstrate here that this approach can indeed be used to label a distinct subset of parasite proteins. Importantly, we show that INA is not incorporated efficiently into the GPI-anchored proteins that dominate the parasite surface and that known integral membrane proteins of the pellicle are INA labeled. We identify a novel 22-kDa INA-labeled protein, named PhIL1, which localizes to the parasite periphery but is concentrated at the apical end. While the function of PhIL1 is not known, it appears to associate with the parasite cytoskeleton.
MATERIALS AND METHODS
Materials.
All chemicals were from Sigma (St. Louis, MO) unless otherwise noted. Protease inhibitor stocks contained 104 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), 0.08 mM aprotinin, 2.1 mM leupeptin, 3.6 mM bestatin, 1.5 mM pepstatin A, and 1.4 mM E-64 in dimethyl sulfoxide (DMSO). All materials used for two-dimensional (2-D) gel electrophoresis were of ultrapure quality.
Parasite culture.
The T. gondii RH(EP) wild-type strain was used in all experiments except where noted. P(LK) and P(LK) B mutant (18) strains were generously provided by L. Kasper. B/SAG1-TM parasites expressing a SAG1 chimera, in which the GPI anchor of SAG1 is replaced with the transmembrane domain of human CD46 (38), were generously provided by J. Boothroyd. All parasites were maintained by serial passage in confluent cultures of human foreskin fibroblasts (HFFs) as previously described (37). Parasites released from freshly lysed HFFs were filtered through 3-μm Nuclepore syringe filters (Whatman, Clifton, NJ) to remove host cell debris and washed twice by centrifugation (5 min, 1,000 × g) in cold phosphate-buffered saline (PBS) before use.
C16-eosin labeling.
Parasites were resuspended at 2 × 107 parasites/ml in PBS. 5-(N-Hexadecanoyl)aminoeosin (C16-eosin; Molecular Probes, Eugene, OR) was added to 2 μg/ml from a 2-mg/ml DMSO stock, and the suspension was rotated for 15 min at 4°C in the dark. After addition of a 1/10 volume of fetal bovine serum, parasites were pelleted, washed once in deoxygenating buffer (0.1 mg/ml glucose oxidase, 0.1 mg/ml catalase, 25 mM glucose in PBS [23]), and incubated in deoxygenating buffer for 20 min on ice in the dark. Parasites were examined by fluorescence microscopy to confirm peripheral labeling before further manipulations. Internal labeling was also sometimes observed; parasite preparations in which >15% of the parasites exhibited internal labeling were not used.
[125I]INA synthesis and parasite labeling.
All manipulations with [125I]INA and INA-labeled parasites were done under low-light conditions. [125I]INA was synthesized as previously described (2) using 1-amino-5-azidonaphthalene as the starting material (Combinix Inc., San Mateo, CA). Briefly, 30 mg of the starting compound was deazotized to the corresponding diazonium ion and solubilized in 1 ml 10% sulfuric acid. An aliquot of 250 μl of the diazonium solution was mixed with 10 μl of a 10-mg/ml solution of sodium iodide (NaI) and 10 mCi of radioactive [125I]NaI (Amersham, Piscataway, NJ). The mixture was incubated at 23°C for 1 h, followed by the addition of 20 μl of a 100-mg/ml solution of NaI, and incubated further for 20 min. The INA precipitate was extracted with 3 ml hexane, passed through a silica gel minicolumn, and dried under a stream of nitrogen. The dry INA was resolubilized in 200 μl of DMSO. The yield was typically 0.5 to 1.0 mCi of INA with a specific radioactivity of 1 Ci/mmol.
Reduced glutathione (15 mM final concentration) was added to 5 × 107 C16-eosin-labeled parasites, followed by approximately 25 μCi INA. Samples were irradiated for 5 min on ice at a distance of 10 cm with either a 254-nm UV lamp (UVP Inc., San Gabriel, CA) or a 115-V, 150-W lamp (Minebea Co., Tokyo, Japan) filtered by three 3-mm-thick Kopp CS 3-72 glass filters (F. J. Gray and Co., Jamaica, NY). The cells were then pelleted at 14,000 × g for 5 min at 4°C. Fluorescence microscopy confirmed that eosin labeling remained peripheral after irradiation.
2-D gel electrophoresis.
Samples were solubilized in one of two ways.
Octylglucoside method.
Pellets (approximately 50 μg parasite protein per sample) were resuspended in 24 μl lysis buffer (10 mM Tris, pH 7.4, 50 μg/ml RNase, 50 μg/ml DNase, 1:100 [vol/vol] protease inhibitors) and subjected to four freeze-thaw cycles in liquid nitrogen. Following addition of 16 μl of sodium dodecyl sulfate (SDS) buffer (0.3% [wt/vol] SDS, 200 mM dithiothreitol [DTT]), the sample was sonicated in a water bath sonicator (Branson, Danbury, CT) for 20 min, heated at 50°C for 10 min, and then incubated at 23°C for 2 h. Octyl buffer (160 μl; 9.9 M urea, 4% [wt/vol] octylglucoside, 100 mM DTT, and 0.5% [vol/vol] pH 3 to 10 or pH 4 to 7 ampholytes [Amersham]) was added, and the sample was incubated for another hour at 23°C. Bromophenol blue was added to 0.01% (wt/vol), and insoluble material was removed by centrifugation for 30 min (15,000 × g) at 23°C.
ASB14 method.
Parasite pellets (approximately 25 μg of total parasite protein) were resuspended in 200 μl amidosulfobetaine-14 (ASB14) 2-D buffer consisting of 7 M urea, 2 M thiourea, 2% (wt/vol) ASB14 (Calbiochem, San Diego, CA), 0.5% (vol/vol) Triton X-100 (TX-100; Pierce, Rockford, IL), 20 mM DTT, and 0.5% (vol/vol) ampholytes. After incubation at 23°C for 2 to 4 h, bromophenol blue was added to 0.01% (wt/vol), and the sample was centrifuged as described above.
Samples were added to 11-cm pH 4 to 7 immobilized pH gradient (IPG) strips (Amersham) or pH 3 to 10 nonlinear (NL) IPG strips (Bio-Rad, Hercules, CA). IPG strips were rehydrated with sample and focused for 35,000 V · hours (Protean-IEF; Bio-Rad). For the second dimension, the strips were equilibrated according to the manufacturer's instructions prior to one-dimensional SDS-polyacrylamide gel electrophoresis (PAGE) and silver staining as previously described (6). Gels were dried and exposed to Bio-Max MS film (Eastman Kodak, Rochester, NY).
Metabolic labeling and immunoprecipitation.
Parasites were metabolically labeled with [35S]methionine-cysteine for 20 to 24 h and extracted as previously described (7), except that the TX-100 concentration was increased to 1% for GAP45 immunoprecipitation. For GAP45 immunoprecipitation, supernatants were incubated for 1 h at 4°C with 1:1,000 anti-GAP45 rabbit serum, followed by incubation with protein A-Sepharose (Zymed, South San Francisco, CA) for 1 h at 4°C. The beads were washed four times in TX-100 buffer, and bound proteins were eluted in reducing SDS-PAGE sample buffer containing 4% (vol/vol) β-mercaptoethanol. For SAG1 immunoprecipitation, the supernatant was incubated for 1 h at 4°C with 5 μg/ml mouse anti-SAG1 monoclonal antibody (MAb; Argene, North Massapequa, NY). Protein G-Sepharose (Amersham) was added to the extract and incubated for 1 h at 4°C, followed by four washes with TX-100 buffer and elution in nonreducing SDS-PAGE sample buffer.
Immunoprecipitated proteins were resolved on 12% (wt/vol) acrylamide-SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA), and exposed to film. Following exposure, the membranes were used for Western blotting as previously described (43) with either GAP45 antiserum (1:2,000) or anti-SAG1 antibody (0.25 μg/ml) and detected by enhanced chemiluminescence (Amersham).
TX-114 phase partitioning.
Parasites (1 × 109) were extracted in Triton X-114 (TX-114), phase partitioned, and digested with phosphatidylinositol-specific phospholipase C (PI-PLC) as previously described (45). The detergent phase was diluted in 10 mM Tris-HCl, pH 7.4, and the proteins were precipitated with 10 volumes of 100% acetone prechilled to −20°C. Insoluble material was pelleted (15,000 × g, 4°C), washed with 90% (−20°C) acetone, and air dried at 23°C. The dried pellet was resuspended in 24 μl 10 mM Tris-HCl (pH 7.4), 1:100 (vol/vol) protease inhibitors, and 16 μl of SDS buffer; sonicated in a bath sonicator (Branson) for 20 min; and incubated for 10 min at 100°C followed by 2 h at 23°C. Octyl buffer (160 μl) was added, and the sample was incubated for another 60 min at 23°C. The sample was then centrifuged (30 min, 15,000 × g) and focused on pH 3 to 10 NL IPG strips as described above.
Purification of PhIL1 and mass spectrometry analysis.
Parasites were solubilized with octylglucoside isofocusing buffer (approximately 300 μg of total parasite protein) and focused on pH 4 to 7 IPG strips as described above. Following the second dimension, the gel was stained with SimplyBlue colloidal Coomassie stain (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The PhIL1 spot was excised, trypsinized, and subjected to microcapillary liquid chromatography-tandem mass spectrometry (Taplin Biological Mass Spectrometry Facility, Harvard, MA).
To determine the site(s) of INA modification, spots were excised from both unlabeled and INA-labeled 2-D gels and analyzed by matrix-assisted laser desorption-ionization mass spectrometry (Yale Cancer Center Mass Spectrometry Resource, New Haven, CT).
Protein analysis was performed using MacVector v. 6.5.3 (Accelrys, San Diego, CA) and PredictProtein (http://cubic.bioc.columbia.edu/predictprotein). Prediction of amphipathic helices was done using HelixWheel (http://www.site.uottawa.ca/∼turcotte/resources/HelixWheel), and prediction of β-strand transmembrane domains was done using MINNOU (http://minnou.cchmc.org).
Cloning of PhIL1.
Total tachyzoite RNA was extracted using TRI reagent (Sigma) according to the manufacturer's instructions. The SuperScript first-strand synthesis system (Invitrogen) was used to generate oligo(dT)-primed first-strand cDNA from 2 μg RNA. A 1.1-kb expressed sequence tag (EST) sequence containing the PhIL1 open reading frame (ORF) was amplified, cloned into TOPO-TA pCRII (Invitrogen), and sequenced. DNA sequence alignments and analysis were performed using Sequencher v.3.1.1 (Gene Codes Corporation, Ann Arbor, MI), and ClustalW protein sequence alignments were performed using MacVector.
To N-terminally tag PhIL1 with myc, the PhIL1 ORF was amplified with PhIL1NcoIfor and PhIL1AvrIIrev and cloned in frame into the NcoI and AvrII sites of pTet4myc (24), creating pTet4myc-PhIL1. myc-PhIL1 was then amplified with BglIImycfor and PhIL1stopAvrIIrev and ligated into the BglII and AvrII sites of pCNA-GFP (33). C-terminally green fluorescent protein (GFP)-tagged PhIL1 was generated by amplifying the PhIL1 ORF using PhIL1BglIIfor and PhIL1AvrIIrev and ligated in frame into the BglII and AvrII sites of PCNA-GFP upstream of GFP. C-terminally yellow fluorescent protein (YFP)-tagged PhIL1 was generated by cloning into the BglII and AvrII sites of TUBIMC1YFP/sagCAT (16). PhIL11-81 was cloned into TUBIMC1YFP/sagCAT to generate Nterm-YFP, and YFP-PhIL182-165 was cloned downstream of YFP in TUBIMC1YFP/sagCAT to generate YFP-Cterm.
PhIL1-glutathione S-transferase (GST) was generated by cloning in frame into the BamHI and NotI sites of pGEX (Invitrogen).
PhIL1 polyclonal rabbit antibody.
BL21 cells expressing PhIL1-GST were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 1 h at 37°C. Inclusion bodies were purified by first lysing bacterial pellets in B-PER protein extraction reagent (Pierce), followed by centrifugation at 15,000 × g for 15 min at 4°C. The insoluble pellet was washed sequentially with (i) 50 mM Tris (pH 8.0)-1 mM EDTA containing 200 μg/ml lysozyme, (ii) 50 mM Tris (pH 8.0)-1% (vol/vol) TX-100, and twice with (iii) 50 mM Tris (pH 8.0)-500 mM guanidinium. The final pellet was dissolved in 8 M urea and 5 mM DTT by being shaken for 1 hour (23°C), followed by centrifugation (15,000 × g, 15 min, 4°C). PhIL1-GST was refolded by adding the supernatant dropwise to refolding buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 1 M l-arginine, 1 mM reduced glutathione, 0.8 mM oxidized glutathione) and stirring the mixture overnight at 4°C. The solution was dialyzed against 50 mM Tris (pH 8.0), insoluble material was pelleted, and the supernatant was incubated with glutathione-Sepharose for 1 hour at 4°C. The beads were washed twice with 50 mM Tris, pH 8.0; PhIL1-GST was eluted with 15 mM reduced glutathione in 50 mM Tris, pH 8.0, and used as an antigen to generate rabbit polyclonal serum (Cocalico, Reamstown, PA) as previously described (9).
Polyclonal rabbit serum was purified using protein A-Sepharose as previously described (7) and used at 4 μg/ml for immunofluorescence assay and 400 ng/ml for Western blotting.
Immunofluorescence.
For deoxycholate (DOC) extraction, parasites were first allowed to adhere to untreated coverslips for 30 min at 23°C in a humidified chamber; after extraction (10 min, 23°C) in PBS containing 10 mM DOC and 0.5 mM MgCl2, the parasites were fixed in 4% formaldehyde for 10 min at room temperature.
For the alpha-toxin experiments, parasites were treated with 20 nM Clostridium septicum alpha-toxin (generously provided by R. Tweten, University of Oklahoma) for 4 hours and processed as previously described (45), except that fixed and permeabilized parasites were incubated first with primary antibody diluted at 1:2,000, followed by incubation with a 1:400 dilution of Alexa Fluor-conjugated secondary antibody (Molecular Probes).
Approximately 24 h prior to use, parasites were added to confluent HFF monolayers on glass coverslips. Extracellular parasites were allowed to adhere to coverslips coated with Cell Tak (Becton Dickinson, Franklin Lakes, NJ) for 30 to 40 min on ice as described previously (43). Following fixation (2.5% [vol/vol] formaldehyde-0.02% [vol/vol] glutaraldehyde for 20 min on ice or 100% methanol for 10 min on ice), parasites were blocked and permeabilized in 1% (wt/vol) bovine serum albumin (BSA)-0.25% (vol/vol) TX-100 in PBS for 10 to 20 min on ice, followed by incubation with primary and secondary antibodies as previously described (6). Primary antibodies used were MAb 45-15 (anti-IMC1) (45), MAb 88-70 (anti-ROP4) (7), and MAb 9E10 (anti-myc; developed by J. M. Bishop and obtained from the Developmental Studies Hybridoma Bank, University of Iowa).
Differential detergent extraction.
Parasites were extracted at 1 × 108/ml for 45 min on ice in one of the following buffers: TX-100 (1% [vol/vol] TX-100, 150 mM NaCl, 50 mM Tris, pH 7.4, or 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.5), DOC (0.5% or 1% [vol/vol] DOC, 50 mM Tris, pH 7.4, 150 mM NaCl), radioimmunoprecipitation assay (RIPA) buffer (1% [vol/vol] TX-100, 0.5% [vol/vol] DOC, 0.1% [vol/vol] SDS, 50 mM Tris, pH 8.0, 150 mM NaCl), and TX-114 (10 mM Tris, pH 7.4, 150 mM NaCl, 0.5% [vol/vol] precondensed TX-114). Extraction in SDS buffer (1% [vol/vol] SDS, 50 mM Tris, pH 8.0, 150 mM NaCl) was done by boiling for 10 min. Extracts were then centrifuged (15,000 × g) for 20 min. Equivalent amounts of supernatant and pellet were run on 12% acrylamide-SDS gels, transferred to nitrocellulose, and analyzed by Western blotting with anti-PhIL1, MAb 45-15 (anti-IMC1), MAb B-5-1-2 (antitubulin), and anti-GAP45.
Immunoelectron microscopy.
Approximately 2 × 107 PhIL1-GFP parasites were resuspended in 50 μl PBS. Carbon-coated nickel grids (M400-Ni; EMS, Hatfield, PA) were inverted on top of 10-μl drops of parasites and incubated in a humid chamber for 40 min. The grids were then transferred to 30-μl drops of extraction buffer (PBS with 0.5 mM MgCl2, 10 mM DOC), incubated for 10 min, and rinsed three times with PBS. The grids were then fixed for 10 min with 3.7% (vol/vol) formaldehyde in PBS and washed three times for 5 min each with PBS. Free aldehyde groups were blocked by incubation with 50 mM glycine in PBS for 15 min followed by 15 min of incubation with 0.1% NaBH4 in PBS.
Grids were blocked with 5% (wt/vol) BSA in PBS for 40 min, washed three times with incubation buffer (0.8% BSA, 10 mM NaN3), incubated overnight at 4°C with anti-PhIL1 antibody (or prebleed serum) diluted 1:1,000 in incubation buffer, and washed six times for 5 min each in incubation buffer. The grids were then inverted on 10-μl drops of secondary antibody (anti-rabbit immunoglobulin G conjugated with 1:160 1.4-nm gold [Nanoprobe Inc., Yaphank, NY] or 1:80 ultrasmall gold [EMS] diluted in incubation buffer), incubated for 8 h at 23°C and 12 h at 4°C, and washed six times for 1 min each and six times for 5 min each with incubation buffer.
The grids were postfixed for 1.5 h with 1% (vol/vol) glutaraldehyde in PBS at 23°C, stored in distilled water at 4°C for 48 h, and washed in distilled water three times for 1 min each and three times for 5 min each. Silver enhancement was carried out using the HQ silver enhancement kit (Nanoprobes Inc.) by floating grids on mixtures of the initiator, activator, and modulator for 3 min in a light-tight chamber and then briefly washing the grids once with distilled water, followed by 5-min and 10-min washes. Grids were negatively stained using 2% phosphotungstic acid (pH 7.0) mixed with cytochrome c.
Nucleotide sequence accession number.
The open reading frame of PhIL1 can be accessed under GenBank accession number DQ186991.
RESULTS
Photosensitized labeling of parasite proteins.
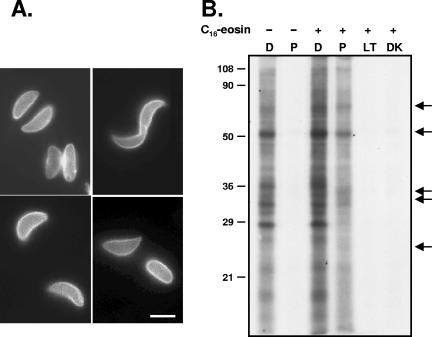
Selective photosensitized labeling with INA requires the restriction of the photosensitizing fluorochrome to the membrane compartment of interest, in our case the plasma membrane and tightly associated IMC membranes of the parasite pellicle. We established conditions under which C16-eosin, which has previously been shown to be an efficient photosensitizer (23), can be concentrated in the parasite pellicle as determined by fluorescence microscopy (Fig. 1A).
FIG. 1.
Photosensitized labeling of parasites with C16-eosin. (A) Fluorescence microscopy of T. gondii tachyzoites labeled with the photosensitizing lipid analog C16-eosin demonstrates that the C16-eosin is highly concentrated in the parasite pellicle. Bar, 2.5 μm. (B) Autoradiogram of INA-labeled parasite proteins resolved by SDS-PAGE. In the absence of C16-eosin, INA can be activated directly only at wavelengths less than 360 nm. When parasites are labeled with C16-eosin, activation of INA at longer wavelengths results in a subset of proteins being labeled with INA (arrows). D, direct activation; P, photosensitization. Controls for inadvertent direct or indirect activation of INA by ambient light showed that high-intensity irradiation was necessary for significant INA incorporation (LT, samples left uncovered in ambient light; DK, samples wrapped in foil). Numbers on the left indicate molecular mass in kDa.
We examined the pattern of proteins labeled by INA after either direct activation or photosensitization with C16-eosin. SDS-PAGE analysis showed that a large number of proteins can be labeled through direct activation; these proteins are predicted to be integral membrane proteins throughout the cell (Fig. 1B). In the absence of C16-eosin, INA is not activated at the higher wavelength of light used for photosensitization, and consequently no proteins are labeled. However, in eosin-labeled parasites, a distinct subset of proteins is labeled through photosensitization (Fig. 1B, arrows). Because the photosensitizing fluorochrome is concentrated in the parasite pellicle, these proteins are predicted to be integral membrane proteins of the pellicle. While other fluorochromes such as C18-fluorescein (5-octadecanoylaminofluorescein) and DiOC16 (3,3′-dihexadecyloxacarbocyanine perchlorate) also showed peripheral labeling (data not shown), C16-eosin was found to be the most selective photosensitizer (i.e., it showed the largest differences between proteins labeled by direct and photosensitized activation) and was used for all subsequent experiments.
Selective labeling of non-GPI-linked pellicle proteins.
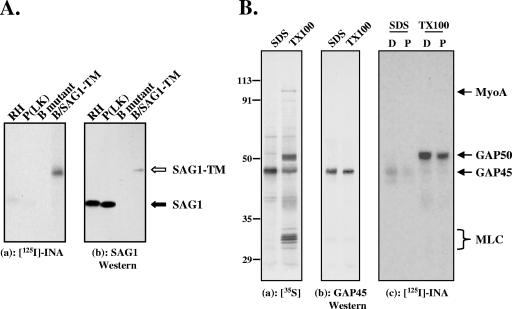
Because a goal of this work was to identify novel, non-GPI-linked integral membrane proteins at the parasite periphery, it was important to confirm that INA is not incorporated to a significant extent into the GPI-linked proteins that dominate the parasite surface. We first examined the extent of INA incorporation into the most abundant GPI-anchored surface protein, SAG1 (18), in comparison to a recombinant SAG1-TM chimera, in which the GPI anchor addition signal sequence of SAG1 was replaced with the transmembrane domain and 33-amino-acid cytoplasmic tail of human CD46. This SAG1-TM chimera folds correctly and is targeted to the plasma membrane (38). After photosensitized labeling with INA, SAG1 was immunoprecipitated from wild-type parasites [RH and P(LK) strains], P(LK) parasites lacking SAG1 (B mutants), and B mutants expressing the SAG1-TM chimera (B/SAG1-TM). Both wild-type strains contain significant amounts of SAG1 by Western blotting, but this SAG1 incorporates very little INA (Fig. 2A, closed arrow). Transgenic parasites expressing the chimeric protein contain less SAG1-TM than the amount of SAG1 in the wild-type strains, but this SAG1-TM is heavily INA labeled (Fig. 2A, B/SAG1-TM; empty arrow). As expected, no INA incorporation is detected in immunoprecipitates from the SAG1-deficient B mutants. Thus, while very little INA is incorporated into GPI-anchored SAG1, INA can readily be incorporated into the same protein if it contains a transmembrane domain, demonstrating that INA does not efficiently bind to the GPI anchor. In addition, the labeling of SAG1-TM confirms the ability of C16-eosin at the parasite surface to activate INA and to label a plasma membrane transmembrane protein by photosensitization.
FIG. 2.
INA preferentially labels non-GPI-linked integral membrane proteins of the pellicle. (A) (a) SAG1 was immunoprecipitated from parasites labeled with INA through photosensitization and analyzed by SDS-PAGE/autoradiography. The parasite strains used were RH and P(LK) (wild-type strains), a SAG1-deficient P(LK) strain (B mutant), and the SAG1-deficient strain expressing a transmembrane version of SAG1 (B/SAG1-TM). (b) Corresponding Western blot with anti-SAG1 antibody. GPI-anchored SAG1 shows weak labeling with INA (panel a, filled arrow). In contrast, much smaller amounts of SAG1-TM [compare Western blotting signals of RH and P(LK) to that of B/SAG1-TM in panel b] show significantly more INA incorporation (panel a, open arrow). The lanes shown were from the same blot and were exposed and adjusted for contrast and brightness identically. (B) (a) Autoradiogram showing that immunoprecipitation of SDS-extracted 35S-labeled parasites with an anti-GAP45 antibody yields primarily GAP45 (SDS), whereas the entire glideosome complex can be immunoprecipitated with the same antibody under nondenaturing conditions (TX-100). The four major proteins in the immunoprecipitated complex are myoA, myosin light chain (MLC), GAP45, and GAP50 (11). (b) Corresponding Western blot with anti-GAP45. (c) Autoradiogram of the INA-labeled proteins immunoprecipitated with anti-GAP45 antibody shows that both GAP45 and GAP50 are labeled, through either direct or photosensitized INA activation, whereas neither myosin light chain nor myoA is detectably labeled. Substoichiometric amounts of myoA were transferred to the nitrocellulose in this particular experiment; other experiments in which the iodination profile of the immunoprecipitate was analyzed directly (i.e., without transfer for Western blotting) confirmed the lack of detectable INA incorporation into myoA (data not shown). D, direct activation; P, photosensitization. Numbers on the left indicate molecular mass in kDa.
We next examined the ability of INA to label other parasite GPI-anchored proteins, which include more than 150 members of the SAG family (17). Triton X-114 phase partitioning can be used to enrich for membrane-associated proteins, including GPI-anchored proteins, which partition into the detergent phase (3). Digestion with PI-PLC cleaves the GPI anchor, shifting GPI-anchored proteins from the detergent phase to the aqueous phase (14, 45). To determine if INA labels GPI-anchored proteins other than SAG1, we therefore compared the 2-D INA labeling profile of proteins recovered in the Triton X-114 detergent phase of control extracts (membrane-associated proteins, including GPI-anchored proteins) to that of those recovered in the detergent phase after PI-PLC treatment (which will deplete GPI-anchored proteins from the detergent fraction). The profiles are virtually identical (data not shown), indicating that the majority of INA-labeled proteins are not GPI anchored.
While GPI-anchored proteins are located in the plasma membrane, the multiprotein motor complex essential for parasite motility and invasion, consisting of myosin A (myoA), myosin light chain, GAP45, and GAP50, is associated with the IMC membrane (11). This membrane association is thought to be mediated by GAP50, a transmembrane protein, and/or GAP45, which is both myristoylated and palmitoylated. To test whether any of the motor complex proteins could be labeled with INA, we immunoprecipitated the complex from parasites that had been INA labeled through either direct activation or photosensitization. Neither myoA nor myosin light chain was detectably labeled with INA (Fig. 2B and data not shown), indicating that they are not membrane embedded. In contrast, GAP45 showed faint INA labeling, and the fourth member of the complex, GAP50, was heavily INA labeled through both direct activation and photosensitization (Fig. 2B).
Collectively, these data demonstrate that photosensitization can be used to label non-GPI-linked membrane proteins of the pellicle, and they provide the first direct experimental evidence that both GAP45 and GAP50 are indeed membrane embedded.
Analysis of INA-labeled proteins by 2-D gel electrophoresis.
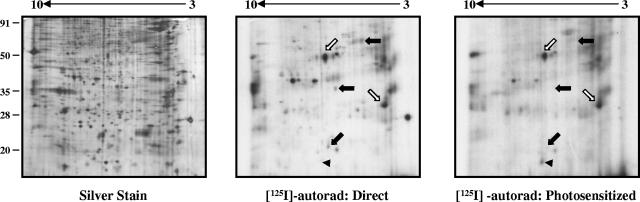
In order to isolate and characterize novel INA-labeled proteins, extracts from labeled parasites were separated by 2-D gel electrophoresis. A large number of parasite proteins can be resolved on broad-pH-range gels (pH 3 to 10), as demonstrated by silver staining (Fig. 3, left panel). A subset of these proteins is labeled with INA through direct activation (Fig. 3, middle panel). After photosensitized labeling (Fig. 3, right panel), three groups of proteins can be identified: those that are labeled by direct but not photosensitized labeling (filled arrows), those that incorporate similar amounts of INA by direct and photosensitized labeling (empty arrows), and those that show increased INA labeling through photosensitization (arrowhead). The first group corresponds to predicted integral membrane proteins of intracellular compartments; the latter two groups correspond to potential integral membrane proteins of the parasite pellicle.
FIG. 3.
2-D gel electrophoresis of INA-labeled proteins. After either direct or photosensitized activation of INA, parasite proteins were extracted in isoelectric focusing buffer containing ASB14 and resolved on pH 3 to 10 NL 2-D gels. Approximately 25 μg of parasite protein was loaded per gel. The INA-labeled proteins resolved under these conditions can be classified into three groups: those that are labeled by direct but not photosensitized activation (filled arrows), those that show similar amounts of INA incorporation by both direct and photosensitized activation (empty arrows), and those that show increased INA incorporation through photosensitization (arrowhead). Numbers on the left indicate molecular mass in kDa.
Identification of PhIL1.
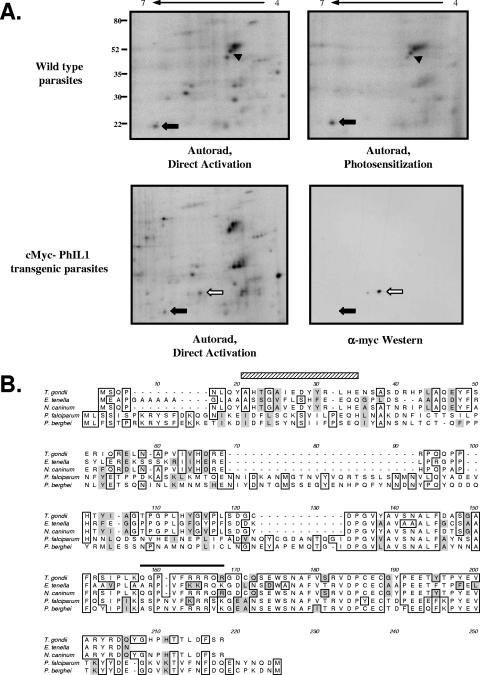
Our early attempts to resolve INA-labeled proteins on 2-D gels utilized octylglucoside as the major detergent; in these gels, two prominent photosensitized proteins are seen in the pH 4 to 7 range. The first protein, which resolves as a charge train containing four to five spots (Fig. 4A, arrowhead), was identified as different isoforms of GAP45 by Western blotting (data not shown). The second protein (Fig. 4A, closed arrow) was purified on preparative 2-D gels, digested with trypsin, and analyzed by mass spectrometry; the three identified tryptic peptides matched a single EST found in the T. gondii EST database. The full-length ORF was amplified by reverse transcription-PCR and sequenced, predicting a protein of 19 kDa, which we named PhIL1 (for photosensitized INA-labeled protein 1; Fig. 4B). Southern blotting indicated that the PhIL1 gene is a single-copy gene (data not shown).
FIG. 4.
Identification of PhIL1. (A) Proteins labeled with INA by either direct (top left panel) or photosensitized (top right panel) activation were solubilized in octylglucoside isoelectric focusing sample buffer, resolved on pH 4 to 7 2-D gels, and visualized by autoradiography. One of the major photosensitized proteins solubilized and resolved under these conditions, of approximately 22 kDa (arrow), was purified from preparative 2-D gels and identified by mass spectrometry; this protein was named PhIL1. The other major labeled protein, which resolves as a charge train (arrowhead), was identified as GAP45 by Western blotting (data not shown). The greater amount of GAP45 present on the 2-D gels compared to that recovered in immunoprecipitates likely accounts for the greater GAP45 labeling intensity seen here, compared to Fig. 3. Numbers on the left indicate molecular mass in kDa. Transgenic parasites expressing N-terminally myc-tagged PhIL1 were labeled with INA and analyzed by 2-D gel electrophoresis, followed by autoradiography and anti-myc Western blotting. A new INA-labeled spot (empty arrow) appears in the transgenic parasites with the predicted molecular weight and pI shift relative to untagged PhIL1; this spot is also recognized in the anti-myc Western blot. Closed arrow, endogenous PhIL1; open arrow, myc-PhIL1. (B) Multiple sequence alignment of predicted PhIL1 proteins from related apicomplexan parasites. Identical amino acid residues are boxed; similar residues are shaded gray. The peptide from T. gondii PhIL1 identified by mass spectrometry as INA labeled is indicated by a black line above the sequence. A predicted amphipathic helix in T. gondii is indicated by the hatched box. Sequences are from Toxoplasma gondii GenBank accession number DQ186991; Eimeria tenella contig of GenBank BM321695, CD659439, and CD346904; Neospora caninum CF273551; Plasmodium falciparum NP_703291; and Plasmodium berghei CAH97932.
To confirm that the cloned sequence corresponds to the protein labeled with INA, we generated transgenic parasites expressing myc-tagged PhIL1. The myc-tagged PhIL1 localized to the parasite periphery (data not shown), with a distribution indistinguishable from that of the untagged protein (see below). On 2-D gels of INA-labeled transgenic parasites, a new spot with the predicted shift in molecular weight and pI, relative to untagged PhIL1, was both labeled with INA and recognized by anti-myc Western blotting (Fig. 4A, open arrow), confirming the identity of PhIL1. Immunoprecipitation with an antibody generated against recombinant PhIL1 also confirmed that PhIL1 is labeled with INA through photosensitization (data not shown).
PhIL1 does not contain any identifiable protein domains or homologs in organisms outside the phylum Apicomplexa. However, homologs are found in the EST and genomic databases of other apicomplexan parasites (Fig. 4B). Although the N-terminal portions of the predicted proteins are divergent, the C-terminal portions share a high degree of sequence identity between species, suggesting a conserved function.
PhIL1 secondary structure and identification of INA-binding sites.
While the labeling of PhIL1 with INA suggests that it is an integral membrane protein, it does not contain a predicted transmembrane domain or a recognizable signal for addition of lipid anchors. A growing number of integral membrane proteins have been shown to horizontally insert themselves into one leaflet of the lipid bilayer via amphipathic helices (27, 40). Secondary structure prediction of PhIL1 suggests the presence of a potential amphipathic helix at the N terminus (Fig. 4B, striped box). To gain a better understanding of how PhIL1 could be membrane associated, we attempted to identify the sites of INA addition by mass spectrometry. Of the recovered peptides (approximately 40% coverage), one was found to be labeled with a single INA molecule (Fig. 4B, solid line). This peptide does not exhibit any obvious secondary structural features (e.g., an amphipathic helix) that would predict membrane association, although it is highly conserved among the different apicomplexan PhIL1 homologs.
PhIL1 localizes to the parasite periphery.
In parasites expressing PhIL1 tagged with GFP, PhIL1-GFP localizes to the parasite periphery (Fig. 5A). In addition, it is concentrated at both ends of the parasite, but especially at the extreme apical end, as seen by dual localization with the apical rhoptry protein ROP4. A gap in PhIL1 staining is visible at the apical end of the parasite (arrows, Fig. 5B; see also Fig. 6A and 8A), a pattern reminiscent of the IMC and subpellicular network, which are not contiguous around the parasite; however, this gap is smaller than that typically seen for IMC1 staining (compare PhIL1-GFP to α-IMC1 in Fig. 5A; see also Fig. 8A). When the parasites are oriented with their apical end towards the viewer, it is clear that the gap in PhIL1 fluorescence corresponds to a tight ring at the extreme apical tip (see Fig. 8C). A similar distribution of PhIL1-GFP is seen using methanol-fixed parasites (data not shown).
FIG. 5.
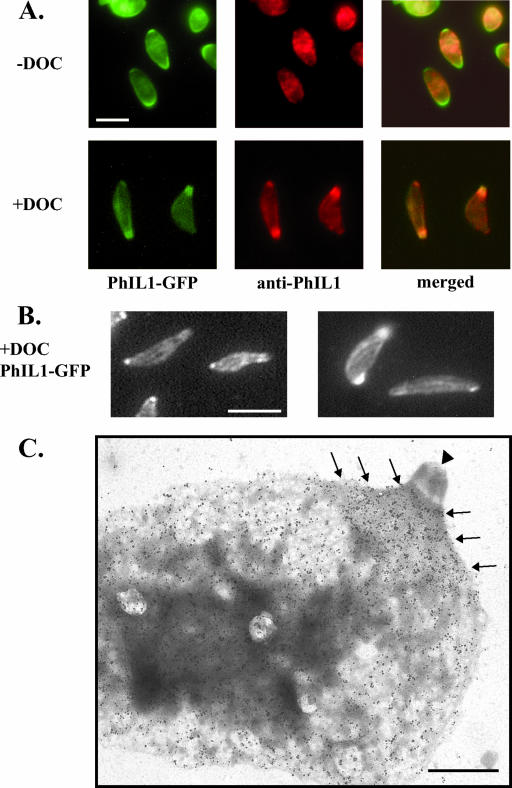
Localization of PhIL1-GFP. (A) PhIL1-GFP localizes to the parasite periphery. It is also concentrated at both ends, but in particular the apical end, as seen by dual fluorescence localization with the apical rhoptry protein ROP4. A gap can be seen in the PhIL1 staining at the extreme anterior end (arrows; also Fig. 5B, 6A, and 8A); this gap is smaller than that seen for IMC1 (arrowheads). In parasites oriented towards the reader, this gap is seen as a tight circle (Fig. 8C). Some faint nuclear staining is also occasionally observed (upper panels, PhIL1-GFP). Bars, 2.5 μm. (B) C. septicum alpha-toxin causes separation of the plasma membrane from the IMC in extracellular parasites (45). The top panels show the near-coincident distribution of SAG1 and PhIL1-GFP in untreated parasites. In toxin-treated parasites (bottom panels), PhIL1-GFP remains with the IMC and/or the underlying cytoskeleton, whereas SAG1 is found in the plasma membrane blebs that separate from the IMC. Bar, 5 μm. DIC, differential interference contrast.
FIG. 6.
Localization of PhIL1 in DOC-extracted parasites. (A) In fixed, TX-100-permeabilized parasites expressing PhIL1-GFP (− DOC), both anti-PhIL1 (middle panels) and anti-GFP (not shown) localize to the parasite periphery by immunofluorescence microscopy, but neither shows the apical and posterior concentration seen by direct fluorescence microscopy of PhIL1-GFP (left panels). However, after extraction with deoxycholate (+ DOC), both antibodies label the apical and posterior ends. Bar, 5 μm. (B) In DOC-extracted parasites, PhIL1 is also found in spiraling “stripes” along the body of the parasite. Two independent examples of this labeling pattern are shown. Bar, 5 μm. (C) DOC-extracted PhIL1-GFP parasites were incubated with either preimmune sera or anti-PhIL1 antibody, followed by gold labeling and negative staining of the parasite “ghosts” with phosphotungstic acid. Very little labeling is seen in the controls labeled with preimmune serum (data not shown). Using anti-PhIL1, labeling is observed along the body of the parasite and in a concentrated “cape” (arrows) at the apical end, just basal to the conoid (arrowhead). No labeling of the conoid is seen. Labeling of the basal end of the parasite is also observed, although this labeling is less pronounced than that at the apical end (data not shown). Bar, 1 μm.
FIG. 8.
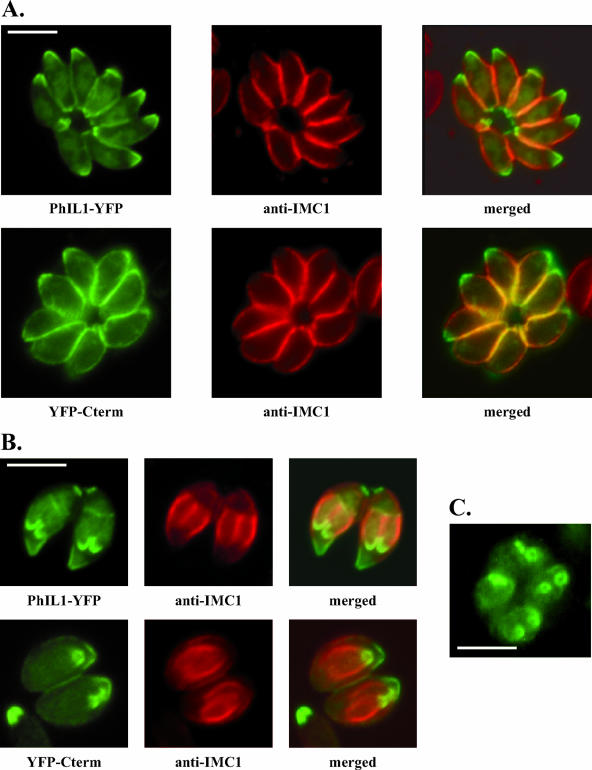
The conserved C terminus targets PhIL1 to the periphery and to the apex of developing daughter parasites during endodiogeny. (A) The C-terminal portion of PhIL1 (YFP-Cterm) localizes to the parasite periphery and is concentrated at the apical end. The distributions of PhIL1-YFP and YFP-Cterm (direct fluorescence) are shown, relative to IMC1 (indirect immunofluorescence). In contrast, an N-terminal YFP fusion of PhIL1 does not target itself to the periphery of the parasite, although it is found at the apical end (data not shown). (B) During parasite replication, where daughter parasites form within the mother, full-length PhIL1-YFP localizes to the apical ends of the forming daughters, anterior to the gap in IMC1. YFP-Cterm is similarly targeted to daughter parasites. (C) When daughter parasites are oriented with their apical end towards the reader, it is clear that the gap in PhIL1 apical fluorescence (here PhIL1-GFP) corresponds to a tight circle. All bars, 2.5 μm.
To determine if PhIL1 is associated with the plasma membrane or with the IMC, we examined the localization of PhIL1-GFP in parasites treated with C. septicum alpha-toxin (11, 45). Alpha-toxin binds to GPI-anchored proteins on the parasite surface and inserts pores, resulting in the separation of the plasma membrane from the IMC and the formation of large plasma membrane blebs. In alpha-toxin-treated parasites, PhIL1 is clearly not associated with these plasma membrane blebs (Fig. 5B, bottom panels), indicating an association with the IMC and/or the underlying cytoskeleton.
While PhIL1-GFP localizes to the periphery and is concentrated at the apical end, anti-PhIL1 immunofluorescence of formaldehyde-fixed and TX-100-permeabilized parasites results in only peripheral staining (Fig. 6A). However, when parasites are extracted with DOC, which removes the plasma membrane and IMC (22), the antibody now also strongly labels the apical end (Fig. 6A), suggesting that, in the absence of DOC extraction, the apical epitope is somehow masked. In addition, spiraling stripes can sometimes be seen along the length of DOC-extracted parasites (Fig. 6B). PhIL1 localization by immunoelectron microscopy on DOC-extracted parasites confirms and extends the observations seen by immunofluorescence assay: PhIL1 is found at the periphery of the parasite but is concentrated at the apical end just basal to the conoid (Fig. 6C). The conoid itself is not labeled.
PhIL1 is associated with the parasite cytoskeleton.
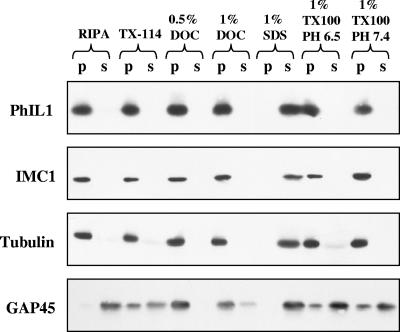
The observation that PhIL1 remains associated with DOC-extracted parasites suggests an interaction with the cytoskeleton underlying the IMC (22). To examine this further, we tested the extractability of PhIL1 in a variety of detergents, pH conditions, and salt concentrations. As can be seen in Fig. 8, detergents typically used to extract membrane proteins (e.g., TX-100, TX-114, and RIPA buffer) did not extract PhIL1, nor did high salt (up to 500 mM; data not shown). PhIL1 is extracted efficiently only when boiled in SDS; partial extraction occurs at basic pH in combination with TX-100 (pH 11, 1% TX-100; data not shown). This extraction profile is indistinguishable from that of the T. gondii cytoskeletal proteins tubulin and IMC1 and different from that of the membrane protein GAP45 (Fig. 7), suggesting that PhIL1 is associated with the parasite cytoskeleton.
FIG. 7.
The detergent extraction profile of PhIL1 suggests that it is associated with the cytoskeleton. Parasites were extracted under a variety of detergent and pH conditions, centrifuged at 15,000 × g, and analyzed by Western blotting using antibodies against PhIL1, IMC1, tubulin, and GAP45. The extraction profiles of PhIL1, IMC1, and tubulin are identical; each is efficiently extracted only when boiled in SDS. RIPA, radioimmunoprecipitation assay buffer; p, pellet; s, supernatant.
The conserved C terminus is necessary and sufficient for proper targeting and cytoskeletal association of PhIL1.
The C-terminal half of PhIL1 is highly conserved (Fig. 4B). A fusion between the PhIL1 C terminus and YFP (YFP-Cterm) showed the same localization as full-length PhIL1, i.e., peripheral with concentration at the ends (Fig. 8A). In addition, both full-length PhIL1-YFP and YFP-Cterm localize to daughter parasites during parasite replication, a process in which two daughter parasites are assembled inside the mother parasite (Fig. 8B and C). In contrast to the C-terminal portion of PhIL1, the N-terminal portion fused to YFP does not localize to the periphery, although localization to the apical ends of both mother and daughter parasites is observed (data not shown).
Like untagged PhIL1 (Fig. 7), PhIL1-YFP is not extracted in TX-100 (data not shown). YFP-Cterm is also insoluble in TX-100, while the N-terminal portion is soluble (data not shown). These data indicate that the conserved C-terminal portion of PhIL1 is sufficient for both cytoskeletal association and proper apical and peripheral localization of the protein.
DISCUSSION
Few of the proteins that act at the T. gondii-host cell interface are known, in particular integral membrane proteins of the parasite pellicle that might function in motility, host cell recognition, or signal transduction. To identify such proteins, we have developed a method that utilizes the photoactivatable aryl azide INA. INA partitions into the core of lipid bilayers and can be activated via short-range energy transfer mechanisms from photosensitizing fluorochromes. Photoactivated INA is a highly reactive nucleophile that binds covalently to lipids and proteins within the bilayer, inserting itself into sulfhydryls, amines, and phenol groups, with an apparent amino acid preference for tryptophan and cysteine (5, 19, 35). If a photosensitizing fluorochrome can be restricted to a specific membrane compartment in the cell, it can be used to selectively target INA labeling to the membrane-embedded proteins in that compartment (36).
INA binds covalently to unsaturated carbons in the acyl chains of membrane lipids, but it has not previously been determined whether INA binds efficiently to lipid-anchored membrane proteins. GPI-anchored proteins dominate the surface of the parasite; SAG1 alone may constitute several percent of total cell protein (18). Because our goal was to identify novel, non-GPI-linked pellicle proteins, it was important to establish that the GPI-anchored proteins would not be present in our labeled protein profiles. The lipid moiety of the T. gondii GPI anchor has been characterized as sn-1,2-diacylglyceride, where sn-2 is palmitate and sn-1 either is shorter than palmitate or contains a double bond(s), reducing its hydrophobicity (41). Our data on the labeling of both SAG1 and GPI-anchored proteins as a whole indicate that INA incorporates very inefficiently into T. gondii GPI-anchored proteins.
Both GAP50 and, to a lesser extent, GAP45 are labeled with INA by photosensitization. The recent demonstration that these two proteins are associated with the IMC, rather than the plasma membrane (11), suggests that C16-eosin added extracellularly to parasites partitions into the membrane(s) of the IMC as well as the plasma membrane. The plasma membrane and IMC are very tightly associated (45); C16-eosin may be able to equilibrate across the narrow (<20-nm) aqueous gap that separates the plasma membrane from the outermost membrane of the IMC (8). Alternatively, the membranes may be directly connected in some unknown way that allows the C16-eosin added to the outside of the parasite access to the membranes of the IMC. GAP50 contains a predicted transmembrane domain, but GAP45 does not; however, GAP45 appears to be both myristoylated and palmitoylated (unpublished observations) and may insert itself into the membrane(s) of the pellicle through these acyl chains. There are no data available on INA labeling of myristate or palmitate anchors, and it is possible that INA can incorporate, albeit inefficiently, into one or both of these acyl chains. However, it is also possible that another hydrophobic domain of GAP45 inserts itself into the membrane and that INA binds to this portion of the protein rather than the lipid anchor(s).
By SDS-PAGE, a subset of the parasite proteins labeled via direct activation of INA was seen to be labeled through photosensitization. Analysis of photosensitized proteins on 2-D gels led to the identification of PhIL1, a novel protein that is highly conserved in apicomplexan parasites, including various species of Plasmodium, the causative agents of malaria. When Plasmodium falciparum extracts were probed by Western blotting with the anti-TgPhIL1 antibody, a single protein of approximately 25 kDa was recognized, in agreement with the predicted size (25.5 kDa) of the P. falciparum homolog (S. D. Gilk and G. E. Ward, unpublished data). In addition, the transcriptome data of DeRisi and colleagues indicate an increase in P. falciparum PhIL1 expression during schizogeny, the replicative stage of malaria parasites (4). These data are consistent with our results showing that T. gondii PhIL1 localizes to the apical end of daughter parasites early in the process of their assembly within the mother cell.
The fact that PhIL1 is labeled with INA suggests that it is an integral membrane protein. However, PhIL1 does not have an identifiable hydrophobic transmembrane domain(s), a predicted β-strand transmembrane domain(s), or a predicted site(s) of lipid modification, although the C-terminal domain does contain two conserved cysteines that could potentially be palmitoylated. While transmembrane and lipid-anchored proteins are the most common types of integral membrane proteins, others (“monotopic” proteins) insert themselves into the membrane using amphipathic helices (27, 31, 40, 44). The best-characterized of these proteins is prostaglandin synthase, where two short amphipathic helices dimerize and insert themselves, parallel to the membrane plane, into one leaflet (27). In T. gondii, two dense granule proteins, GRA2 and GRA3, are secreted in a soluble form into the parasitophorous vacuole, where they become tightly membrane bound (25, 28); both contain predicted amphipathic helices, which are postulated to insert themselves into the membranes of the tubulovesicular network. PhIL1 contains one potential amphipathic helix at its N terminus. It is unknown if this portion of the protein inserts itself into the membrane; no N-terminal INA-labeled peptides were recovered in our mass spectrometry analysis, but coverage of the predicted helix was incomplete.
GFP fusions demonstrated that the C-terminal half of the protein is sufficient to confer proper localization of PhIL1. This half of the protein contains no predicted amphipathic helices, but a C-terminal INA-modified peptide was identified by mass spectrometry. This peptide does not contain the potential sites of palmitoylation mentioned above. PhIL1 is predicted to be a globular protein, and INA has been known to bind to hydrophobic pockets of peripheral or even soluble proteins (36). It is therefore possible that PhIL1 is not, in fact, membrane associated but contains a hydrophobic pocket that is labeled with INA. Alternatively, other proteins, such as influenza virus hemagglutinin and glutathione S-transferase, do not contain predicted transmembrane domains but have been shown to reversibly insert themselves into membranes where they become accessible to INA labeling (26, 29). Further studies are needed to determine how PhIL1 is associated with the membrane and if it is modified on additional sites with INA.
While the ability to label PhIL1 with INA suggests that the protein is membrane associated, detergents commonly used to extract membrane proteins do not solubilize PhIL1. High-salt conditions used to strip peripheral proteins also do not remove PhIL1; in fact, PhIL1 requires harsh ionic detergents such as SDS for efficient solubilization. The extraction profile of PhIL1 is indistinguishable from that of two known T. gondii cytoskeletal proteins, tubulin and IMC1. Retrospective analysis of proteomic data from partially purified conoid/apical complex preparations (15) revealed that PhIL1 was strongly represented in the data set (15 partially overlapping peptides, for a total of 109 out of the 165 amino acids [J. M. Murray and K. Hu, unpublished data]). PhIL1 peptides were detected more often in the “conoid-depleted” than the “conoid-enriched” fraction, consistent with our immunolocalization data and the hypothesis that PhIL1 is a cytoskeletal, nonconoid protein.
Intriguingly, when the pellicle membranes were removed with DOC, PhIL1 sometimes appeared as spiraling “stripes” down the body of the parasite. These PhIL1 stripes do not colocalize with the subpellicular microtubules (Gilk and Ward, unpublished). However, they are reminiscent of the longitudinal “sutures” found at the junctions between the flattened rectangular plates of the IMC (32), although no stripes of PhIL1 staining were detected perpendicular to the longitudinal stripes, i.e., corresponding to the sutures on the other edges of the IMC plates. At both the apical and posterior ends, the longitudinal sutures between the IMC plates converge in a spiral; this could account for the increased amount of PhIL1 found at the ends of the parasites.
Little is known about the proteins that tie the tachyzoite cytoskeleton (i.e., the subpellicular network and subpellicular microtubules) to the tightly associated membranes of the pellicle. Our observation that PhIL1 has properties of both a cytoskeletal protein and a membrane-embedded protein of the IMC suggests that PhIL1 could play a role in mediating this association. Further studies will be required to determine the precise localization of PhIL1 relative to other structures at the parasite periphery and to explore the possibility that PhIL1 plays a role in linking the pellicle membranes to the underlying cytoskeleton.
Acknowledgments
We thank Lloyd Kasper and John Boothroyd for generously providing parasite strains; Mark Rould for assistance analyzing secondary structure; Mathias Viard for analyzing INA mass spectrometry data; and Doug Johnson, Mariana Matrajt, and members of the Ward laboratory for thoughtful discussions and critical reading of the manuscript.
This work was supported by The Burroughs Wellcome Fund (G.E.W.), PHS grants AI42355 and AI01719 (G.E.W.) and AI049301 (J.M.M.), and the Vermont EPSCoR program under NSF grant EPS-9874685 and in part by grant P30 CA22435 from the National Cancer Institute.
REFERENCES
- 1.Bayley, H., and J. R. Knowles. 1980. Photogenerated, hydrophobic reagents for intrinsic membrane proteins. Ann. N. Y. Acad. Sci. 346:45-58. [DOI] [PubMed] [Google Scholar]
- 2.Bercovici, T., and C. Gitler. 1978. 5-[125I]Iodonaphthyl azide, a reagent to determine the penetration of proteins into the lipid bilayer of biological membranes. Biochemistry 17:1484-1489. [DOI] [PubMed] [Google Scholar]
- 3.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 4.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunner, J., and F. M. Richards. 1980. Analysis of membranes photolabeled with lipid analogues. Reaction of phospholipids containing a disulfide group and a nitrene or carbene precursor with lipids and with gramicidin A. J. Biol. Chem. 255:3319-3329. [PubMed] [Google Scholar]
- 6.Carey, K. L., C. G. Donahue, and G. E. Ward. 2000. Identification and molecular characterization of GRA8, a novel, proline-rich, dense granule protein of Toxoplasma gondii. Mol. Biochem. Parasitol. 105:25-37. [DOI] [PubMed] [Google Scholar]
- 7.Carey, K. L., A. M. Jongco, K. Kim, and G. E. Ward. 2004. The Toxoplasma gondii rhoptry protein ROP4 is secreted into the parasitophorous vacuole and becomes phosphorylated in infected cells. Eukaryot. Cell 3:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Melo, E. J., and W. de Souza. 1997. A cytochemistry study of the inner membrane complex of the pellicle of tachyzoites of Toxoplasma gondii. Parasitol. Res. 83:252-256. [DOI] [PubMed] [Google Scholar]
- 9.Donahue, C. G., V. B. Carruthers, S. D. Gilk, and G. E. Ward. 2000. The Toxoplasma homolog of Plasmodium apical membrane antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Mol. Biochem. Parasitol. 111:15-30. [DOI] [PubMed] [Google Scholar]
- 10.Dzierszinski, F., M. Mortuaire, M. F. Cesbron-Delauw, and S. Tomavo. 2000. Targeted disruption of the glycosylphosphatidylinositol-anchored surface antigen SAG3 gene in Toxoplasma gondii decreases host cell adhesion and drastically reduces virulence in mice. Mol. Microbiol. 37:574-582. [DOI] [PubMed] [Google Scholar]
- 11.Gaskins, E., S. D. Gilk, N. DeVore, T. Mann, L. Peck, G. E. Ward, and C. Beckers. 2004. Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. J. Cell Biol. 165:383-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gitler, C., and T. Bercovici. 1980. Use of lipophilic photoactivatable reagents to identify the lipid-embedded domains of membrane proteins. Ann. N. Y. Acad. Sci. 346:199-211. [DOI] [PubMed] [Google Scholar]
- 13.He, X. L., M. E. Grigg, J. C. Boothroyd, and K. C. Garcia. 2002. Structure of the immunodominant surface antigen from the Toxoplasma gondii SRS superfamily. Nat. Struct. Biol. 9:606-611. [DOI] [PubMed] [Google Scholar]
- 14.Hooper, N. M., and A. Bashir. 1991. Glycosyl-phosphatidylinositol-anchored membrane proteins can be distinguished from transmembrane polypeptide-anchored proteins by differential solubilization and temperature-induced phase separation in Triton X-114. Biochem. J. 280:745-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, K., J. Johnson, L. Florens, M. Fraunholz, S. Suravajjala, C. Dilullo, J. Yates, D. S. Roos, and J. M. Murray. 2006. Cytoskeletal components of an invasion machine—the apical complex of Toxoplasma gondii. PLoS Pathog. 2:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, K., T. Mann, B. Striepen, C. J. Beckers, D. S. Roos, and J. M. Murray. 2002. Daughter cell assembly in the protozoan parasite Toxoplasma gondii. Mol. Biol. Cell 13:593-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung, C., C. Y. Lee, and M. E. Grigg. 2004. The SRS superfamily of Toxoplasma surface proteins. Int. J. Parasitol. 34:285-296. [DOI] [PubMed] [Google Scholar]
- 18.Kasper, L. H. 1987. Isolation and characterization of a monoclonal anti-P30 antibody resistant mutant of Toxoplasma gondii. Parasite Immunol. 9:433-445. [DOI] [PubMed] [Google Scholar]
- 19.Knowles, J. R. 1972. Photogenerated reagents for biological receptor-site labeling. Acc. Chem. Res. 5:155-160. [Google Scholar]
- 20.Lekutis, C., D. J. Ferguson, M. E. Grigg, M. Camps, and J. C. Boothroyd. 2001. Surface antigens of Toxoplasma gondii: variations on a theme. Int. J. Parasitol. 31:1285-1292. [DOI] [PubMed] [Google Scholar]
- 21.Manger, I. D., A. B. Hehl, and J. C. Boothroyd. 1998. The surface of Toxoplasma tachyzoites is dominated by a family of glycosylphosphatidylinositol-anchored antigens related to SAG1. Infect. Immun. 66:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann, T., and C. Beckers. 2001. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol. Biochem. Parasitol. 115:257-268. [DOI] [PubMed] [Google Scholar]
- 23.Meiklejohn, B. I., N. A. Rahman, D. A. Roess, and B. G. Barisas. 1997. 5-Iodonaphthyl-1-azide labeling of plasma membrane proteins adjacent to specific sites via energy transfer. Biochim. Biophys. Acta 1324:320-332. [DOI] [PubMed] [Google Scholar]
- 24.Meissner, M., S. Brecht, H. Bujard, and D. Soldati. 2001. Modulation of myosin A expression by a newly established tetracycline repressor-based inducible system in Toxoplasma gondii. Nucleic Acids Res. 29:E115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercier, C., M. F. Cesbron-Delauw, and L. D. Sibley. 1998. The amphipathic alpha helices of the toxoplasma protein GRA2 mediate post-secretory membrane association. J. Cell Sci. 111:2171-2180. [DOI] [PubMed] [Google Scholar]
- 26.Merezhinskaya, N., G. A. Kuijpers, and Y. Raviv. 1998. Reversible penetration of alpha-glutathione S-transferase into biological membranes revealed by photosensitized labelling in situ. Biochem. J. 335:597-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nina, M., S. Berneche, and B. Roux. 2000. Anchoring of a monotopic membrane protein: the binding of prostaglandin H2 synthase-1 to the surface of a phospholipid bilayer. Eur. Biophys. J. 29:439-454. [DOI] [PubMed] [Google Scholar]
- 28.Ossorio, P. N., J. F. Dubremetz, and K. A. Joiner. 1994. A soluble secretory protein of the intracellular parasite Toxoplasma gondii associates with the parasitophorous vacuole membrane through hydrophobic interactions. J. Biol. Chem. 269:15350-15357. [PubMed] [Google Scholar]
- 29.Pak, C. C., M. Krumbiegel, R. Blumenthal, and Y. Raviv. 1994. Detection of influenza hemagglutinin interaction with biological membranes by photosensitized activation of [125I]iodonaphthylazide. J. Biol. Chem. 269:14614-14619. [PubMed] [Google Scholar]
- 30.Pak, C. C., A. Puri, and R. Blumenthal. 1997. Conformational changes and fusion activity of vesicular stomatitis virus glycoprotein: [125I]iodonaphthyl azide photolabeling studies in biological membranes. Biochemistry 36:8890-8896. [DOI] [PubMed] [Google Scholar]
- 31.Picot, D., P. J. Loll, and R. M. Garavito. 1994. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature 367:243-249. [DOI] [PubMed] [Google Scholar]
- 32.Porchet, E., and G. Torpier. 1977. Freeze fracture study of Toxoplasma and Sarcocystis infective stages. Z. Parasitenkd. 54:101-124. (In French.) [DOI] [PubMed] [Google Scholar]
- 33.Radke, J. R., B. Striepen, M. N. Guerini, M. E. Jerome, D. S. Roos, and M. W. White. 2001. Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Mol. Biochem. Parasitol. 115:165-175. [DOI] [PubMed] [Google Scholar]
- 34.Raviv, Y., T. Bercovici, C. Gitler, and Y. Salomon. 1989. Detection of nearest neighbors to specific fluorescently tagged ligands in rod outer segment and lymphocyte plasma membranes by photosensitization of 5-iodonaphthyl 1-azide. Biochemistry 28:1313-1319. [DOI] [PubMed] [Google Scholar]
- 35.Raviv, Y., T. Bercovici, C. Gitler, and Y. Salomon. 1984. Selective photoinduced uncoupling of the response of adenylate cyclase to gonadotropins by 5-iodonaphthyl 1-azide. Biochemistry 23:503-508. [DOI] [PubMed] [Google Scholar]
- 36.Raviv, Y., Y. Salomon, C. Gitler, and T. Bercovici. 1987. Selective labeling of proteins in biological systems by photosensitization of 5-iodonaphthalene-1-azide. Proc. Natl. Acad. Sci. USA 84:6103-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roos, D. S., R. G. Donald, N. S. Morrissette, and A. L. Moulton. 1994. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27-63. [DOI] [PubMed] [Google Scholar]
- 38.Seeber, F., J. F. Dubremetz, and J. C. Boothroyd. 1998. Analysis of Toxoplasma gondii stably transfected with a transmembrane variant of its major surface protein, SAG1. J. Cell Sci. 111:23-29. [DOI] [PubMed] [Google Scholar]
- 39.Soldati, D., J. F. Dubremetz, and M. Lebrun. 2001. Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. Int. J. Parasitol. 31:1293-1302. [DOI] [PubMed] [Google Scholar]
- 40.Spencer, A. G., E. Thuresson, J. C. Otto, I. Song, T. Smith, D. L. DeWitt, R. M. Garavito, and W. L. Smith. 1999. The membrane binding domains of prostaglandin endoperoxide H synthases 1 and 2. Peptide mapping and mutational analysis. J. Biol. Chem. 274:32936-32942. [DOI] [PubMed] [Google Scholar]
- 41.Tomavo, S., J. F. Dubremetz, and R. T. Schwarz. 1993. Structural analysis of glycosyl-phosphatidylinositol membrane anchor of the Toxoplasma gondii tachyzoite surface glycoprotein gp23. Biol. Cell 78:155-162. [DOI] [PubMed] [Google Scholar]
- 42.Tomley, F. M., and D. S. Soldati. 2001. Mix and match modules: structure and function of microneme proteins in apicomplexan parasites. Trends Parasitol. 17:81-88. [DOI] [PubMed] [Google Scholar]
- 43.Ward, G. E., and K. L. Carey. 1999. 96-well plates providing high optical resolution for high-throughput, immunofluorescence-based screening of monoclonal antibodies against Toxoplasma gondii. J. Immunol. Methods 230:11-18. [DOI] [PubMed] [Google Scholar]
- 44.Wendt, K. U., A. Lenhart, and G. E. Schulz. 1999. The structure of the membrane protein squalene-hopene cyclase at 2.0 A resolution. J. Mol. Biol. 286:175-187. [DOI] [PubMed] [Google Scholar]
- 45.Wichroski, M. J., J. A. Melton, C. G. Donahue, R. K. Tweten, and G. E. Ward. 2002. Clostridium septicum alpha-toxin is active against the parasitic protozoan Toxoplasma gondii and targets members of the SAG family of glycosylphosphatidylinositol-anchored surface proteins. Infect. Immun. 70:4353-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]