Abstract
The green sulfur bacterium Chlorobium tepidum produces chlorobactene as its primary carotenoid. Small amounts of chlorobactene are hydroxylated by the enzyme CrtC and then glucosylated and acylated to produce chlorobactene glucoside laurate. The genes encoding the enzymes responsible for these modifications of chlorobactene, CT1987, and CT0967, have been identified by comparative genomics, and these genes were insertionally inactivated in C. tepidum to verify their predicted function. The gene encoding chlorobactene glucosyltransferase (CT1987) has been named cruC, and the gene encoding chlorobactene lauroyltransferase (CT0967) has been named cruD. Homologs of these genes are found in the genomes of all sequenced green sulfur bacteria and filamentous anoxygenic phototrophs as well as in the genomes of several nonphotosynthetic bacteria that produce similarly modified carotenoids. The other bacteria in which these genes are found are not closely related to green sulfur bacteria or to one another. This suggests that the ability to synthesize modified carotenoids has been a frequently transferred trait.
Carotenoids bearing sugars and acylated sugars have been found in many bacteria and occur in a wide range of phyla in both photosynthetic and nonphotosynthetic species. Typically, glucose is linked to a hydroxyl group on the carotenoid backbone, and the glucose moiety may be acylated. The carotenoid-linked sugar may also be mannose (3) or fucose (27, 33, 34), and the length of the acyl chain can vary from 1 to 20 carbons (9). Glycosylated and acylated carotenoids are often found in the cytoplasmic membrane (11, 20, 22, 35), but their physiological functions remain largely uncharacterized. In some cases, it has been proposed that the modified carotenoids affect membrane rigidity in response to stress (11, 39). Experiments with polar carotenoids in artificial membranes suggest that they reduce the rate of oxygen diffusion through the membrane, thus protecting membrane lipids from oxidative damage (31).
Modified carotenoids are synthesized by such a variety of organisms that it is impossible to infer a function(s) from their distribution. They are found in both aerobic (33) and anaerobic (32, 36, 37, 38) photosynthetic bacteria as well as in the flowers and fruits of a few plant species (9). Among nonphotosynthetic bacteria, glycosylated and acylated carotenoids have also been identified in aerobes (22) and facultative aerobes (25) as well as in halophiles (23), psychrophiles (11), and thermophiles (41, 42).
The thermophilic filamentous anoxygenic phototroph (FAP) Chloroflexus aurantiacus produces OH-γ-carotene glucoside esters, for which the major fatty acids are hexadecanoate and hexadecenoate (37). Thermus thermophilus has also been shown to produce thermozeaxanthins, which are zeaxanthin glucoside esters (42). Deinococcus spp., which are phylogenetically related to Thermus spp., are red pigmented as a result of carotenoid production, and at least two species of glycosylated xanthophyll carotenoids are produced by these organisms (4). Finally, acylated carotenoids also occur in Myxococcus xanthus, which synthesizes these carotenoids in response to blue light (6, 8), nutrient status (12), or copper stress (28).
The enzymatic pathway for modification of carotenoids has been elucidated for a few species. In Erwinia herbicola and related bacteria, the glucosyltransferase is CrtX, a membrane-associated protein that transfers glucose from UDP-glucose to zeaxanthin (20, 26). Staphylococcus aureus has an unrelated carotenoid glycosyltransferase, which converts the C30 carotenoid 4,4′-diaponeurosporenic acid to glycosyl 4,4′-diaponeurosporenoate (29). Acylation of this compound produces staphyloxanthin, or β-D-glucopyranosyl 1-O-(4,4′-diaponeurosporen-4-oate)-6-O-(12-methyltetradecanoate). The genes encoding the glycosyltransferase and acyltransferase in S. aureus have been named crtQ and crtO, respectively (29). However, in this paper, we will refer to the S. aureus enzymes by their locus tags, SA2350 and SA2352, in order to avoid confusion with ζ-carotene desaturase, which is also named CrtQ (2), and β-carotene ketolase, also named CrtO (19).
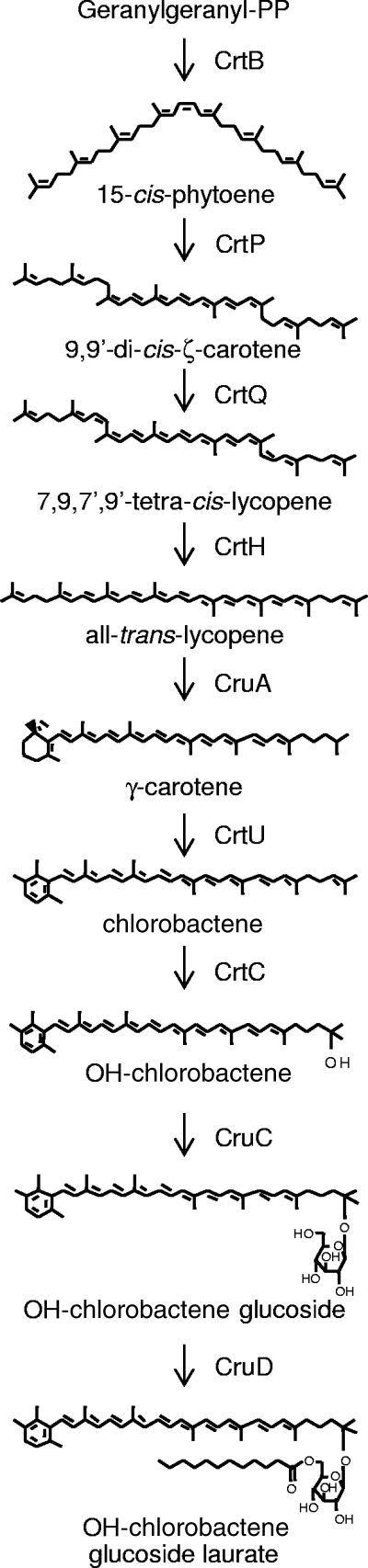
The green sulfur bacterium Chlorobium tepidum produces a small amount of OH-chlorobactene glucoside laurate (35, 38), which is found in the cytoplasmic membrane rather than associated with the chlorosomes (35). The genes in the chlorobactene biosynthetic pathway up to OH-chlorobactene have previously been identified (Fig. 1) (14, 24), but no homologs of the E. herbicola glycosyltransferase, CrtX, could be found in the predicted open reading frames (ORFs) in the C. tepidum genome (10). In this study, a comparative genomics approach was used to identify possible carotenoid-specific glycosyltransferases and acyltransferases in C. tepidum and other carotenogenic organisms. This approach identified single genes as candidates to encode the glycosyltransferase and the acyltransferase, both of which were insertionally inactivated in C. tepidum to confirm the roles of their products in carotenogenesis. The identification of these genes, which have now been named cruC and cruD, completes the identification of genes encoding the enzymes of the carotenoid biosynthetic pathway in C. tepidum (Fig. 1).
FIG. 1.
Biosynthetic pathway of carotenoids in C. tepidum. All genes identified in this pathway have been insertionally inactivated, and the resulting mutant strains have been biochemically and physiologically characterized (14, 24; this work).
MATERIALS AND METHODS
Strains and growth conditions.
The wild-type strain of C. tepidum is the plating strain WT2321 (40). All C. tepidum strains were grown at 46°C either in liquid CL medium or on CP plates (13); mutants were selected on CP plates amended with 300 μg spectinomycin ml−1 and 150 μg streptomycin ml−1. For measurement of growth rates, all strains were grown in CL medium without antibiotics on a rotating shaker (40 rpm) at a light intensity of 70 or 350 μmol photons m−2 s−1. Light intensity was measured with a QSL-100 light meter from Biospherical Instruments (San Diego, CA).
Identification of candidate genes and phylogenetic comparisons.
All ORFs in the C. tepidum genome that had been annotated as either glycosyltransferases or acyltransferases were compared to proteins in the nonredundant databases in the Comprehensive Microbial Resources (CMR; The Institute for Genomic Research, Rockville, MD; http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi) and the Integrated Microbial Genomes (IMG; Joint Genome Institute, Walnut Grove, CA; http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) sites by use of the BLASTP algorithm (1). The genome neighborhoods of the best hits were inspected for the presence of nearby ORFs predicted to be involved in carotenoid biosynthesis. For construction of phylogenetic trees, amino acid sequences were aligned by use of the ClustalW tool in MacVector 7.2.3 (Accelrys, San Diego, CA) and neighbor-joining trees were generated from the alignments with Paup 4.0 (Sinauer Associates, Inc., Sunderland, MA).
Inactivation of cruC and cruD.
The left and right flanks of genes selected for inactivation were amplified by PCR from wild-type C. tepidum genomic DNA with the primers described in Table 1. The reverse primers for the left flanks and forward primers for the right flanks contained KpnI and SalI restriction sites, respectively. PCR products were digested with the appropriate restriction enzymes and purified from agarose gels by use of an Eppendorf Perfectprep gel cleanup kit (catalog number 0032 007.759; Westbury, NY). The aadA gene, conferring resistance to streptomycin and spectinomycin, was excised from plasmid pSRA81 (14) with SalI and KpnI and purified in the same manner. The fragments were mixed in a 1:1:1 ratio and ligated. The ligation products were separated on an agarose gel, and the products of the appropriate size were excised, purified, and used to transform wild-type C. tepidum as described previously (13, 15). Antibiotic-resistant colonies appeared after approximately 1 week and were transferred to fresh plates amended with spectinomycin and streptomycin. After three successive transfers, PCR across the site of insertion, using primers CT1987 F1 and CT1987 R1 for CT1987 and primers CT0967 F3 and CT0967 R3 for CT0967 (Table 1), confirmed that the wild-type and mutant alleles for both mutants had completely segregated. Two representative transformants from each mutant strain were selected for further analysis.
TABLE 1.
Sequences of primers used for construction of mutants and confirmation of segregation
| Primer name | Primer sequencea | Purpose |
|---|---|---|
| CT1987 F1 | CAG GAA GCT TAT GAA AAG TGC | Confirming segregation |
| CT1987 R1 | CGC GAG CTA CCG GAG GGT TGG | Confirming segregation |
| CT1987 F2 | TGG AGT CGA CGA AAG CAT CC | Amplifying the right flank of inactivation construct |
| CT1987 R2 | CTG CCT TAG GTA CCG CGG TTT CG | Amplifying the left flank of inactivation construct |
| CT1987 F3 | CCA GAT TTT TCA CCA CAG GCT CC | Amplifying the left flank of inactivation construct |
| CT1987 R3 | TTC CAC TTT TCC GTC CAG AGC C | Amplifying the right flank of inactivation construct |
| CT0967 F2 | GAA TGC ACT GTC GAC TTT TCC G | Amplifying the right flank of inactivation construct |
| CT0967 R2 | AGT GAA GAA CTG GTA CCT GAA GAG | Amplifying the left flank of inactivation construct |
| CT0967 F3 | TCA TCA ATG CGT GCG ACA ACT | Amplifying the left flank of inactivation construct |
| CT0967 R3 | CAT CGT TTC TTC TAC GGA TTT ACC C | Amplifying the right flank of inactivation construct |
KpnI sites are underlined, and SalI sites are underlined and in italics.
Pigment analyses.
Mutants were grown without antibiotics in CL medium until the optical density at 600 nm was approximately 1.0. Pigments were extracted from cell pellets by sonication in acetone-methanol (7:2, vol/vol), and cellular debris was removed by centrifugation. The supernatant was filtered through a 0.2-μm polytetrafluoroethylene syringe filter (6783-0402; Whatman, Clifton, NJ), and the solvent was dried under a stream of N2 gas. The dried pigments were resuspended in 100 μl of filtered (0.2 μm) acetone-methanol (7:2, vol/vol) prior to injection into the high-performance liquid chromatography (HPLC) system. The HPLC system consisted of a 25-cm-by-4.6-mm 5-μm Discovery C18 column (Supelco, Bellefonte, Pa.) fitted to a binary pump (model G1312A) and solvent degasser (model 1379A) (1100 Series; Agilent Technologies, Palo Alto, CA). Eluates were monitored with a 1,024-element diode array detector (model G1315B, 1100 series; Agilent Technologies, Palo Alto, CA), and the system was controlled with Agilent ChemStation software for HPLC. The solvents were as follows: solvent A (methanol-acetonitrile-water [21:16.5:62.5, vol/vol/vol], containing 10 mM ammonium acetate) and solvent B (methanol-acetonitrile-ethyl acetate [50:20:30, vol/vol/vol]). The elution gradient began with 20% solvent B at a flow rate of 0.75 ml min−1 and increased to 70% solvent B by 10 min. After this, the flow rate was increased to 1 ml min−1, and the solvent was increased to 100% solvent B by 40 min and was held constant at 100% solvent B for an additional 20 min. Elution of carotenoids was monitored at 491 nm. Mass spectrometry was performed by A. Daniel Jones, Department of Chemistry, The Pennsylvania State University.
RESULTS
Identification of candidate genes for the terminal steps of carotenogenesis in C. tepidum.
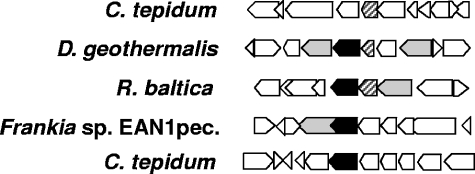
The C. tepidum genome encodes nearly 40 glycosyltransferases and several acyltransferases. Rather than systematically attempt to inactivate each gene by interposon mutagenesis to identify the genes whose products are involved in carotenogenesis, we employed a comparative bioinformatics approach to identify a subset of candidate genes. As shown in Fig. 2, the genomic contexts of CT1987 and CT0967 in C. tepidum provide no clues that the proteins encoded by these genes might be involved in carotenogenesis, since each is flanked by hypothetical and conserved hypothetical proteins. Visual inspection of the genome neighborhoods of orthologs of annotated C. tepidum glycosyl- and acyltransferases identified one candidate OH-chlorobactene glycosyltransferase (CT1987) and one candidate OH-chlorobactene glucoside acyltransferase (CT0967). Each of these candidate genes had apparent orthologs in Deinococcus geothermalis, Rhodopirellula baltica, and Frankia sp. strain EAN1pec, and in these species, the acyl- or glycosyltransferases are clustered with one or more genes encoding proteins annotated as carotenoid dehydrogenases or phytoene synthase (Fig. 2). Based upon these observations, CT1987 and CT0967 were selected for insertional inactivation and further characterization.
FIG. 2.
Genome neighborhood comparisons. CT0967 (cruD) in C. tepidum and orthologs in other organisms are striped; CT1987 (cruC) in C. tepidum and homologs in other organisms are solid black. Other genes predicted to be involved in carotenoid biosynthesis are light gray; all other genes are white. In two species, orthologs of both CT1987 and CT0967 appear in a genomic region with at least one other carotenoid biosynthesis gene. This arrangement of genes suggests that these two ORFs might encode proteins that function in carotenoid biosynthesis.
Insertional inactivation of CT1987 and CT0976.
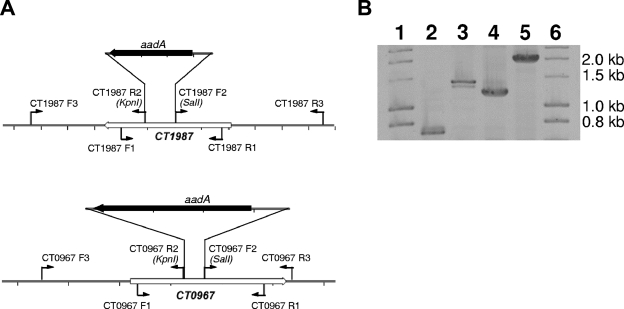
A DNA fragment encoding the aadA gene, which encodes aminoglycoside acetyltransferase and confers resistance to spectinomycin and streptomycin, was ligated to the DNA fragments encoding the upstream and downstream flanks of CT1987 and CT0967 (Fig. 3A). The resulting constructions were purified and used to transform wild-type C. tepidum cells. Transformants were selected and streaked to obtain isolated colonies and to allow segregation of alleles. PCR across these two genes showed that each transformant had a 0.9-kb insertion in the inactivated gene, which corresponds to the size of the aadA cassette (Fig. 3B). No product corresponding to the size of the wild-type product was observed in either mutant; therefore, the wild-type and mutant alleles were completely segregated in the mutant strains.
FIG. 3.
Restriction maps of constructions used to inactivate CT1987 and CT0967 and confirmation of segregated mutants. (A) Restriction maps and primer locations of constructions used to inactivate CT1987 and CT0967. (B) Electrophoretic analysis of PCR products to evaluate putative mutants. The CT1987 and CT0967 loci were amplified by PCR with the primers indicated in Table 1. The DNA templates were derived from wild-type C. tepidum (lanes 2 and 4), from a CT1987::aadA transformant (lane 3), and from a CT0967::aadA transformant (lane 5). The amplicons from the two transformants are 0.9 kb larger than the corresponding amplicon from the wild type. This demonstrates that wild-type and mutant alleles had segregated fully in both transformants. Lanes 1 and 6 contain DNA size markers (Ladder I; GeneChoice, Frederick, MD).
Characterization of mutant strains.
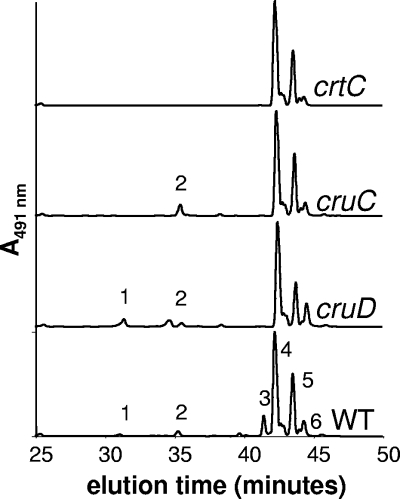
When carotenoids from wild-type C. tepidum were analyzed by HPLC, six major species were resolved (Fig. 4). Based upon previous characterization of various crt mutants as well as mass spectrometric analyses (14), these peaks correspond to OH-chlorobactene glucoside (peak 1), OH-chlorobactene (peak 2), OH-chlorobactene glucoside laurate (peak 3), chlorobactene (peak 4), γ-carotene (peak 5), and 1′,2′-dihydrochlorobactene (peak 6). As shown in Fig. 4, peaks 1, 2, and 3 were all missing when the crtC gene, encoding chlorobactene 2′-hydroxylase, is inactivated. The CT1987::aadA mutant no longer produced OH-chlorobactene glucoside or OH-chlorobactene glucoside laurate but still produced a carotenoid (peak 2) with an absorption spectrum identical to that for chlorobactene (absorption maxima at 461 and 491 nm; data not shown) and with the mass of OH-chlorobactene (550.4 Da). The CT0967::aadA mutant could no longer synthesize OH-chlorobactene glucoside laurate (peak 3) but could still produce OH-chlorobactene glucoside (peak 1) and OH-chlorobactene (peak 2). The carotenoid analyses of the two mutants confirm that CT1987 encodes the OH-chlorobactene glucosyltransferase while CT0967 encodes the OH-chlorobactene glucoside lauroyltransferase. These two ORFs have been renamed cruC and cruD, respectively.
FIG. 4.
HPLC elution profiles of the C. tepidum wild type (WT) and mutants unable to synthesize various modified carotenoids. Peak 1, OH-chlorobactene (550.4 Da); peak 2, OH-chlorobactene glucoside (729.4 Da); peak 3, OH-chlorobactene glucoside laurate (894.6 Da); peak 4, chlorobactene (532.4 Da); peak 5, γ-carotene (536.4 Da); peak 6, 1′,2′-dihydrochlorobactene (534.5 Da). Elution of carotenoid species was monitored at 491 nm.
Growth rates of the mutants.
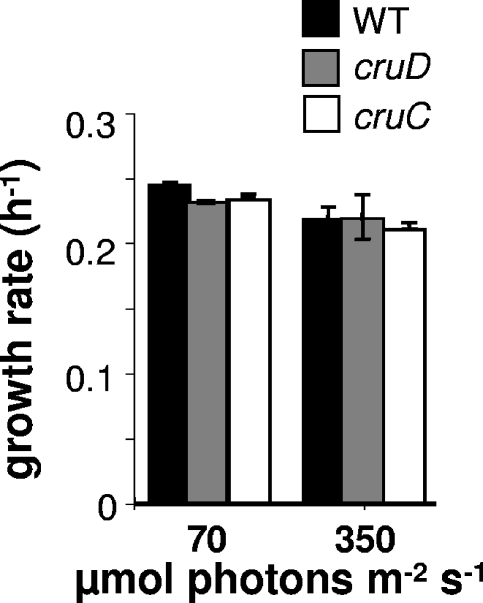
Previous studies had shown that a C. tepidum crtB mutant lacking all carotenoids is viable and that mutants producing predominantly lycopene, γ-carotene, or chlorobactene (Fig. 1) can grow at rates similar to that of the wild type (14). The crtC mutant, which cannot hydroxylate chlorobactene and therefore makes no glycosylated carotenoids (Fig. 4), grows at a rate ∼90% of that of the wild type at 70 μmol photons m−2 s−1 and at the same rate as the wild type at high light intensity (14). In contrast, the cruC and cruD mutants grow at the same rate as the wild type at both intermediate and high light intensities (Fig. 5).
FIG. 5.
Growth rates for the wild type (WT) and cruC and cruD mutant strains of C. tepidum at different light intensities. The rates are the averages from three independent determinations, and standard errors are indicated.
Sequence comparisons and phylogenetic analyses.
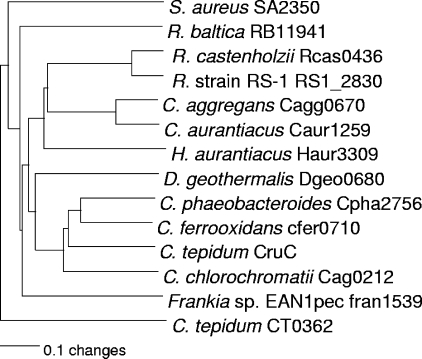
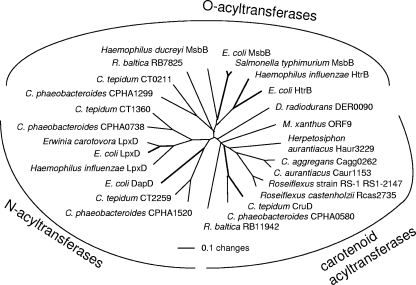
The apparently orthologous glycosyltransferases, encoded in the genomic neighborhoods of other predicted carotenoid biosynthesis genes, were aligned with CruC and analyzed (Fig. 6). Among these carotenoid-specific glycosyltransferases, those from green sulfur bacteria (GSB) form a distinct clade within a larger group that includes all Chloroflexi (Herpetosiphon aurantiacus, Chloroflexus aurantiacus, and Chloroflexus aggregans, Roseiflexus sp. strain RS-1, and Roseiflexus castenholzii). The recently characterized diaponeurosporene glycosyltransferase from Staphylococcus aureus, SA2350 (29), can be aligned with the CruC orthologs, but it consistently groups outside the cluster of C40 carotenoid glycosyltransferases (Fig. 6). The acyltransferases found in the same or similar genomic regions and apparently encoding carotenoid biosynthetic enzymes orthologous to CruD were aligned with selected acyltransferases from lipid and amino acid biosynthetic pathways. The carotenoid acyltransferases form a phylogenetically distinctive clade that is separate from the other two (Fig. 7).
FIG. 6.
Neighbor-joining tree showing phylogenetic relationships among carotenoid glycosyltransferases. Amino acid sequences were aligned and used to reconstruct the phylogeny; all labels are the automatically assigned locus tags for that ORF. Only C. tepidum CruC and S. aureus SA2350 have been genetically characterized. Another class II glycosyltransferase, CT0362, from C. tepidum was used as the outgroup for this comparison. Numbers for open reading frames in C. aggregans, H. aurantiacus, R. castenholzii, and Roseiflexus sp. strain (R. strain) RS-1 are the numbers currently assigned in the draft genome sequences.
FIG. 7.
Neighbor-joining tree showing phylogenetic relationships among various acyltransferases. Amino acid sequences were aligned and used to reconstruct the phylogeny. Bold lines indicate proteins whose functions have been genetically or biochemically confirmed. Numbers for open reading frames in C. aggregans, H. aurantiacus, R. castenholzii, and Roseiflexus sp. strain RS-1 are the numbers currently assigned in the draft genome sequences. E. coli, Escherichia coli.
DISCUSSION
This study demonstrates the utility of using gene neighborhood comparisons of orthologous genes to identify candidate genes when the genes in question lack contextual information showing that they could be involved in a particular biosynthetic pathway. In the examples shown here, this approach identified single candidate genes for both a carotenoid glucosyltransferase and a carotenoid acyltransferase in the genome of C. tepidum. Inactivation of these two candidate genes and biochemical analyses of the corresponding mutants confirmed that the product of CT1987 is the OH-chlorobactene glucosyltransferase and that CT0967 encodes the OH-chlorobactene glucoside lauroyltransferase. The identification of these genes, now named cruC and cruD, respectively, completes the identification of the genes responsible for the enzymes of OH-chlorobactene glucoside laurate in C. tepidum (Fig. 1).
For C. tepidum, the presence of a population of carotenoids anchored in the cytoplasmic membrane most likely protects the membrane from radicals generated by the reaction centers, from reactive oxygen species, from chlorophyll triplet states, or from all of these. Although GSB are not oxygenic, the strong reductants generated by their reaction center can readily react with O2 to form reactive oxygen species (16), which have the potential to damage membrane lipids. Given their absolute dependence upon light for growth, many GSB inhabit the upper parts of anoxic zones, where periodic disturbance of the chemocline by wind or tides can result in transient exposure to molecular oxygen. Because homologs of both CruC and CruD are found in the genomes of all sequenced GSB, it is likely that these specialized carotenoids play an important role(s) in these organisms. Additionally, although inactivation of crtC, which encodes the enzyme that acts immediately before CruC and CruD, reduces the growth rate of C. tepidum under saturating-light intensity, neither the cruC nor the cruD mutant has a growth defect under optimal laboratory conditions. This suggests that the modified carotenoid may be more important under stress conditions such as low temperature or oxic conditions.
Because glycosylated and acylated carotenoids are widespread among both photosynthetic and nonphotosynthetic bacteria, it is unlikely that they are principally involved in light harvesting and the conversion of light energy to chemical energy. Instead, they most likely serve other structural or protective functions in the membrane, absorbing and dissipating the energy from reactive oxygen species, UV light, or chlorophyll triplet states. Polar modifications of carotenoids, such as the introduction of hydroxyl or keto groups, tend to facilitate interactions with the phosphate head groups of lipids, thereby orienting the carotenoids within membranes (7, 31). The acyl group should anchor these molecules within the membrane and could also provide structural interactions with particular membrane proteins. In some cases, specific lipids are known to play important roles in the biogenesis of some membrane proteins (5) or to have specific binding sites on membrane proteins associated with photosynthesis (17, 21). In light of the observations that chlorobactene glucoside esters are found in the cytoplasmic membrane rather than the chlorosome and that they copurify with reaction centers (35), it is also possible that in GSB, these modified carotenoids play some role, albeit nonessential, either in biogenesis of the photosystem proteins or in photoprotection of the reaction centers in the membrane.
The S. aureus carotenoid glycosyltransferase SA2350 can be aligned with CruC glycosyltransferases, but its sequence is not more similar to the carotenoid glycosyltransferases than to other glycosyltransferases; several outgroup sequences were tested, and SA2350 always grouped with that sequence (data not shown). The large sequence differences between SA2350 and CruC may be due to the fact that the substrates for SA2350 are C30 carotenoids rather than the C40 compounds synthesized by other species. Because the S. aureus acyltransferase, SA2352, also seems to be unrelated to other putative carotenoid acyltransferases (see below), it appears likely that the similarity of the SA2350 carotenoid glycosyltransferase to the other carotenoid glycosyltransferases may have arisen by convergent evolution from a nonorthologous glycosyltransferase subfamily.
The putative carotenoid acyltransferases orthologous to CruD form a coherent phylogenetic group when aligned and compared with lipid O-specific or N-specific acyltransferases. This grouping, in conjunction with gene neighborhood analysis, allows us to predict confidently that the ORFs RB11942 (in Rhodopirellula baltica) and Der0090 (in Deinococcus radiodurans), as well as ORF 9 in the M. xanthus carotenoid biosynthesis operon, also encode carotenoid acyltransferases. However, the S. aureus acyltransferase SA2352 cannot be aligned with these carotenoid acyltransferases. Thus, CruD appears to be the first characterized member of a new family of carotenoid acyltransferases.
The five sequenced FAP species, Chloroflexus aurantiacus, Chloroflexus aggregans, Roseiflexus sp. strain RS-1, and R. castenholzii, and the nonphotosynthetic Chloroflexus strain Herpetosiphon aurantiacus also have homologs of both cruC and cruD. As shown in Fig. 6, the glycosyltransferases of these Chloroflexi cluster together within the group of carotenoid glycosyltransferases, but their sequences are not very similar to those of the GSB and their lineage is deeply divergent from the GSB clade (Fig. 6). The CruD sequences, which form a distinct clade among the larger family of acyltransferases, exhibit a similar pattern, in which the sequences derived from the Chloroflexi and Chlorobi form distinct clades that are deeply divergent. Interestingly, the CruD sequence most divergent from the Chloroflexi species is that of the nonphototrophic H. aurantiacus; the sequences from the two Chloroflexus spp. and Roseiflexus spp. are more similar (Fig. 7). These relationships are similar to those inferred from comparisons of the 16S rRNA sequences of these organisms (data not shown).
The identification of cruC and cruD, and the distribution of these genes in other bacteria, highlights the chimeric nature of the carotenoid biosynthetic pathway in GSB. The enzymes for the conversion of phytoene to γ-carotene in Chlorobi are similar to those for the conversion of phytoene to β-carotene in cyanobacteria, and they differ from the enzymes found in Chloroflexi and most other bacteria (14, 24). However, the enzymes involved in modification of the cyclized carotenoids (ring modification, ψ-end hydroxylation, glycosylation, and acylation) are more similar to those in FAPs and nonphotosynthetic bacteria (14). This raises interesting questions about the acquisition and evolution of carotenoid biosynthesis genes in GSB. It seems plausible that the majority of the carotenoid biosynthesis pathway was inherited vertically from a common ancestor of GSB and cyanobacteria, while the genes encoding the final three steps for chlorobactene modification may have been acquired laterally at a later stage from another organism. Considering the prevalence of lateral gene transfer in the evolutionary history of carotenoid biosynthesis (18, 30), this is perhaps the rule rather than the exception.
Acknowledgments
We thank A. Daniel Jones for the mass spectrometric analyses of pigments from the carotenoid mutants.
This research was funded by grant DE-FG02-94ER20137 from the U.S. Department of Energy to D.A.B.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, G. A. 1997. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu. Rev. Microbiol. 51:629-659. [DOI] [PubMed] [Google Scholar]
- 3.Arpin, N., J.-L. Fiasson, and S. Liaaen-Jensen. 1972. Bacterial carotenoids XXXIX. C50 carotenoids 10. Bacterioruberin mono- and diglycosides. Acta Chem. Scand. 26:2526-2528. [DOI] [PubMed] [Google Scholar]
- 4.Battista, J. R., and F. A. Rainey. Deinococcaceae, p. 395-403. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.
- 5.Bogdanov, M., J. Sun, H. R. Kaback, and W. Dowhan. 1996. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J. Biol. Chem. 271:11615-11618. [DOI] [PubMed] [Google Scholar]
- 6.Botella, J. A., F. J. Murillo, and R. Ruiz-Vazquez. 1995. A cluster of structural and regulatory genes for light-induced carotenogenesis in Myxococcus xanthus. Eur. J. Biochem. 233:238-348. [DOI] [PubMed] [Google Scholar]
- 7.Britton, G. 1995. Structure and properties of carotenoids in relation to function. FASEB J. 9:1551-1558. [PubMed] [Google Scholar]
- 8.Burchard, R. P., and M. Dworkin. 1966. Light-induced lysis and carotenogenesis in Myxococcus xanthus. J. Bacteriol. 91:535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dembitsky, V. M. 2005. Astonishing diversity of natural surfactants. 3. Carotenoid glycosides and isoprenoid glycolipids. Lipids 40:535-557. [DOI] [PubMed] [Google Scholar]
- 10.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelon, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong, N. J. C., M. L. Burgess, K. D. Barrow, and D. R. Glenn. 2001. Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 56:750-756. [DOI] [PubMed] [Google Scholar]
- 12.Fontes, M., R. Ruiz-Vazquez, and F. J. Murillo. 1993. Growth phase dependence of the activation of a bacterial gene for carotenoid synthesis by blue light. EMBO J. 12:1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frigaard, N.-U., and D. A. Bryant. 2001. Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl. Environ. Microbiol. 67:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frigaard, N.-U., J. A. Maresca, C. E. Yunker, A. D. Jones, and D. A. Bryant. 2004. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 186:5210-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frigaard, N.-U., Y. Sakuragi, and D. A. Bryant. 2004. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274:325-340. [DOI] [PubMed] [Google Scholar]
- 16.Frigaard, N.-U., S. Takaichi, M. Hirota, K. Shimada, and K. Matsuura. 1997. Quinones in chlorosomes of green sulfur bacteria and their role in the redox-dependent fluorescence studied in chlorosome-like bacteriochlorophyll c aggregates. Arch. Microbiol. 167:343-349. [Google Scholar]
- 17.Fromme, P., P. Jordan, and N. Krauss. 2001. Structure of photosystem I. Biochim. Biophys. Acta 1507:5-31. [DOI] [PubMed] [Google Scholar]
- 18.Giraud, E., L. Hannibal, J. L. Fardoux, M. Jaubert, P. Jourand, B. Dreyfus, J. N. Sturgis, and A. Vermeglio. 2004. Two distinct crt gene clusters for two different functional classes of carotenoid in Bradyrhizobium. J. Biol. Chem. 279:15076-15083. [DOI] [PubMed] [Google Scholar]
- 19.Harker, M., and J. Hirschberg. 1997. Biosynthesis of ketocarotenoids in transgenic cyanobacteria expressing the algal gene for β-C-4-oxygenase, crtO. FEBS Lett. 404:129-134. [DOI] [PubMed] [Google Scholar]
- 20.Hundle, B. S., D. A. O'Brien, M. Alberti, P. Meyer, and J. E. Hearst. 1992. Functional expression of a zeaxanthin glucosyltransferase from Erwinia herbicola and a proposed uridine diphosphate binding site. Proc. Natl. Acad. Sci. USA 89:9321-9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, M. R., P. K. Fyfe, A. W. Roszak, N. W. Isaacs, and R. J. Cogdell. 2002. Protein-lipid interactions in the purple bacterial reaction center. Biochim. Biophys. Acta 1565:206-214. [DOI] [PubMed] [Google Scholar]
- 22.Kleinig, H. 1972. Membranes from Myxococcus fulvus (Myxobacteriales) containing carotenoid glucosides. Biochim. Biophys. Acta 274:489-498. [DOI] [PubMed] [Google Scholar]
- 23.Lutnaes, B. F., A. Oren, and S. Liaaen-Jensen. 2002. New C40-carotenoid acyl glycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J. Nat. Prod. 65:1340-1343. [DOI] [PubMed] [Google Scholar]
- 24.Maresca, J. A., N.-U. Frigaard, and D. A. Bryant. 2005. Identification of a novel class of lycopene cyclases in photosynthetic organisms, p. 884-886. In A. van der Est and D. Bruce (ed.), Photosynthesis: fundamental aspects to global perspectives. Allen Press, Lawrence, Kans.
- 25.Marshall, J. H., and G. J. Wilmoth. 1981. Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J. Bacteriol. 147:900-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misawa, N., M. Nakagawa, K. Kobayashi, S. Yamano, Y. Izawa, K. Nakamura, and K. Harashima. 1990. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172:6704-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohamed, H. E., and W. Vermaas. 2004. Slr1293 in Synechocystis sp. strain PCC 6803 is the C-3′,4′ desaturase (CrtD) involved in myxoxanthophyll biosynthesis. J. Bacteriol. 186:5621-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moraleda-Muñoz, A., J. Perez, M. Fontes, F. J. Murillo, and J. Muñoz-Dorado. 2005. Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus. Mol. Microbiol. 56:1159-1168. [DOI] [PubMed] [Google Scholar]
- 29.Pelz, A., K.-P. Wieland, K. Putzbach, P. Henschel, K. Albert, and F. Götz. 2005. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280:32493-32498. [DOI] [PubMed] [Google Scholar]
- 30.Phadwal, K. 2005. Carotenoid biosynthetic pathway: molecular phylogenies and evolutionary behavior of crt genes in eubacteria. Gene 345:35-43. [DOI] [PubMed] [Google Scholar]
- 31.Subczynski, W. K., E. Markowska, and J. Sielewiesiuk. 1991. Effect of polar carotenoids on the oxygen diffusion-concentration product in lipid bilayers. An EPR spin label study. Biochim. Biophys. Acta 1068:68-72. [DOI] [PubMed] [Google Scholar]
- 32.Takaichi, S., T. Maoka, S. Hanada, and J. F. Imhoff. 2001. Dihydroxylycopene diglucoside diesters: a novel class of carotenoids from the phototrophic purple sulfur bacteria Halorhodospira abdelmalekii and Halorhodospira halochloris. Arch. Microbiol. 175:161-167. [DOI] [PubMed] [Google Scholar]
- 33.Takaichi, S., T. Maoka, and K. Masamoto. 2001. Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2′S)-myxol 2′-(2,4-di-O-methyl-α-l-fucoside), not rhamnoside. Plant Cell Physiol. 42:756-762. [DOI] [PubMed] [Google Scholar]
- 34.Takaichi, S., M. Mochimaru, T. Maoka, and H. Katoh. 2005. Myxol and 4-ketomyxol 2′-fucosides, not rhamnosides, from Anabaena sp. PCC 7120 and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol. 46:497-504. [DOI] [PubMed] [Google Scholar]
- 35.Takaichi, S., and H. Oh-Oka. 1999. Pigment composition in the reaction center complex from the thermophilic green sulfur bacterium Chlorobium tepidum: carotenoid glucoside esters, menaquinone, and chlorophylls. Plant Cell Physiol. 40:691-694. [Google Scholar]
- 36.Takaichi, S., H. Oh-oka, T. Maoka, D. O. Jung, and M. T. Madigan. 2003. Novel carotenoid glucoside esters from alkaliphilic heliobacteria. Arch. Microbiol. 179:95-100. [DOI] [PubMed] [Google Scholar]
- 37.Takaichi, S., K. Tsuji, K. Matsuura, and K. Shimada. 1995. A monocyclic carotenoid glucoside ester is a major carotenoid in the filamentous bacterium Chloroflexus aurantiacus. Plant Cell Physiol. 36:773-778. [Google Scholar]
- 38.Takaichi, S., Z.-W. Wang, M. Umetsu, T. Nozawa, K. Shimada, and M. T. Madigan. 1997. New carotenoids from the thermophilic green sulfur bacterium Chlorobium tepidum: 1′,2′-dihydro-γ-carotene, 1′,2′-dihydrochlorobactene, and OH-chlorobactene glucoside ester, and the carotenoid composition of different strains. Arch. Microbiol. 168:270-276. [DOI] [PubMed] [Google Scholar]
- 39.Varkonyi, Z., K. Masamoto, M. Debreczeny, O. Zsiros, B. Ughy, Z. Gombos, I. Domonkos, T. Farkas, H. Wada, and B. Szalontai. 2002. Low-temperature-induced accumulation of xanthophylls and its structural consequences in the photosynthetic membranes of the cyanobacterium Cylindrospermopsis raciborskii: an FTIR spectroscopic study. Proc. Natl. Acad. Sci. USA 99:2410-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahlund, T. M., and M. T. Madigan. 1995. Genetic transfer by conjugation in the thermophilic green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 177:2583-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoyama, A., G. Sandmann, T. Hoshino, K. Adachi, M. Sakai, and Y. Shizuri. 1995. Thermozeaxanthins, new carotenoid-glycoside-eseters from thermophilic eubacterium Thermus thermophilus. Tetrahedron Lett. 36:4901-4904. [Google Scholar]
- 42.Yokoyama, A., Y. Shizuri, T. Hoshino, and G. Sandmann. 1996. Thermocryptoxanthins: novel intermediates in the carotenoid biosynthetic pathway of Thermus thermophilus. Arch. Microbiol. 165:342-345. [DOI] [PubMed] [Google Scholar]