Abstract
The ppk gene of Streptomyces lividans encodes an enzyme catalyzing, in vitro, the reversible polymerization of the γ phosphate of ATP into polyphosphate and was previously shown to play a negative role in the control of antibiotic biosynthesis (H. Chouayekh and M. J. Virolle, Mol. Microbiol. 43:919-930, 2002). In the present work, some regulatory features of the expression of ppk were established and the polyphosphate content of S. lividans TK24 and the ppk mutant was determined. In Pi sufficiency, the expression of ppk was shown to be low but detectable. DNA gel shift experiments suggested that ppk expression might be controlled by a repressor using ATP as a corepressor. Under these conditions, short acid-soluble polyphosphates accumulated upon entry into the stationary phase in the wild-type strain but not in the ppk mutant strain. The expression of ppk under Pi-limiting conditions was shown to be much higher than that under Pi-sufficient conditions and was under positive control of the two-component system PhoR/PhoP. Under these conditions, the polyphosphate content of the cell was low and polyphosphates were reproducibly found to be longer and more abundant in the ppk mutant strain than in the wild-type strain, suggesting that Ppk might act as a nucleoside diphosphate kinase. In light of our results, a novel view of the role of this enzyme in the regulation of antibiotic biosynthesis in S. lividans TK24 is proposed.
Streptomyces is a genus of gram-positive filamentous soil bacteria that are well known for their ability to produce antibiotics which are important for human and animal health. The genes responsible for the biosynthesis of an antibiotic are clustered on the chromosome, and their coordinated expression is most often under the direct control of “specific” activators linked to each biosynthetic pathway (4, 12, 14). The expression of these pathways is usually switched on when growth slows down or stops, suggesting that the expression of the corresponding specific activators might be triggered by some nutritional deprivation (4). The expression of these specific regulators is thought to be under the control of global regulators sensing the nutritional status of the cell. The most efficient and well-documented trigger of antibiotic biosynthesis in streptomycetes is a nutritional limitation in inorganic phosphate (Pi) (3, 17, 21, 24) that was shown to correlate with a low adenylate energy charge (22, 23). However, the regulatory cascade starting from sensing of a Pi limitation to the switching on or off of the expression of different pathway-specific activators/repressors is only just beginning to be unraveled.
Recently the disruption of two loci (phoR/phoP and ppk), both related to phosphate metabolism, was shown to enhance greatly antibiotic production in the normally very weak producer Streptomyces lividans TK24 (7, 10, 28). The genes phoR and phoP encode a sensory kinase and a response regulator, respectively, constituting a two-component system for signaling and promoting the adaptation of the bacteria to Pi limitation (10, 28). Since the supply of Pi is reduced in a phoP mutant, the latter is prematurely starved of Pi, and this starvation is thought to be responsible for the triggering of antibiotic biosynthesis (28). Strong antibiotic production was also observed upon disruption of the ppk gene of S. lividans (7). This gene encodes an enzyme catalyzing the reversible polymerization of the γ phosphate of ATP into polyphosphate (polyP). This enzyme thus acts as a polyphosphate kinase when the ATP/ADP ratio in the in vitro reaction mixture is high and as a nucleoside diphosphate kinase (NDPK), regenerating ATP from ADP and polyphosphate, when this ratio is low (7). Since the function of this enzyme, established in vitro, does not necessarily reflect its in vivo function, in this study, we attempted to determine the activity of Ppk in vivo, via analysis of the polyphosphate content of the cell. We also established some of the regulatory features of the expression of ppk in response to Pi availability and characterized some cis- and trans-acting elements playing a role in the regulation of ppk expression. Finally, in light of our results, we propose a novel view of the role of this enzyme in the regulation of antibiotic biosynthesis in S. lividans TK24.
MATERIALS AND METHODS
Bacterial strains.
The Streptomyces strains used in this study were all derivatives of S. lividans TK24 (13): S. lividans TK24 ppk::Ωhygro (7), S. lividans TK24 EcoRI::Ωaac ppk (this work), and the S. lividans TK24 phoP::Ωaac strain (this work).
Construction of S. lividans TK24 phoP::Ωaac and S. lividans TK24 EcoRI::Ωaacppk mutant strains.
In order to construct the S. lividans TK24 phoP::Ωaac mutant strain, a 3.2-kb fragment encompassing the genes phoP and phoR from S. lividans TK24 was amplified by PCR using the primers phoRP5′ and phoRP3′ (see Table 2) derived from the putative phoP and phoR genes of Streptomyces coelicolor carried by the cosmid SCD8A and total chromosomal DNA from S. lividans TK24 as template. The PCR fragment was cloned into pIJ2925 cut with SmaI to yield pSG110. A 1,783-bp SmaI DNA fragment carrying the cassette Ωaac (5), conferring resistance to apramycin (Aprar), was cloned into pSG110 cut by NotI internal to phoP and blunt ended with the Klenow fragment to produce pSG111. The BglII fragment carrying the insert of pSG111 was then cloned into the thermosensitive vector pGM160Δ (Table 1) digested by BamHI, yielding pSG112. pSG112 was used to replace the wild-type phoP gene by its disrupted version using a previously described procedure (25).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pIJ2925 | pUC18 derivative | 2, 16 |
| pGM160Δ | pGM160 derivative, obtained by deletion of HindIII of pGM160 fragment carrying aac gene | 25 (gift of J. L. Pernodet) |
| pHP45 Ωaac | E. coli vector carrying Ωaac cassette | 5 |
| pSG100 | pIJ2925 derivative carrying PCR fragment encompassing ppk gene and 1-kb region located upstream of ppk | This study |
| pSG101 | pSG100 with EcoRI site of polylinker filled in with Klenow fragment | This study |
| pSG102 | pSG101 containing newly created EcoRI site | This study |
| pSG103 | Cloning of Ωaac resistance cassette from pHP45 Ωaac into newly created EcoRI site of pSG102 | This study |
| pSG104 | Cloning of insert of pSG103 into pGM160Δ | This study |
| pSG110 | Cloning of 3.2-kb PCR fragment carrying phoR and phoP genes of S. lividans into SmaI site of pIJ2925 | This study |
| pSG111 | Cloning of Ωaac resistance cassette from pHP45 Ωaac into NotI site of pSG110, filled in by Klenow fragment | This study |
| pSG112 | Cloning of insert of pSG111 into pGM160Δ | This study |
TABLE 1.
Sequences of the primers used for PCR amplification of various fragments
| Primer | Sequence | Purpose |
|---|---|---|
| ppk1 | 5′ CTGCACGACGTCGTCGACCACGGC 3′ | Creation of an EcoRI site (underlined) upstream of the ppk −35 promoter region |
| ppkL | 5′ GGGGAATTCTGGAGGGTGGTGGCTGGG 3′ | |
| ppk2 | 5′ GACCATCGAGTTGACCTTGA 3′ | |
| ppkR | 5′ CCAGAATTCCCCCGCTATCCCACACGT 3′ | |
| phoRP5′ | 5′ CTTCGCCATCAGGCGCTGCG 3′ | Cloning of 3.2-kb DNA fragment carrying phoR and phoP |
| phoRP3′ | 5′ TCTCCGCCACCCTGTTCAACAG 3′ | |
| PhoP1 | 5′ TGTCGTACATGCTCCGCAAG 3′ | Amplification of fragment internal to phoP used as probe in Northern blot |
| PhoP2 | 5′ ACGGCTCGAACTTGTAACCC 3′ | |
| AC5′ | 5′ CGACCCACCCACCGGACAA 3′ | Amplification of fragment AC (Fig. 3A) |
| AC3′ | 5′ GAAGGCTGTGGCTCGGTCGGCTT 3′ | |
| AC5′ | 5′ CGACCCACCCACCGGACAA 3′ | Amplification of fragment AB (Fig. 3A) |
| AB3′ | 5′ CCTCTGGAGGGTGGTGGC 3′ | |
| BC5′ | 5′ CCCCGCTATCCCACACGT 3′ | Amplification of fragment BC (Fig. 3A) |
| AC3′ | 5′ GAAGGCTGTGGCTCGGTCGGCTT 3′ |
In order to construct the S. lividans TK24 EcoRI::Ωaacppk mutant strain, the CA-rich region located upstream of the −35 promoter sequence of the ppk gene was displaced from the promoter region by the introduction of an EcoRI site 6 bp upstream of the −35 motif, using the procedure described below. First, the EcoRI site of the polylinker of pSG100 (Table 1), a pIJ2925 derivative carrying a 3.2-kb PCR fragment, encompassing the ppk gene, and a region of 1 kb located upstream of ppk, was filled in with the Klenow fragment, yielding pSG101. Then, the following two PCRs were performed using pSG100 as a matrix. In reaction 1, ppk1 and ppkL, harboring an EcoRI site, were used as primers (Table 2). In reaction 2, ppk2 and ppkR, harboring an EcoRI site, were used as primers. PCR fragment 1 was cleaved by AgeI (located upstream of the CA-rich region) and EcoRI, and PCR fragment 2 was cleaved by EcoRI and StuI (site internal to ppk). Both the resulting 757-bp AgeI-EcoRI fragment and the 872-bp EcoRI-StuI fragment were cloned in pSG101, replacing the original 1,629-bp AgeI-StuI DNA fragment, giving pSG102 containing the newly created EcoRI site. A 1,783-bp EcoRI DNA fragment from pHp45Ωaac (5), carrying the Ωaac cassette conferring resistance to apramycin (G418), was cloned into pSG102 cut by EcoRI, yielding pSG103. The new mutant ppk promoter region was transferred into the S. lividans TK24 chromosome as follows. The 4,720-bp BglII fragment carrying the insert of pSG103 was cloned into the thermosensitive vector pGM160Δ cut by BamHI, yielding pSG104. This plasmid was used to replace the wild-type ppk gene by its modified version using the procedure previously described in reference 25.
In both cases, the chromosomal structure of the wild-type strain and of three Aprar nosiheptide-sensitive (Nosis) colonies which had lost the replicative plasmid was compared in the modified regions by appropriate restriction analysis of chromosomal DNA, Southern blotting, and hybridization of the restricted DNAs with the appropriate probes labeled with [α-32P]dCTP, using the T7 Quick Prime kit (Pharmacia). The PCR conditions used to amplify the various fragments used were as follows: 5 cycles of 60 s at 96°C, 60 s at 54°C, and 280 s at 72°C followed by 25 cycles of 60 s at 96°C, 60 s at 51°C, and 280 s at 72°C in the presence of 10% glycerol, using Turbo Pfu polymerase from Stratagene.
Media and growth conditions.
Spores of the various S. lividans strains were spread on the surface of cellophane disks (Cannings Packaging Limited, Bristol, United Kingdom) laid down on the surface of agar plates of the solid modified minimal medium (see Fig. 1 and 2) and of the rich solid medium R2YE supplemented or not with various concentrations of K2HPO4 (see Fig. 3 to 6) (11). The concentration of Pi in R2YE not supplemented with K2HPO4 is 1 mM (condition of Pi limitation), as determined with a phosphorus assay kit from Sigma Diagnosis. The liquid growth media used were HT, TSB (tryptic soy broth), and YEME (11).
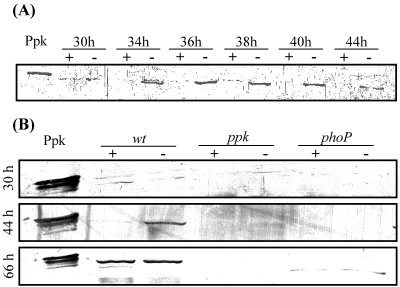
FIG. 1.
(A) Western blot analysis of ppk expression in S. lividans TK24 grown on plates of modified minimal medium under phosphate-sufficient (+; 2.2 mM K2HPO4) or phosphate-limiting (−; 0.44 mM K2HPO4) conditions. Twenty-microgram portions of protein extract from mycelial samples taken after 30, 34, 36, 38, 40, and 44 h of growth were loaded into the wells. (B) Western blot analysis of ppk expression in S. lividans TK24 (wild type [wt]), S. lividans TK24 ppk::Ωhygro (ppk) and S. lividans TK24 phoP::Ωaac (phoP) grown under the conditions described above. Twenty-microgram portions of protein extracts from mycelial samples taken after 30, 44, and 66 h of growth were loaded into the wells. In panels A and B, 60 ng of purified His-tagged Ppk was loaded into the first well as control.
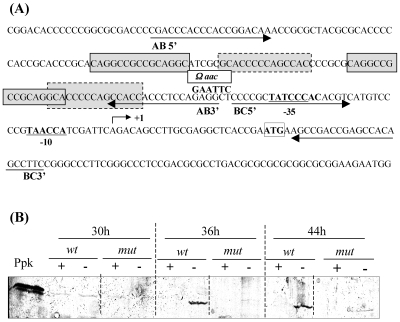
FIG. 2.
(A) Organization of the ppk promoter region. The ppk start codon is enclosed in a box. The −10 and −35 regions of ppk are underlined. The ppk transcriptional start site is indicated by an arrow. The region located upstream of the ppk promoter region was displaced from the promoter region by the insertion of an Ωaac cassette conferring resistance to apramycin at an EcoRI site created by PCR. The positions of the primers used for the amplification of the fragments, AB (131 bp) and BC (91 bp), used in band shift experiments shown in Fig. 3, are indicated by arrows. Some repeated motifs are enclosed in gray boxes. (B) Western blot analysis of ppk expression in S. lividans TK24 (wild type [wt]) and S. lividans TK24 EcoRI-Ωaacppk (mut) grown on modified minimal medium plates under Pi-sufficient (+; 2.2 mM K2HPO4) or Pi-limited (−; 0.44 mM K2HPO4) conditions. Twenty-microgram portions of protein extracts from mycelial samples taken after 30, 36, and 42 h of growth were loaded into the wells. Sixty nanograms of purified His-tagged Ppk was loaded into the first well as control.
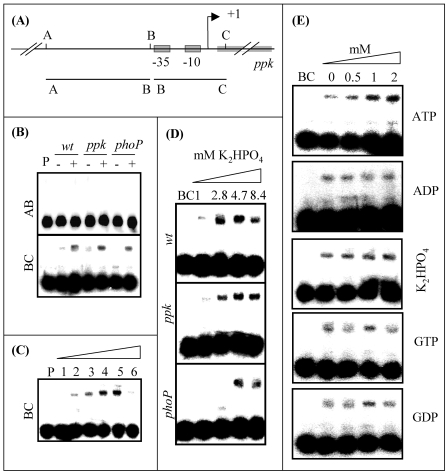
FIG. 3.
(A) Schematic diagram of the ppk promoter region (B) Gel shift experiments carried out with DNA fragments of the ppk gene 5′ region. 32P-labeled PCR fragments AB and BC (0.5 ng/5,000 to 10,000 cpm) were incubated for 15 min at 30°C with 30-μg portions of protein extract of S. lividans TK24 (wild type [wt]), S. lividans TK24 ppk::Ωhygro (ppk), and S. lividans TK24 phoP::Ωaac (phoP) grown on solid R2YE medium under Pi-limited (−; 1 mM) or Pi-sufficient (+; 8.4 mM) conditions. (C) Gel-shift experiments carried out with fragment BC. 32P-labeled PCR fragment BC (0.5 ng/5,000 to 10,000 cpm) was incubated for 15 min at 30°C with 10, 20, 30, 40, or 50 μg of protein extracts (lanes 1 to 5, respectively) prepared from cultures of S. lividans TK24 grown for 44 h on solid R2YE medium containing 8.4 mM KH2PO4. For lane 6, 100 ng of the unlabeled fragment BC was added to 50 μg of protein extract. (D) Gel-shift experiments carried out with fragment BC in the presence of protein extracts prepared from cultures of S. lividans TK24 (wild type), S. lividans TK24 ppk::Ωhygro (ppk mutant) and S. lividans TK24 phoP::Ωaac (phoP mutant) grown for 44 h on solid R2YE medium containing 1, 2.85, 4.7, and 8.4 mM Pi, respectively. 32P-labeled PCR fragment BC (0.5 ng/5,000 to 10,000 cpm) was incubated for 15 min at 30°C with 30 μg of the various protein extracts. Free DNA and protein-bound DNA were separated in a 4% polyacrylamide gel as described in Materials and Methods. (E) Gel shift experiments carried out with 32P-labeled PCR fragment BC (0.5 ng/5,000 to 10,000 cpm) incubated for 15 min at 30°C with 30 μg of protein extract of S. lividans TK24 (wild type) grown on solid R2YE medium proficient in Pi (+; 4.7 mM) in the presence of various concentrations of ATP, ADP, GTP, GDP, or K2HPO4.
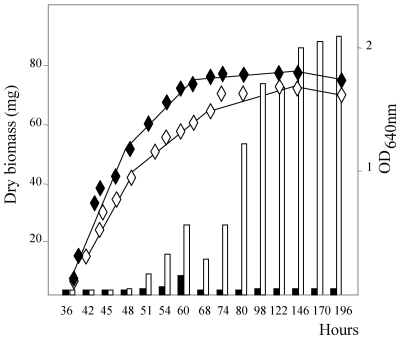
FIG. 6.
Growth curve and blue-pigmented antibiotic production (OD640) for S. lividans TK24 (black lozenges and solid bars) and S. lividans TK24 ppk::Ωhygro (white diamonds and open bars) grown on R2YE medium containing 1 mM phosphate (Pi-limiting conditions). A similar assay was done with the same strains grown on R2YE medium containing 4.7 mM phosphate (Pi-sufficient conditions), and no production of the blue-pigmented antibiotics was detected.
Immunodetection of Ppk by Western blotting.
Twenty micrograms of total protein was loaded in each well and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (BDH, 40% liquid Electran acrylogel 2.6), with a Tris-glycine electrophoresis buffer (25 mM Tris, 250 mM glycine, 0.1% sodium dodecyl sulfate). The gel was transferred to an Immobilon P membrane using a semidry Owl transfer apparatus from Hellmann. Nonspecific binding of the antibodies was prevented by soaking the membrane in TBS buffer (25 mM Tris, 0.8% NaCl, 0.02% KCl, pH 7.4) containing 5% dried skim milk for 1 h at 37°C. The membrane was then incubated overnight at 4°C with rabbit polyclonal Ppk antiserum diluted 1/200 in TBS containing 1% dried skim milk. Immunoreactive bands were revealed using alkaline phosphatase-conjugated anti-rabbit immunoglobulin G antibodies (Promega) diluted to 1/30,000 and the 5-bromo-4-chloro-3-indoxyl-1-phosphate (BCIP)/nitroblue tetrazolium-colored substrate.
Preparation of crude protein extracts.
One hundred milligrams of mycelium of S. lividans TK24 was collected at various times and washed with ice-cold water containing 5 mM EDTA, pH 8. The mycelium was resuspended in 1 ml of sonication buffer (20 mM Tris, pH 7, 5 mM EDTA, containing protease inhibitors from Sigma (final concentrations, benzamidine, 313 μg · ml−1; pepstatin A, 1.36 μg · ml−1; leupeptin, 0.26 μg · ml−1; antipain, 2 μg · ml−1; and chymostatin, 2 μg · ml−1), disrupted by sonication on ice (total time of 30 min with 30 s at amplitude of 22 μm and 100-s pause). Cell debris was then removed by centrifugation for 30 min at 30,000 × g. Protein concentration was quantified with the Bradford reagent (Bio-Rad) using a bovine serum albumin standard curve. Cell extracts for band shift and Western blot experiments were stored at −70°C in 50-μl aliquots.
Gel shift experiments with the 5′ region of the ppk gene.
The DNA fragments AB and BC from the 5′ region of ppk were labeled by PCR using [α-32P]dCTP. Primers AB5′ and AB3′ (Table 2) were used to amplify fragment AB. Primers BC5′ and BC3′ were used to amplify fragment BC. The conditions for the radiolabeled PCR used were as follows: 30 cycles of 60 s at 96°C, 60 s at 57°C, and 30 s at 72°C. Amplification was carried out using the Taq polymerase from Qiagene in the presence of 0.2 mM dATP, dTTP, and dGTP; 0.02 mM unlabeled dCTP; and 40 μCi of [α-32P]dCTP (10 mCi/ml). The conditions of the band shift used were those described by Ausubel et al. (2). 32P-labeled DNA fragments (0.5 ng, 5,000 ± 10,000 cpm) were incubated with 30 μg of cell extract for 15 min at 30°C in a total volume of 15 μl of the binding buffer (BB; 4× BB is 50 mM Tris, pH 7.9, 240 mM KCl, 4 mM EDTA, 4 mM dithiothreitol, and 48% glycerol) with 2 μg of sonicated salmon sperm DNA and 4.5 μg of bovine serum albumin. After incubation, protein-bound DNA and free DNA were separated in nondenaturing polyacrylamide gels (4% acrylamide, 0.05% bisacrylamide, 2.5% glycerol), run in a high-ionic-strength buffer (50 mM Tris, 380 mM glycine, and 2 mM EDTA, pH 8.5) at 4°C. The gels were dried and revealed with a PhosphorImager.
Measurement of cell growth and extraction of polyphosphates.
Polyphosphates were extracted at different time points throughout growth of cultures of S. lividans TK24 (wild type) and S. lividans TK24 ppk::Ωhygro (ppk mutant) on R2YE agar plates containing either 1 mM or 4.7 mM Pi and covered with a cellophane disk. Two different methods were used: the acidic method (19) and the silica gel method (1). In both cases, half of the wet biomass (0.5 to 1 g) obtained from one or several pooled plates was used to estimate growth (dry biomass or protein content) and the other half was used to extract polyphosphates. In the acidic method, 1 g of wet mycelium was extracted with 10 ml of 0.5 N HClO4 at 4°C for 30 min with continuous stirring and then centrifuged at 4°C. The supernatant contained acid-soluble polyphosphates (ASPPs) as well as free Pi and nucleoside phosphate. The latter were removed by absorption on active coal Norit A from Prolabo. The amount of Pi units in the ASPP was determined by the estimation of the difference in the Pi content of the samples before and after the complete hydrolysis of the ASPP, following incubation of the samples at 100°C for 10 min. The concentration of free Pi was determined using a phosphorus assay kit from Sigma Diagnosis. The results obtained from three independent experiments were highly reproducible.
Polyphosphates were also extracted from 36-, 56-, and 72-h cultures of S. lividans TK24 (wild type) and S. lividans TK24 ppk::Ωhygro (ppk mutant) grown on R2YE agar plates containing 1 mM Pi and covered with a cellophane disk, using a modification of the silica gel method described in reference 1. The collected mycelial samples (0.2 g) were spun down in a 1.5-ml Eppendorf tube, frozen in liquid nitrogen, stored at −70°C, and treated as follows: 0.5 ml of lysis buffer (4 M guanidine isothiocyanate in 50 mM Tris-HCl, pH 7.0) prewarmed at 95°C was added to each tube. Mycelial samples were homogenized, incubated for 2 to 5 min in a dry bath at 95°C, and sonicated briefly in order to obtain a clear solution. A 10-μl sample was removed from each tube for protein estimation with the Bradford reagent. This estimation is critical as it allows the standardization of the polyP deposits. Then 30 μl of 10% SDS, 0.5 ml of 95% ethanol, and 10 μl of the silica suspension were added to each tube. The silica suspension was prepared as follows: 0.1 g of silica (Sigma S-5631) was suspended in 1 ml of phosphate-buffered saline buffer (0.5% KH2PO4, pH 7) and allowed to settle for 2 h at 4°C in the dark. The supernatant was discarded, and the operation was repeated twice. After the last wash, the silica was spun down and the pellet was resuspended in 1 ml of lysis buffer. This suspension was stored at 4°C in the dark. After addition of the silica suspension, the tubes were homogenized by agitation and spun down briefly to pellet the silica. The supernatant was discarded, and the silica pellet was resuspended in 0.5 ml of wash buffer (5 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM EDTA, 50% ethanol) prewarmed at 37°C. This operation was repeated twice. After the last centrifugation, polyPs were eluted from the silica pellet with 50 μl of elution buffer (50 mM Tris-HCl, pH 8.0) at 95°C for 2 min; recovery of polyPs was complete with two additional elutions. A sample of the extracted products, corresponding to 20 μg of protein, was incubated for 4 h at 37°C in the presence of 3 U/μl RNase-free DNase I from Boehringer, 1.25 μg/μl RNase A from Roche, 0.25 μg/μl pronase from Sigma, and 2.5 U/μl CIPA from Roche. After addition of loading dye (10× loading dye is 50% sucrose; 0.125% bromophenol blue; 450 mM Tris borate, pH 8.3, and 13.5 mM EDTA), polyP samples were run overnight at 300 V and at maximal current intensity on a denaturing polyacrylamide gel (8% acrylamide; 0.3% bisacrylamide, 8 M urea) with a Tris-borate running buffer (90 mM Tris-borate, pH 8.3, 2.7 mM EDTA). The gel was prerun for 1 h at 300 V and maximal current intensity before the loading of the samples. The samples were made to enter quickly into the gel by applying a voltage of 1,000 V and maximal current intensity to the gel for 20 min. Polyphosphates of various sizes from Sigma were used as size standards. After migration, the gel was stained for 1 h in solution A (0.05% toluidine blue, 25% methanol, 5% glycerol), destained for 2 to 3 h in solution B (25% methanol; 5% glycerol), and then dried.
Assay of blue-pigmented antibiotics.
In order to assay the production of blue-pigmented antibiotics excreted by S. lividans TK24 and S. lividans TK24 ppk::Ωhygro grown on R2YE containing 1 mM phosphate (Pi limitation) or 4.7 mM phosphate (Pi sufficiency), these strains were grown on a cellophane disc as described above. At different time points during growth, the cellophane was lifted and three sterile agar cylinders of the growth medium were taken using an inverted Pasteur pipette and incubated at 4°C for at least 24 h in 1 ml of distilled water in order to allow the diffusion of the blue pigment. The latter was assayed at an optical density of 630 nm (OD630) using a Beckmann spectrophotometer.
RESULTS
PhoR/PhoP two-component system positively regulates ppk expression under Pi-limiting conditions.
In order to assess regulatory features of ppk expression in response to Pi availability, protein extracts of S. lividans grown on the solid minimal medium MM containing 2.2 mM (Pi sufficiency) or 0.44 mM (Pi limitation) K2HPO4 were prepared as described in Materials and Methods. The presence of Ppk in these extracts was revealed using polyclonal antibodies raised against the purified His-tagged Ppk. The amount of Ppk detected was consistently much higher in cultures grown in a Pi-limited medium than in those grown in a Pi-proficient medium (Fig. 1A). However, ppk expression was detected in the wild-type strain grown in the Pi-sufficient medium for 66 h (Fig. 2A), suggesting that no Pi was left in the growth medium at this late time and that Pi exhaustion was responsible for the triggering of ppk expression. In contrast, neither carbon nor nitrogen limitation led to the triggering of ppk expression (data not shown). These results indicate that the expression of ppk in S. lividans TK24 is triggered when Pi becomes limiting in the growth medium, as previously demonstrated in other bacteria (8, 9).
In bacteria, genes whose expression is induced under conditions of Pi limitation constitute the pho regulon and their expression is usually under the direct or indirect control of two-component systems, PhoR/PhoP in Bacillus subtilis (15) and PhoR/PhoB (32) in Escherichia coli. In order to test whether ppk of S. lividans was a member of the pho regulon, we made a knockout mutant of the phoP gene of the two-component system PhoR/PhoP of S. lividans (28) and assessed ppk expression in this genetic background. The Western blot shown in Fig. 1B clearly demonstrates that in the phoP mutant strain, the induction of ppk expression under Pi limitation is strongly reduced. This result indicates that ppk expression is, either directly or indirectly, under the positive control of PhoR/PhoP. ppk thus belongs to the pho regulon. However, the detection of a small amount of Ppk in the phoP mutant after 66 h of growth (conditions of severe Pi limitation) suggests that the expression of ppk might be controlled by another regulator in addition to PhoR/PhoP.
Since the siege of the positive regulation by PhoP-like regulators (pho boxes) is usually located upstream of the promoter region of the regulated genes, the region located upstream of the −35 promoter sequence of ppk was displaced from the −35 promoter sequence by insertion of an Ωaac cassette (5) into an EcoRI site created by PCR 6 bp upstream of the −35 sequence (Fig. 2A). The Western blot of Fig. 2B clearly shows that, in this mutant as in the phoP mutant, the expression of ppk is strongly reduced under conditions of Pi limitation compared to the wild-type strain. This effect is not due to the cassette itself, since when the latter was inserted upstream of the negatively regulated sblA promoter, for example, it enhanced rather than reduced sblA transcription (unpublished data). The reduction of ppk expression in the mutant (mut) strain of Fig. 2B thus indicates that sequences located upstream of the −35 promoter region play a positive role in the regulation of ppk expression by Pi limitation. However, no putative consensus Pho boxes, G(G/T)TCAYYYR(G/C)G, as proposed by Sola-Landa et al. (29) were found in this region, suggesting that the positive regulation of ppk expression by PhoR/PhoP might be indirect, involving another regulatory component under the control of PhoR/PhoP. Furthermore, the detection of a very small amount of Ppk after 44 h of growth, under Pi limitation (but not under Pi sufficiency), suggested that this altered promoter region might still be weakly regulated and that some cis-acting regulatory elements are present downstream of the created EcoRI site.
ppk promoter region is shifted by a putative repressor under the condition of Pi sufficiency.
In order to test whether some putative regulatory proteins were interacting with the ppk promoter region, band-shifting experiments were set up with fragments AB and BC originating from the ppk promoter region (Fig. 3A). Fragment AB encompasses the region located upstream of the −35 promoter sequence that was shown to play a positive role in the regulation of ppk expression. Fragment BC encompasses the ppk promoter region, from 65 bp upstream to 23 bp downstream of the ATG start codon (Fig. 2A). These two fragments were incubated in the presence of crude protein extracts prepared from the wild-type strain and the ppk and phoP mutant strains, grown under conditions of Pi proficiency or limitation, for 44 h. The results are shown in Fig. 3B. Surprisingly, no band shifting was seen under any conditions in any of the three strains with fragment AB, whereas protein extracts prepared from cultures of the three strains grown under conditions of Pi proficiency gave strong band shifting with fragment BC (Fig. 3B). The intensity of the shifting of fragment BC increased with increasing amounts of protein extract, whereas it was strongly reduced upon the addition of 100 ng of cold fragment BC (Fig. 3C), indicating that the binding was specific. These results demonstrated that a binding site for a retarding component is present on fragment BC. Since ppk is poorly expressed when Pi is in excess, this retarding component is likely to be a transcriptional repressor. Furthermore, the amount of fragment BC shifted increases in the three strains when the concentration of Pi in the growth medium increases from 1 to 2.8 mM (Fig. 3D), suggesting that this putative repressor might be more abundant, and/or its affinity for fragment BC, might be higher, when Pi is sufficient than when Pi is limiting in the growth medium. This observation suggests repression of ppk expression under conditions of Pi excess and relief of this repression under Pi limitation. It is also noticeable that the phoP mutant has to be grown in the presence of a higher Pi concentration than the other two strains in order to produce cell extracts able to delay fragment BC (Fig. 3D). This observation is consistent with the knowledge that the phoP mutant transports Pi less efficiently than the wild-type strain does (28).
Shifting of the ppk promoter region is stimulated by the presence of ATP.
Reports in the literature indicate that the intracellular concentration of ATP is in the millimolar range, and this level increases and decreases when the concentration of Pi in the growth medium increases and decreases, respectively (22, 23). Therefore, we examined whether the addition of millimolar amounts of ATP, ADP, GTP, GDP, and Pi had an effect on the ability of the retarding component to bind to fragment BC in vitro. Figure 3E clearly shows that addition of GTP, ADP, GDP, and Pi had no effect on the putative repressor shifting ability, whereas the addition of ATP had a clear stimulatory effect. These observations suggest that the increase in the shifting intensity observed when the concentration of Pi in the growth medium increases (Fig. 3D) might be at least partly due to an increase in the intracellular concentration of ATP.
Intracellular content of short acid-soluble polyphosphate in S. lividans and its ppk mutant grown in the rich R2YE medium proficient in Pi.
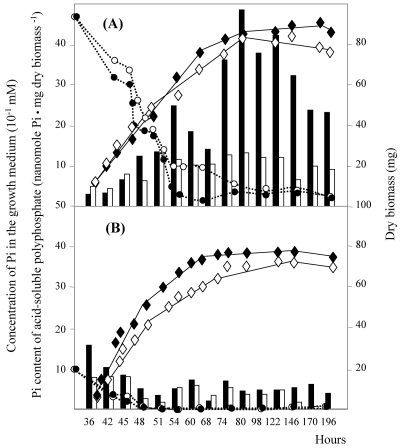
We attempted to prepare ASPPs from cultures of S. lividans TK24 and of the ppk mutant grown on the solid minimal medium, MM, used for the regulatory studies, but under these conditions, these polymers were hardly detectable. Thus, ASPPs were prepared at different time points from cultures grown on the solid carbon- and nitrogen-rich R2YE medium, containing 4.7 mM Pi (Pi excess), as described by Kulaev et al. (19). The average chain length of ASPPs is thought to be approximately 30 to 70 residues, and the Pi content was determined as described by Kulakovskaya et al. (20). Results shown in Fig. 4A revealed that, in the wild-type strain, ASPP content rose sharply when the bacteria reach the stationary phase around the 74th h of growth, whereas this sharp increase in the ASPP content was not observed in the ppk mutant strain. Similar levels of ASPPs were present at the beginning of the growth in both strains, suggesting that Ppk was not involved in their biosynthesis. The origin of the biosynthesis of these polymers is totally unknown (27). Under these conditions of Pi sufficiency, no production of blue-pigmented antibiotics was detected in either of the two strains (data not shown).
FIG. 4.
Growth curves for S. lividans TK24 (black diamonds) and S. lividans TK24 ppk::Ωhygro (white diamonds), phosphate contents of extracted acid-soluble polyphosphates from S. lividans TK24 (solid bars) and S. lividans TK24 ppk::Ωhygro (open bars), and concentrations of KH2PO4 in the growth medium throughout growth of S. lividans TK24 (black circles) and S. lividans TK24 ppk::Ωhygro (white circles) (A) Pi limited (R2YE medium containing 1 mM Pi). (B) Pi sufficient (R2YE medium containing 4.7 mM Pi).
Intracellular content of short acid-soluble polyphosphate in S. lividans and its ppk mutant grown in the rich R2YE medium limited in Pi.
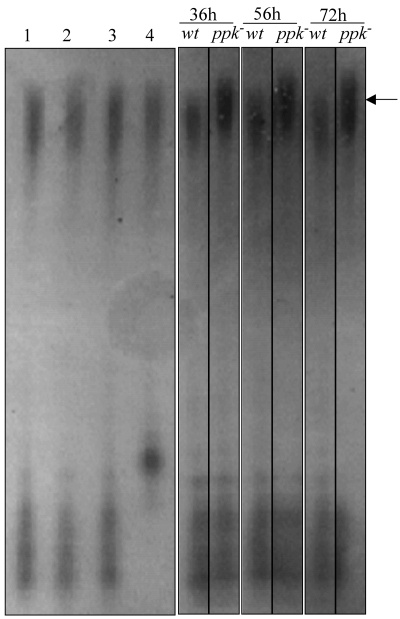
ASPPs were also prepared from the wild-type strain and the ppk mutant strain of S. lividans grown on the rich medium R2YE containing 1 mM Pi. In this medium, which initially had a low Pi content, Pi was rapidly exhausted. Under these conditions (Pi scarcity), the ASPP content was much lower than it was under Pi-sufficient conditions and no significant difference was noticeable between the two strains (Fig. 4B). Since we thought that the acidic conditions of extraction of these small amounts of polyphosphate might be potentially damaging for these short polymers, we extracted them with the more gentle silica gel method adapted from reference 1 and described in Materials and Methods. Purified polyphosphates were then run on a polyacrylamide gel stained with toluidine blue for visualization and qualitative analysis. The results of Fig. 5 show that polyphosphates were reproducibly longer and more abundant in the ppk mutant strain than in the wild-type strain. Under these conditions of Pi scarcity, the production of the blue-pigmented antibiotic is detectable from the 54th h of growth in the ppk mutant strain but not in the wild-type strain (Fig. 6).
FIG. 5.
Polyphosphates (indicated by an arrow) were purified on a silica gel as described in Materials and Methods from lawns of S. lividans TK24 (wild type [wt]) and S. lividans TK24 ppk::Ωhygro (ppk−) grown for 36, 56, and 72 h on solid R2YE medium under Pi-limiting conditions (1 mM Pi). Polyphosphates purified from 36-h lawns of S. lividans TK24 either were not treated (lane 1) or were incubated for 4 h at 37°C with 0.25 μg/μl pronase (lane 2), 3 U/μl of RNase-free DNase I (lane 3), 1.25 μg/μl of RNase A (lane 3), and 2.5 U/μl of CIPA (lane 4). All polyphosphate samples were electrophoresed overnight on a 60-cm-long denaturating polyacrylamide gel stained with toluidine blue.
DISCUSSION
In this study, we demonstrated that when S. lividans TK24 is grown under conditions of Pi excess, the expression of ppk is weak but detectable. Our band-shifting experiments (Fig. 3) suggest that, under these conditions, the expression of ppk might be repressed by a regulator interacting with sequences located in the ppk promoter region. Figure 3E suggests that the binding of this putative repressor is stimulated by ATP. ATP might thus be considered as a corepressor of ppk expression. Since the intensity of the band shifting of the fragment BC increases when the concentration of the Pi in the growth medium increases from 1 to 2.8 mM (Fig. 3D), this increase could be at least partly attributed to an augmentation of the intracellular ATP concentration that is known to increase in parallel with the concentration of Pi in the growth medium (23) (22). Conversely, when Pi becomes limiting in the growth medium, the intracellular ATP concentration is expected to drop, leading to a partial relief of ppk repression. Under these conditions of Pi limitation, the concomitant partial relief of ppk repression and its PhoR/PhoP-dependent induction might result in the strong expression of ppk observed in Fig. 1A. The absence of any obvious pho box in the ppk region suggests that the positive regulation of ppk expression by PhoR/PhoP might be indirect. Ppk expression might be induced by an activator under the positive control of Pho/PhoP. This putative activator, not detected in band-shifting experiments, might loosely interact with the region located upstream of the ppk promoter region that was shown to play a positive role in the regulation of ppk expression (Fig. 2B). However, at this stage of the work, we cannot exclude that PhoR/PhoP is also down regulating the expression of the putative repressor of ppk expression, contributing to the alleviation of ppk repression.
In order to assess the role of Ppk in polyP metabolism, we prepared short ASPPs from S. lividans TK24 and its ppk mutant grown in R2YE medium proficient (4.7 mM) or limited (1 mM) in Pi. As expected, the ASPP content of the cell was higher in both strains when cells were grown in a Pi-proficient medium than in a Pi-limited medium (Fig. 4). When the cells are grown in a Pi-proficient medium, polyPs are likely to be longer and more abundant than when the cells are grown in a Pi-limited medium. When S. lividans TK24 was grown in the R2YE medium with sufficient Pi (4.7 mM), the ASPP content of the cell was rather low during active growth and rose sharply upon entry into the stationary phase (Fig. 4A), as previously described for E. coli (18, 26, 27). This sharp increase in the ASPP content of the cell, upon entry into the stationary phase, was not observed in the ppk mutant strain. In the wild-type strain, this abrupt increase in the ASPP content of the cell could either result from the “de novo” synthesis of short ASPPs or from the degradation of longer polyP into shorter ones. If these ASPPs indeed result from a “de novo” synthesis, this synthesis necessitates the concomitant presence of abundant ATP and of Ppk acting as a polyphosphate kinase. The concentration of ATP within the cell is known to result principally from the balance of production by the central metabolic pathways (glycolysis, Krebs cycle, and pentose phosphate pathways) coupled to the respiratory chain and consumption by anabolic reactions necessary for cell growth. During active growth, the ATP produced might be consumed for anabolic reactions. At the stationary phase, when growth slows down or stops, a surplus of ATP might result from continuation of active metabolism combined with reduced consumption by anabolic reactions. This ATP surplus might be used for a Ppk-dependent ASPP biosynthesis (Fig. 4A). However, at this stage of the work, we cannot exclude that the accumulation of ASPPs upon entry into the stationary phase in the wild-type strain does not result from polyP synthesis but from the degradation of long polyPs into shorter ones. Storage of phosphate and energy in long-chain polymers is known to be important for long-term survival in the stationary phase (27). Free Pi could easily be made available again from these polymers by the action of polyphosphate phosphatases (6), and ATP could be regenerated from ADP and polyP by the action of the polyphosphate kinase (7, 30). Both of these processes are leading to the shortening of the polyP. At the stationary phase, the relative poverty in short ASPPs of the ppk mutant compared to the wild-type strain, grown in a Pi-proficient medium, might simply indicate a slower rate of degradation of long polyPs into short ASPPs, due to the absence of Ppk. These polyPs would simply remain too long to be purified by the method used to prepare the short ASPPs. As a matter of fact, when the wild-type strain and the ppk mutant strain of S. lividans are grown on the R2YE medium with a low concentration of Pi (1 mM), the polyPs prepared by the silica gel method (they are slightly longer than the ASPPs) were found to be reproducibly longer and more abundant in the ppk mutant than in the wild type (Fig. 5). These results suggest that in extreme Pi scarcity, when the ATP/ADP ratio within the cell is low (22, 23), Ppk might act, as previously demonstrated in vitro (7), as an NDPK, regenerating ATP from ADP and polyphosphates. Since, the ppk mutant strain is thought to be devoid of an important ATP-regenerating enzyme, the polyphosphates are not used to regenerate ATP in that strain, explaining why they were found to be longer and more abundant in the ppk mutant than in the wild-type strain. As previously mentioned, a similar process might explain the apparent poverty in short ASPPs noticed in the ppk mutant strain grown in a Pi-proficient medium, upon entry into the stationary phase.
Our results suggest that when the Pi from the growth medium is exhausted, the more or less abundant polyP reserves made (depending on the initial concentration in Pi of the growth medium) are likely to be used to provide free Pi as well as to regenerate ATP in a Ppk-dependent manner.
When the initial concentration of Pi of the growth medium is low, the polyP reserves (Fig. 4) and the energetic charge of the cell are low (22, 23). Under these conditions, the ppk mutant is likely to have a even weaker energetic charge than the wild-type strain since it is lacking an important ATP-regenerating enzyme. This important deficit in the energetic charge might be the real trigger for antibiotic biosynthesis as previously suggested by Martin et al. (23) and Lounes et al. (22). Thus, we propose that, in order to compensate for the deficit in its energetic balance and to rapidly regenerate the necessary ATP from ADP, the ppk mutant, under conditions of Pi scarcity, somehow triggers a strong activation of its central metabolic pathways that are the main ATP-producing routes within the cell. This stimulation requires enhanced uptake of carbon and phosphate sources, the fuels of metabolism. We indeed demonstrated that the expression of the two-component system PhoR/PhoP governing the expression of genes of the Pho regulon (a phosphate provider) is greatly enhanced in the ppk mutant strain (10). Similarly, the rate of glucose uptake is also enhanced in that strain (unpublished results). The result of this stimulation should be production of carbon skeletons, reduced cofactors (NADH and NADPH), and ATP. Since antibiotic production starts when growth slows down or stops, conditions under which these key compounds are not being consumed for anabolism, the latter might thus be available for antibiotic biosynthesis. We think that all these adaptive procedures in response to Pi scarcity occur in the wild-type strain but are greatly amplified in the ppk mutant strain. Experiments are currently in progress to test this novel model of the triggering of antibiotic biosynthesis by the energetic balance of the cell in Streptomyces species.
Acknowledgments
This work was supported by the CNRS and the University Paris XI.
We thank P. R. Jensen, J. L. Pernodet, M. Guérineau, F. Francou, and E. Darbon-Rongère for stimulating discussions and are most grateful to B. Holland for critical reading and correction of the manuscript.
REFERENCES
- 1.Ault-Riché, D., C. D. Fraley, C.-M. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Avignone Rossa, C., J. White, A. Kuiper, P. W. Postma, M. Bibb, and M. J. Teixeira de Mattos. 2002. Carbon flux distribution in antibiotic-producing chemostat cultures of Streptomyces lividans. Metab. Eng. 4:138-150. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. [DOI] [PubMed] [Google Scholar]
- 5.Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 6.Bolesch, D. G., and J. D. Keasling. 2000. Polyphosphate binding and chain length recognition of Escherichia coli exopolyphosphatase. J. Biol. Chem. 275:33814-33819. [DOI] [PubMed] [Google Scholar]
- 7.Chouayekh, H., and M. J. Virolle. 2002. The polyphosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans. Mol. Microbiol. 43:919-930. [DOI] [PubMed] [Google Scholar]
- 8.Gavigan, J. A., L. M. Marshall, and A. D. Dobson. 1999. Regulation of polyphosphate kinase gene expression in Acinetobacter baumannii 252. Microbiology 145:2931-2937. [DOI] [PubMed] [Google Scholar]
- 9.Geißdorfer, W., A. Ratajczak, and W. Hillen. 1998. Transcription of ppk from Acinetobacter sp. strain ADP1, encoding a putative polyphosphate kinase, is induced by phosphate starvation. Appl. Environ. Microbiol. 64:896-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghorbel, S., J. Kormanec, A. Artus, and M. J. Virolle. 2006. Transcriptional studies and regulatory interactions between the phoR/phoP operon and the phoU, mtpA, and ppk genes of Streptomyces lividans TK24. J. Bacteriol. 188:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 12.Hopwood, D. A., K. F. Chater, and M. J. Bibb. 1995. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Bio/Technology 28:65-102. [DOI] [PubMed] [Google Scholar]
- 13.Hopwood, D. A., T. Kieser, H. M. Wright, and M. J. Bibb. 1983. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J. Gen. Microbiol. 129:2257-2269. [DOI] [PubMed] [Google Scholar]
- 14.Huang, J., J. Shi, V. Molle, B. Sohlberg, D. Weaver, M. J. Bibb, N. Karoonuthaisiri, C. J. Lih, C. M. Kao, M. J. Buttner, and S. N. Cohen. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276-1287. [DOI] [PubMed] [Google Scholar]
- 15.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 16.Janssen, G. R., and M. J. Bibb. 1993. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene 124:133-134. [DOI] [PubMed] [Google Scholar]
- 17.Kang, S. G., W. Jin, M. Bibb, and K. J. Lee. 1998. Actinorhodin and undecylprodigiosin production in wild-type and relA mutant strains of Streptomyces coelicolor A3(2) grown in continuous culture. FEMS Microbiol. Lett. 168:221-226. [DOI] [PubMed] [Google Scholar]
- 18.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 19.Kulaev, I. S., V. M. Vagabov, and T. V. Kulakovskaya. 2004. The biochemistry of inorganic polyphosphates. J. Wiley, Chichester, United Kingdom.
- 20.Kulakovskaya, T. V., N. A. Andreeva, A. V. Karpov, I. A. Sidorov, and I. S. Kulaev. 1999. Hydrolysis of tripolyphosphate by purified exopolyphosphatase from Saccharomyces cerevisiae cytosol: kinetic model. Biochemistry (Moscow) 64:990-993. [PubMed] [Google Scholar]
- 21.Liras, P., J. A. Asturias, and J. F. Martin. 1990. Phosphate control sequences involved in transcriptional regulation of antibiotic biosynthesis. Trends Biotechnol. 8:184-189. [DOI] [PubMed] [Google Scholar]
- 22.Lounes, A., A. Lebrihi, C. Benslimane, G. Lefebvre, and P. Germain. 1996. Regulation of spiramycin synthesis in Streptomyces ambofaciens: effects of glucose and inorganic phosphate. Appl. Microbiol. Biotechnol. 45:204-211. [DOI] [PubMed] [Google Scholar]
- 23.Martin, J. F., P. Liras, and A. L. Demain. 1978. ATP and adenylate energy charge during phosphate-mediated control of antibiotic synthesis. Biochem. Biophys. Res. Commun. 83:822-828. [DOI] [PubMed] [Google Scholar]
- 24.McDowall, K. J., A. Thamchaipenet, and I. S. Hunter. 1999. Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding sites of the OmpR family. J. Bacteriol. 181:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muth, G., B. Nussbaumer, W. Wohlleben, and A. Pühler. 1989. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol. Gen. Genet. 219:341-348. [Google Scholar]
- 26.Rao, N. N., and A. Kornberg. 1999. Inorganic polyphosphate regulates responses of Escherichia coli to nutritional stringencies, environmental stresses and survival in the stationary phase. Prog. Mol. Subcell. Biol. 23:183-195. [DOI] [PubMed] [Google Scholar]
- 27.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sola-Landa, A., R. S. Moura, and J. F. Martin. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 100:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sola-Landa, A., A. Rodriguez-Garcia, E. Franco-Dominguez, and J. F. Martin. 2005. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 56:1373-1385. [DOI] [PubMed] [Google Scholar]
- 30.Tzeng, C.-M., and A. Kornberg. 2000. The multiple activities pf polyphosphate kinase of Escherichia coli and their subunit structure determined by radiation target analysis. J. Biol. Chem. 275:3977-3983. [DOI] [PubMed] [Google Scholar]
- 31.Voelker, F., and S. Altaba. 2001. Nitrogen source governs the patterns of growth and pristinamycin production in ‘Streptomyces pristinaespiralis.’ Microbiology 147:2447-2459. [DOI] [PubMed] [Google Scholar]
- 32.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon. American Society for Microbiology, Washington, D.C.