Abstract
The obligate intracytoplasmic pathogen Rickettsia prowazekii relies on the transport of many essential compounds from the cytoplasm of the eukaryotic host cell in lieu of de novo synthesis, an evolutionary outcome undoubtedly linked to obligatory growth in this metabolite-replete niche. The paradigm for the study of rickettsial transport systems is the ATP/ADP translocase Tlc1, which exchanges bacterial ADP for host cell ATP as a source of energy, rather than as a source of adenylate. Interestingly, the R. prowazekii genome encodes four open reading frames that are highly homologous to the well-characterized ATP/ADP translocase Tlc1. Therefore, by annotation, the R. prowazekii genome encodes a total of five ATP/ADP translocases: Tlc1, Tlc2, Tlc3, Tlc4, and Tlc5. We have confirmed by quantitative reverse transcriptase PCR that mRNAs corresponding to all five tlc homologues are expressed in R. prowazekii growing in L-929 cells and have shown their heterologous protein expression in Escherichia coli, suggesting that none of the tlc genes are pseudogenes in the process of evolutionary meltdown. However, we demonstrate by heterologous expression in E. coli that only Tlc1 functions as an ATP/ADP transporter. A survey of nucleotides and nucleosides has determined that Tlc4 transports CTP, UTP, and GDP. Intriguingly, although GTP was not transported by Tlc4, it was an inhibitor of CTP and UTP uptake and demonstrated a Ki similar to that of GDP. In addition, we demonstrate that Tlc5 transports GTP and GDP. We postulate that Tlc4 and Tlc5 serve the primary function of maintaining intracellular pools of nucleotides for rickettsial nucleic acid biosynthesis and do not provide the cell with nucleoside triphosphates as an energy source, as is the case for Tlc1. Although heterologous expression of Tlc2 and Tlc3 was observed in E. coli, we were unable to identify substrates for these proteins.
Rickettsia prowazekii is the etiological agent of epidemic typhus in humans and is designated as a select agent based on its potential use as an agent of bioterrorism. R. prowazekii is an obligate intracellular bacterium and grows only within the cytoplasm of a eukaryotic host cell, unbounded by a host-derived membrane vesicle. As a consequence of growth in this nutrient-rich niche, R. prowazekii has evolved several unique transport systems specific for large, charged metabolites, such as UDP-glucose, S-adenosylmethionine, ATP, NAD, UMP, GMP, and AMP (11, 12, 45, 52, 54, 55). The ability to transport substrates that are present in the host cell cytosol and rarely available in the extracellular milieu has likely contributed to the evolution of the small-sized rickettsial genome (834 open reading frames [ORFs]); many of the de novo biosynthetic pathways characteristic of free-living bacteria are no longer present in rickettsiae (8). The ATP/ADP translocase Tlc1 is the most thoroughly characterized of these transport systems (1-5, 13, 14, 16, 18, 27, 36, 50, 53, 56). Tlc1 is an obligate-exchange transport system that catalyzes the equilibration of bacterial ATP/ADP with host cell ATP/ADP and thus is a means to acquire high-energy phosphate, not adenylate. The rickettsial Tlc1 belongs to the family of 12 transmembrane-containing ATP/ADP antiporters in the classification scheme of Paulsen et al. (35), in which five members of this family (Tlc1, Tlc2, Tlc3, Tlc4, and Tlc5) were annotated for R. prowazekii (9). The five tlc homologues are present and appear intact in all spotted fever group (R. rickettsii, R. akari, R. conorii, R. montanensis, R. sibirica, and R. felis) and typhus group (R. prowazekii and R. typhi) rickettsiae sequenced to date. Two Tlc homologues have been annotated for all sequenced species of obligate intracellular parasitic chlamydiae, and transport activities have been characterized for Chlamydia trachomatis and Chlamydophila pneumoniae (23, 42, 44). Tlc1 homologues have also been identified in the endosymbiotic (nonpathogenic) bacteria Holospora obscura and Caedibacter caryophilus and in activities characterized by heterologous expression in and assay of Escherichia coli (16, 29). However, the Tlc1 family of transporters is not ubiquitous within obligate intracellular organisms; it is absent from the genomes of Coxiella, Anaplasma, and Ehrlichia spp., which grow inside host-derived vesicles. Finally, the Tlc1 transport system has been identified as a nucleus-encoded protein in plant plastids (26, 31, 32, 40), a curiosity considering that Tlc1 homologues are conspicuously absent in cyanobacteria, the postulated ancestors of these plant organelles.
Several recently published phylogenetic analyses have discussed the origins of the nonmitochondrial ATP/ADP transporters and postulate that the progenitor ATP/ADP translocase protein originated in a chlamydial ancestor and was acquired by the rickettsial ancestor through lateral transfer (6, 21, 39). Intriguingly, pathogenic chlamydiae (in the family Chlamydiaceae) differ from rickettsiae in that they encode two, not five, Tlc homologues. C. trachomatis nucleotide phosphate transporter 1 (Npt1) is similar to the rickettsial Tlc1 in both amino acid sequence (68% similarity) and function (ATP/ADP exchange), whereas Npt2 transports all four ribonucleoside triphosphates (44). Thus, it is postulated that rickettsiae evolved to possess five tlc gene copies subsequent to acquiring the ancestral Tlc. However, this model is complicated by the recent elucidation of the genome of “Candidatus Protochlamydia amoebophila” (15) strain UWE25, an endosymbiont of Acanthamoeba spp., which revealed the presence of five ORFs annotated as nucleotide transporters (Ntt1 through Ntt5) (24). Ntt1 is a bona fide ATP/ADP obligate-exchange system (39), and Ntt4 is a novel NAD+ transport system (22). Phylogenetic analysis does not indicate a close evolutionary link between the five parachlamydial Ntt proteins and the five rickettsial Tlc proteins. The close evolutionary relationship between chlamydiae and parachlamydiae (21, 39) and the identification of five translocase homologues in UWE25 raise an interesting question regarding the evolution of the nonmitochondrial ATP/ADP transporter gene copy number in chlamydiae and rickettsiae.
Although rickettsiae and mitochondria are thought to share a common ancestor (6, 9, 33, 41, 47, 49), there is no significant homology between mitochondrial and nonmitochondrial ATP/ADP transport proteins (57). This divergence could be linked to the altruistic nature of mitochondria, which provide energy to cells, versus the parasitic nature of rickettsiae. It appears that nonmitochondrial ATP/ADP transport systems have evolved in response to the cytoplasmic growth environment and are widespread in organisms and some ATP-requiring organelles that obligatorily occupy this niche but are absent in free-living or facultative intracellular bacteria.
Considering that intracellular bacteria tend to have single copies of genes that are present in multiple copies in the genomes of other bacteria (7, 10, 34, 43), it seems unlikely that the Tlc1 through Tlc5 homologues possess redundant functions, as the current annotation suggests. We proffer three possible models that will be tested here: (i) all five Tlc homologues are ATP/ADP obligate-exchange transport systems that have been maintained because they are differentially expressed in response to changing environmental conditions; (ii) all five Tlc homologues have different substrate specificities and fulfill different metabolic requirements (and may or may not be differentially regulated); and (iii) Tlc1 is the only active protein, and the other four homologues are pseudogenes in the process of evolutionary meltdown. All models intermediate to these are implied by discussing the three extremes presented above. We have determined by using quantitative reverse transcriptase PCR (RT-PCR) that the mRNAs of all four previously uncharacterized members of the tlc family are expressed in L-929 mouse fibroblast-grown R. prowazekii. Subsequent cloning and heterologous expression of these putative rickettsial transporters in E. coli has allowed us to determine that only Tlc1 is an obligate-exchange ATP/ADP transporter. We demonstrate that Tlc4 transports CTP, UTP, and GDP and that GTP is a competitive inhibitor of Tlc4 despite the fact that it is not a transportable substrate. We also demonstrate that Tlc5 transports GTP and GDP. Of the compounds tested, we were unable to identify substrates for Tlc2 and Tlc3.
MATERIALS AND METHODS
Growth media and chemicals.
E. coli were routinely grown at 37°C with aeration in LB broth or M9 glucose (0.2%) minimal medium supplemented with 100 μg/ml of ampicillin for plasmid maintenance. Chemicals were purchased from Sigma, enzymes were purchased from New England Biolabs, and radiolabeled nucleotides were purchased from PerkinElmer or MP Biomedicals. All PCR primers were synthesized by IDT.
Cloning of the tlc2, tlc3, tlc4, and tlc5 genes.
The R. prowazekii genes encoding Tlc2 through Tlc5 were amplified from genomic DNA by PCR under standard conditions (38) and cloned into pBluescript II KS+ (Stratagene), and their DNA sequences were verified. The PCR primers included an engineered NdeI site at the start codon and a BamHI site downstream of the stop codon to facilitate insertion into the pET-11a (no histidine tag) and pET-16b (N-terminal decahistidine tag) expression vectors (Novagen). In addition, the tlc2 gene was also engineered to contain a C-terminal decahistidine tag and cloned into pET-11a. The decahistidine tags allowed for protein purification by immobilized metal affinity chromatography. Cloning of the tlc1 gene was previously described (18, 27). The pET plasmids expressing the tlc1 through tlc5 genes were transformed into the BL21(DE3) or the C41 strain of E. coli [a derivative of BL21(DE3)] (30) as indicated below. All experiments were performed on fresh transformants that were used for no longer than 2 weeks because of observed inconsistencies in the transport activities of strains subsequent to being stocked in LB containing 10% glycerol at −80°C.
R. prowazekii infection of L-929 mouse fibroblasts and RNA isolation.
L-929 mouse fibroblasts were infected with R. prowazekii (Madrid E strain) at a multiplicity of infection of 50 in a suspension of Hanks balanced salt solution supplemented with 5 mM monopotassium glutamate and 0.1% gelatin at 34°C for 1 h. The infected L-929 cells were collected by centrifugation, washed three times with and suspended in Eagle minimal essential medium supplemented with 10% newborn bovine serum, transferred to cell culture flasks (185 cm2), and allowed to adhere. The number of rickettsiae per infected cell was monitored over time by microscopy (20). Flasks were grown at 34°C with 5% CO2. After 10 h of growth, emetine (1 μg/ml) was added to inhibit further L-929 cell replication (a manipulation that has no adverse affect on the growth of rickettsiae). After 48 h of incubation, L-929 cells (with approximately 200 to 300 rickettsiae per infected cell) were released by trypsin treatment and collected by centrifugation. Trypsinization and all subsequent steps were performed in the presence of a 20% (vol/vol) concentration of the DNA/RNA Protect reagent from Sierra Diagnostics to preserve nucleic acid integrity. The rickettsia-infected L-929 cell pellet was suspended in a solution of phosphate-buffered sucrose (53), and rickettsiae were released by ballistic shearing with 1-mm glass beads. L-929 cell debris was removed and rickettsiae isolated by differential centrifugation. The purified rickettsiae were suspended in 1 ml of RNAWIZ (Ambion), and RNA was extracted as per the manufacturer's instructions, with the following modification. In lieu of the final RNA precipitation step, a 0.56 volume of 100% ethanol was slowly added to the RNA-containing solution so that it could be added directly to an RNeasy mini column (QIAGEN). Once the RNA was bound to the RNeasy column, QIAGEN RNase-free DNase I was added and allowed to incubate for 15 min at room temperature as per the manufacturer's directions. The column was then washed according to the manufacturer's instructions, and RNA was eluted twice for a total of 60 μl. Total RNA (L-929 cell and rickettsial) quality and quantity were determined with an Agilent bioanalyzer.
Quantitative RT-PCR.
The relative amount of mRNA corresponding to each of the tlc1 through tlc5 genes was analyzed using real-time RT-PCR performed in a Cepheid SmartCycler. Real-time RT-PCR analysis was internally controlled and allowed for a relative comparison of all five tlc mRNAs present in the extracted total RNA. All five tlc cDNAs were amplified in a single RT reaction containing total RNA as the template and the five specific RT primers (Table 1). In all RT reaction mixtures, RNA loading was normalized such that 250 ng of 16S rRNA (based on the Agilent bioanalysis) was added. All RT reactions were performed using SuperScript II (Invitrogen) as per the manufacturer's directions. Upon completion of cDNA synthesis, treatment with RNase H (37°C for 5 min) digested the RNA, leaving only the tlc-specific cDNA.
TABLE 1.
Quantitative RT-PCR primer sequences and reaction conditionsa
| Gene and primer | Primer sequence | PCR conditions |
|---|---|---|
| tlc1 | ||
| RT primer | 5′-CTATTTTTCATTTTTATTTACCAAAACTTGATACTC-3′ | 95°C, 15 s |
| Q-PCR forward primer | 5′-GTACTACCTTCTGCTGTAATTGCTATG-3′ | 55°C, 15 s |
| Q-PCR reverse primer | 5′-GATCAGGGTGGACTAAATCAGGATATG-3′ | 72°C, 15 s |
| tlc2 | ||
| RT primer | 5′-TTAACGTTTTATCATTTTATTATAATTTTGTGATAAAAC-3′ | 95°C, 15 s |
| Q-PCR forward primer | 5′-ATGGGTGAGTTATGGCCTGTTATAG-3′ | 56°C, 15 s |
| Q-PCR reverse primer | 5′-CAGTGCCTGAGAAGAGCAAATTAGT-3′ | 72°C, 15 s |
| tlc3 | ||
| RT primer | 5′-TTATTGACACAGCTTTATATATTCCTTATTTA-3′ | 95°C, 15 s |
| Q-PCR forward primer | 5′-GGTAGTAAATGGAGTTATGCGCTAATG-3′ | 56°C, 15 s |
| Q-PCR reverse primer | 5′-GATATTACCAACCATCCCAAGAACAGG-3′ | 72°C, 15 s |
| tlc4 | ||
| RT primer | 5′-TTATTTTTGAGAAAATTTAATAGAATTTTTATATTC-3′ | 95°C, 15 s |
| Q-PCR forward primer | 5′-GTTAGAAGAATGGGCTGGTTTACTTCAG-3′ | 55°C, 15 s |
| Q-PCR reverse primer | 5′-GTTATAGCAACTAAAGCAGGATCGG-3′ | 72°C, 15 s |
| tlc5 | ||
| RT primer | 5′-TCAGGCTATTTTTTGATATTCAAAATATATTTTTC-3′ | 95°C, 15 s |
| Q-PCR forward primer | 5′-GTATGCATAATTGGTTTGTAGCCGCAG-3′ | 58°C, 15 s |
| Q-PCR reverse primer | 5′-CTGAATACCGCCAATAGAAACAGCAAG-3′ | 72°C, 15 s |
Q-PCR, quantitative PCR.
The RNase-digested reaction mixture containing the tlc-specific cDNAs was serially diluted, and 2 μl of each dilution was used as the template for real-time PCR (conditions are listed in Table 1) using Epicentre's TAQurate Green real-time PCR master mix as per the manufacturer's directions. All reactions were run in duplicate using pairs of nested primers specific for each tlc cDNA that were designed to yield products of approximately 150 bp. All reactions started with an initial cycle of 95°C for 120 s, followed by 40 cycles under the conditions described in Table 1. The fourfold-serial-dilution series served as an internal control, providing a measure of reaction efficiency for each primer set. In theory, a PCR with a one-quarter amount of template should be two cycles behind if the amplification is linear and the products are doubled with every cycle. When the cycle threshold (CT) is plotted against the logarithm of template concentration, a linear regression generates a theoretical slope of 3.3 for a PCR running at maximum efficiency. This technique allowed us to make relevant comparisons of the levels of all five tlc cDNAs within the extracted RNA samples. In all cases, control RT reactions containing no SuperScript II were run to confirm the absence of contaminating DNA. These samples showed reaction profiles similar to those of primer controls where no template was added.
RPAs.
As an independent confirmation of the presence of mRNA from the five rickettsial translocase genes, RNase protection assays (RPAs) were performed on RNA isolated from infected L-929 cells, as previously described (14). The RNA probes required for RPAs were generated by in vitro transcription using fragments of each tlc gene amplified by PCR and cloned into the EcoRV site of pBluescript II KS+.
Transport assays for uptake of nucleotides.
Plasmids (pET11a derivatives) engineered to express each of the native tlc1 through tlc5 genes (no affinity tags) were introduced into the C41 strain of E. coli [a derivative of BL21(DE3)] (30) by electroporation. Overnight cultures grown in LB broth supplemented with 100 μg/ml of ampicillin were diluted (1:200) into 50 ml of fresh medium and grown to an optical density at 600 nm of 0.4, followed by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce the expression of tlc genes. After 1 h of induction, bacterial cells were collected by centrifugation, washed in 60 ml of 50 mM potassium phosphate buffer (KPi; pH 7.5), and suspended in 2.5 ml of KPi (all on ice). Cells were diluted 10-fold into the reaction buffer (at 23°C) to initiate the uptake assay. The reaction buffer contained KPi and the various nucleotides (concentrations are indicated in the figure legends) with 1 to 2 μCi/ml of an α-32P-labeled nucleotide. Transport activity was measured by filtering 0.1-ml aliquots on wetted membrane filters (HAWP02500; Millipore), washing the filters once with 5 ml of KPi, and measuring incorporated radioactivity using liquid scintillation spectrometry as previously described (51). Radiolabeled nucleotide monophosphate and diphosphate compounds were synthesized using the α-32P-containing triphosphate and, when necessary, purified by polyethyleneimine chromatography (37). As a negative control, we verified that none of the labeled substrates tested in this study were transported by a vector-controlled E. coli strain.
Competitive inhibition analysis of Tlc4 and Tlc5.
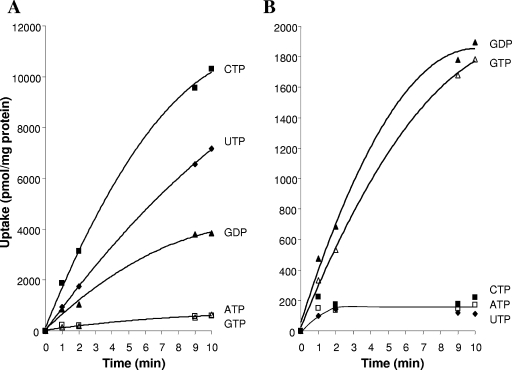
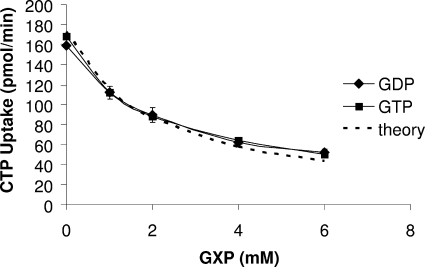
Based on the initial screen, we determined that Tlc4 transported CTP, UTP, and GDP and that Tlc5 transported GTP and GDP (see Fig. 2). The protocol described above was used for competitive inhibition assays to survey other compounds as potential inhibitors of the uptake activity of Tlc4 and Tlc5. For Tlc4, CTP (at 0.2 mM) and UTP (at 0.6 mM) were tested as substrates, with potential inhibitors added at a 10-fold excess to the substrate. For Tlc5, GTP (at 0.1 mM) was tested as a substrate, with potential inhibitors added at a 10-fold excess. The cells were exposed to the inhibitor and the substrate simultaneously. The data presented in Table 3 representing the average inhibition of the 1-, 2-, and 3-min time points were combined from at least two experiments (the 2- and 3-min data were converted to data per minute prior to taking the overall average of all three data points). The effects of GTP and GDP as inhibitors of CTP uptake by Tlc4 were subsequently tested using various concentrations (see the legend to Fig. 3).
FIG. 2.
Assay of NXP transport by Tlc4 (A) and Tlc5 (B). The tlc4 and tlc5 genes were cloned into the pET-11a expression vector and introduced into the C41 strain of E. coli by electroporation. Cultures were grown, induced, and assayed for the transport of ATP, CTP, UTP, GTP, and GDP as described in Materials and Methods. All NXPs were added at a concentration of 1 mM (with 1 to 2 μCi/ml of the appropriate α-32P-labeled nucleotide phosphate), and uptake reactions were initiated by the addition of cells. Symbols: ATP, open squares; CTP, filled squares; UTP, filled diamonds; GTP, open triangles; and GDP, filled triangles.
TABLE 3.
Specificity of nucleotide transport by Tlc4 and Tlc5 measured in the presence of putative competitive inhibitorsc
| Putative inhibitora | Tlc4
|
Tlc5 GTP uptake
|
||||
|---|---|---|---|---|---|---|
| UTP uptake
|
CTP uptake
|
|||||
| Avg relative to control value (%)b | ±SD (%) | Avg relative to control value (%) | ±SD (%) | Avg relative to control value (%) | ±SD (%) | |
| None | 100 | 100 | 100 | |||
| ATP | 63 | 6 | 116 | 18 | 100 | 11 |
| ADP | 83 | 3 | 92 | 4 | 126 | 20 |
| AMP | 122 | 10 | 146 | 12 | 100 | 6 |
| Adenosine | 141 | 4 | 152 | 31 | 101 | 1 |
| dATP | 89 | 4 | 116 | 9 | 114 | 7 |
| CTP | 17 | 0 | 20 | 2 | 105 | 3 |
| CDP | 58 | 4 | 93 | 15 | 113 | 10 |
| CMP | 113 | 2 | 135 | 27 | 144 | 45 |
| Cytidine | 139 | 4 | 159 | 31 | 104 | 7 |
| dCTP | 111 | 1 | 133 | 26 | 118 | 11 |
| GTP | 35 | 3 | 67 | 12 | 18 | 1 |
| GDP | 31 | 10 | 52 | 4 | 23 | 2 |
| GMP | 90 | 16 | 131 | 28 | 69 | 4 |
| Guanosine | 115 | 3 | 160 | 38 | 114 | 4 |
| dGTP | 82 | 5 | 113 | 26 | 63 | 4 |
| UTP | 30 | 4 | 56 | 17 | 123 | 1 |
| UDP | 70 | 4 | 91 | 23 | 121 | 6 |
| UMP | 136 | 3 | 150 | 56 | 122 | 22 |
| Uridine | 160 | 12 | 160 | 41 | 114 | 14 |
| dUTP | 98 | 7 | 141 | 31 | 116 | 11 |
As described in Materials and Methods, substrates for Tlc4 were present at 0.6 mM (UTP) and 0.2 mM (CTP). The substrate for Tlc5 was present at 0.1 mM. All inhibitors were added at a 10-fold excess.
Average of the results from the 1-, 2-, and 3-min time points after normalization, taken from at least duplicate experiments.
Underlined numbers denote compounds that provided modest repression (>30 %). Boldface numbers denote compounds that provided substantial repression (>50 %).
FIG. 3.
Effect of increasing concentrations of GTP and GDP on the transport of CTP by Tlc4. The tlc4 gene was cloned into the pET-11a expression vector and introduced into the C41 strain of E. coli by electroporation. Cultures were grown, induced, and assayed for transport as described in Materials and Methods. CTP was added as a substrate at a concentration of 0.2 mM (with 1 to 2 μCi/ml of [α32P]CTP), and transport was assayed in the presence of increasing concentrations of GTP and GDP (0, 1, 2, 4, and 6 mM). Symbols: GTP, filled squares; GDP, filled diamonds. The dotted line represents the theoretical curve for an inhibitor with a Ki of 1 mM. GXP, GDP or GTP.
In vivo [35S]methionine labeling and protein purification.
The pET-16b derivatives of Tlc1 through Tlc5 (including a pET-11a derivative expressing Tlc2 with a C-terminal decahistidine tag) were introduced into BL21(DE3) or C41 strains of E. coli, and protein was labeled with [35S]methionine as described in reference 48. Subsequently, cells were washed, suspended in KPi containing 1× protease inhibitor cocktail (Roche), and broken by sonication. Unbroken cells and debris were removed by centrifugation (7,000 × g, 15 min, 4°C) and membranes collected by centrifugation at 150,000 × g for 90 min at 4°C. Membranes were suspended in 0.5 ml of buffer A (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1× protease inhibitor cocktail) containing 20 mM imidazole and solubilized (30 min on ice) by the addition of n-dodecylmaltoside (DDM) to a final concentration of 2%. Insoluble material was removed by centrifugation at 150,000 × g for 15 min at 4°C. The supernatant was mixed with Ni-nitriloacetic acid resin slurry (QIAGEN) for 2 h at 4°C. The resin was washed twice with 1 bed volume of buffer A containing 100 mM imidazole and 0.2% DDM before being eluted with buffer A containing 300 mM imidazole and 0.2% DDM. Total and purified membrane proteins were suspended in sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and exposure on a Cyclone phosphorimager (Perkin Elmer).
RESULTS AND DISCUSSION
Sequence and hydropathy analysis of Tlc1 through Tlc5.
RP053, RP377, RP477, RP500, and RP739, corresponding to tlc1, tlc2, tlc3, tlc4, and tlc5, respectively, are discrete cistrons with no apparent operonal organization (9). The rickettsial Tlc1 through Tlc5 proteins are similar to all known nonmitochondrial nucleotide transport proteins and are predicted to possess 12 putative transmembrane domains (8, 35, 57). It should be noted that some hydropathicity algorithms predicted 11 transmembrane regions for Tlc1, -3, -4, and -5; however, 12 transmembrane regions was the predominant number, and this has been independently confirmed for Tlc1 (4, 36). The five Tlc proteins range from 498 to 512 amino acids, with predicted molecular masses of ∼57 to 58 kDa. Hydropathy plots (25, 28, 46) demonstrated the substantial similarity between the five Tlc proteins. The Tlc2 through Tlc5 proteins share sequence identity with Tlc1 ranging from 34 to 45% when compared by pairwise alignment within a species (9). In contrast, there is approximately 90% identity between corresponding pairs of Tlc proteins from rickettsial species (e.g., Tlc2 from R. prowazekii compared to Tlc2 from R. typhi), a conservation of amino acid sequence that strongly indicates that these genes code for functional proteins and are not pseudogenes in the process of genetic meltdown. Further, these data suggest that the divergence of the typhus group and spotted fever group rickettsial species occurred after the quadruplication of the ancestral tlc gene (6).
All five R. prowazekii translocase mRNAs are expressed in L-929 fibroblast-grown R. prowazekii, and all five Tlc proteins can be expressed using a heterologous E. coli system.
Are the tlc genes expressed at different levels in rickettsiae grown in L-929 host cells, or are one or more of them pseudogenes in the process of evolutionary meltdown? The R. prowazekii genome contains an unusually large amount of noncoding DNA and several pseudogenes (8), including the metK gene, which has become redundant in function due to the acquisition of an S-adenosylmethionine transport protein (17, 45). Therefore, the amount of mRNA corresponding to each of the four R. prowazekii tlc homologues was compared to that of tlc1 to indicate whether these proteins are expressed (acknowledging the caveat that posttranscriptional regulation may also affect the levels of these proteins) and to determine if they are differentially regulated. Total RNA was isolated from R. prowazekii-infected L-929 mouse fibroblasts and assayed by real-time, quantitative RT-PCR and RPA as described in Materials and Methods. The results presented in Table 2 show data from two biological replicates demonstrating that all five tlc mRNAs are expressed in R. prowazekii grown in L-929 cells under the conditions tested here. Control RT reactions confirmed the absence of contaminating DNA. RNase protection assays confirmed the expression of all five tlc mRNAs (data not shown). Although all five tlc mRNAs are expressed in L-929 cell-grown rickettsiae, it is apparent that the level of expression of each tlc homologue varied from that of tlc1 up to sevenfold (Table 2). Confirmation of mRNA expression by RT-PCR is indirect evidence that the four tlc homologues are not pseudogenes that have accumulated promoter mutations that silenced expression. Also consider the bioinformatics data presented above showing very similar molecular masses for all five Tlc proteins (confirmed empirically [see below]) and gene conservation between rickettsial species. If these were pseudogenes under neutral selection pressure, one could expect (especially in the AT-rich rickettsial genome) the acquisition of stop codons that would truncate the ORF. We are currently investigating the observed differential regulation of the five tlc genes.
TABLE 2.
Quantitative RT-PCR analysis of mRNA levels in tlc1 through tlc5
| Gene | Reaction efficiencya
|
 b b
|
Avg ΔCT from that of tlc1c | Expression relative to that of tlc1d | ||
|---|---|---|---|---|---|---|
| Sample 1 | Sample 2 | Sample 1 | Sample 2 | |||
| tlc1 | 3.1 | 3.6 | 25.5 | 25.7 | 1.0 | |
| tlc2 | 2.9 | 3.3 | 27.1 | 27.9 | −1.9 | 0.3 |
| tlc3 | 3.5 | 4.0 | 27.5 | 27.3 | −1.8 | 0.3 |
| tlc4 | 3.4 | 3.9 | 23.5 | 22.2 | 2.8 | 6.7 |
| tlc5 | 3.1 | 3.9 | 25.8 | 25.3 | 0.1 | 1.1 |
As described in Materials and Methods, quantitative PCR was performed in duplicate on serial dilutions of the tlc-specific cDNA, and the CT was plotted against the log template concentration from two biological replicates. Linear regression analysis determined the reaction efficiency, where a theoretical slope of 3.3 is expected.
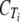 , the ordinate intercept of the linear regression.
, the ordinate intercept of the linear regression.
The 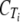 values from the two samples were averaged and used to calculate the difference in CT (ΔCT) from that of tlc1.
values from the two samples were averaged and used to calculate the difference in CT (ΔCT) from that of tlc1.
The ΔCT values were used to determine expression relative to that of tlc1 where change (n = fold) was 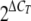 .
.
In our efforts to determine the substrate specificity of each of the rickettsial Tlc homologues, we employed the Novagen series of inducible pET expression vectors. To first verify expression, we cloned each tlc gene into pET-16b, thus incorporating an N-terminal decahistidine affinity tag. The resulting plasmids were introduced into the E. coli BL21(DE3) expression strain, which was grown, induced, and protein labeled with [35S]methionine as described in Materials and Methods. Each Tlc protein was purified from the membrane fraction using immobilized metal affinity chromatography and resolved by denaturing gel electrophoresis. Our analyses verified that heterologous expression of Tlc3, Tlc4, and Tlc5 gave full-length proteins with molecular weights similar to that of Tlc1 (3, 18; data not shown). Analysis of Tlc2 revealed that expression in the BL21(DE3) strain of E. coli resulted in the isolation of the full-length protein and an additional, truncated protein, an indication that proteolytic cleavage had occurred. Incorporation of a C-terminal decahistidine tag into the Tlc2 coding sequence resulted in the observation of only full-length protein. The loss of the truncated form suggests that the processing occurred in the carboxyl end of the Tlc2 protein when expressed in BL21(DE3). When Tlc2 was assayed in the C41 strain of E. coli, only full-length Tlc2 protein was isolated (regardless of the position of the decahistidine tag). Therefore, activity assays presented below were performed with the C41 strain to avoid any problems associated with proteolytic processing of the proteins.
Only Tlc1 is an ATP/ADP transport system.
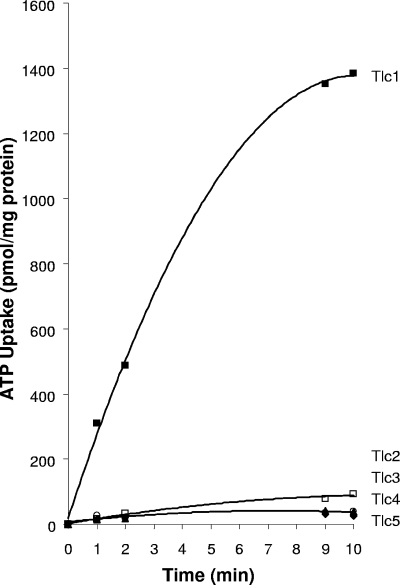
R. prowazekii Tlc1 operates as a very specific obligate-exchange antiport system allowing concurrent exchange of ADP and ATP between bacterium and host cytoplasms as a source of energy (53). To determine the substrate specificities of Tlc2 through Tlc5, the tlc genes cloned into the pET-11a expression vector (with no affinity tags) were expressed and activity was determined using filtration uptake assays. Strikingly, none of the four putative transporters demonstrated significant uptake of ATP compared to that of Tlc1 expressed in E. coli (Fig. 1).
FIG. 1.
Assay of ATP transport by the R. prowazekii Tlc homologues. The tlc1 through tlc5 homologues were cloned into the pET-11a expression vector and introduced into the C41 strain of E. coli by electroporation. Cultures were grown, induced, and assayed for transport of ATP as described in Materials and Methods. ATP was added at a concentration of 0.05 mM (with 1 to 2 μCi/ml of [α-32P]ATP), and uptake reactions were initiated by the addition of cells. Symbols: Tlc1, filled squares; Tlc2, filled diamonds; Tlc3, filled triangles; Tlc4, open squares; and Tlc5, open circles.
Tlc4 transports UTP, CTP, and GDP.
Considering our previous studies demonstrating the transport of other nucleotides and nucleotide-containing compounds by isolated rickettsiae (11, 12, 52, 54, 55), the four Tlc homologues were tested for uptake of other nucleotide substrates. Tlc4 was able to transport CTP, UTP, and GDP (with very similar maximum rates) when expressed in E. coli, and the amount of uptake of each nucleotide corresponded to an estimate of the affinity of the substrate for Tlc4, with CTP being bound most avidly (0.1 mM), followed by UTP (0.3 mM) and GDP (0.6 mM) (Fig. 2A). Other deoxynucleoside triphosphate (dNTP), nucleoside, diphosphate, and monophosphate derivatives were tested as competitive inhibitors of UTP and CTP uptake by Tlc4, and ATP, UDP, and CDP gave only modest inhibition of UTP transport and even less inhibition of the higher-affinity substrate CTP (Table 3). Surprisingly, although GTP was not transported by Tlc4 (Fig. 2A), it was a competitive inhibitor (Fig. 3, and Table 3). Both the nontransportable GTP and the transportable GDP were equally effective as competitive inhibitors of CTP uptake by Tlc4 (Fig. 3). Because Tlc4 has a higher affinity for CTP and UTP than for GTP, the inhibition by and lack of transport of GTP is not due to it binding so avidly that it cannot be easily released from the carrier, as has been suggested for thiodigalactoside transport via the lactose permease (19, 58). These data raise some interesting questions about the mechanism of substrate recognition and release by Tlc4. The fact that UDP and CDP were poor inhibitors of Tlc4 activity relative to UTP and CTP (Table 3) indicates that the Tlc4 transporter is able to discriminate between substrates, with a preference for the triphosphate. However, although both GTP and GDP are substrates of Tlc4, only the diphosphate is efficiently transported. This could indicate that the translocation of GTP by Tlc4 is extremely low relative to that of GDP even though both are able to occupy the same binding site (i.e., they display competitive inhibition).
Tlc5 transports GTP and GDP.
Tlc5 is specific for GTP and GDP (Fig. 2B). As shown in Table 3, the only compounds that demonstrated substantial inhibition of GTP transport when present at a 10-fold excess were GTP and GDP, though dGTP and GMP provided modest inhibition. These data indicate that this system discriminates on the bases of the type of nucleobase (only G), the type of sugar (ribose, not deoxyribose), and the number of phosphates present in the substrate. It is interesting to note the relative affinities of Tlc4 and Tlc5 for GDP. Tlc4, a transporter with a broad substrate specificity range, has a low affinity for GDP (0.6 mM), whereas the more selective Tlc5 has a higher affinity (0.1 mM).
Tlc2 and Tlc3 do not appear to be nucleotide transporters.
Despite our testing a broad range of potential substrates (ATP, ADP, AMP, CTP, CDP, CMP, GTP, GDP, GMP, UTP, and UMP), no transportable substrate was observed for E. coli cells expressing either Tlc2 or Tlc3. The argument made above (mRNA expression, similar molecular weights, and conservation of sequence among all species of rickettsiae) suggests that Tlc2 and Tlc3 have not lost their function and are not in the process of evolutionary meltdown. It remains possible that they require ancillary, trans-activating factors for proper function or insertion into the membrane that are not present in E. coli, thus abrogating activity. Our radiolabeling and purification analyses demonstrated that there were no problems with insertion of Tlc proteins into E. coli membranes, but we cannot comment on whether these proteins adopt a native conformation. Finally, it is possible (and in our view, likely) that the appropriate substrate was not tested.
Concluding remarks.
The mechanism of action of Tlc4 and Tlc5 (recall that Tlc1 is an obligate-exchange antiporter) and their energetic requirements remain unknown. We have used polyethyleneimine cellulose chromatography and phosphorimage analysis to analyze cell extracts after incubation with radiolabeled nucleotides and have determined that these substrates are rapidly metabolized by the E. coli cells to trichloroacetic acid-insoluble material, most likely RNA (data not shown). This rapid metabolism of NTPs during the transport assays has hindered analysis of the energetic requirements of Tlc4 and Tlc5. Unfortunately, our use of the T7 expression system in these experiments precludes the addition of rifampin to block incorporation and allow accumulation of the NTPs in the E. coli cytosol. We are currently working on this problem.
Our demonstration that Tlc4 expressed in E. coli mediates the uptake of CTP is puzzling considering previous studies in which intact rickettsiae purified from the yolk sacs of embryonated chicken eggs did not incorporate cytidine nucleotides into RNA (54). The R. prowazekii genome encodes and expresses the enzyme, PurC, that synthesizes CTP from UTP, and therefore, these organisms do not need to transport cytidine nucleotides when uridylates, which are incorporated by rickettsiae under these conditions, are present (9, 54). Furthermore, previous work demonstrated that the nucleoside monophosphates are preferred by purified, intact rickettsiae over the corresponding NTPs based on their incorporation into RNA (54).
In conclusion, we have demonstrated that, of the five R. prowazekii genes annotated as ATP/ADP translocases, only Tlc1 is an ATP/ADP obligate-exchange antiporter that transports ATP as a source of energy. We have identified substrates for two of the remaining four Tlc homologues. When heterologously expressed in E. coli, Tlc4 transports CTP, UTP, and GDP, whereas Tlc5 is specific for GTP and GDP. These results indicate a role for Tlc4 and Tlc5 in facilitating the accumulation of NTPs across the bacterial membrane to maintain adequate nucleotide pools in the rickettsial cytosol (where, except for PurC, the appropriate biosynthetic enzymes are lacking). These results demonstrate that Tlc4 and Tlc5 are not pseudogenes and suggest that the similarly expressed Tlc2 and Tlc3 have activities yet to be defined.
Acknowledgments
This work was supported by Public Health Service grant AI-15035 from the National Institute of Allergy and Infectious Diseases to H.H.W.
We thank Rosemary Roberts, Mary Patrick, Robin Daugherty, Andrew Woodard, and David Clark for expert technical assistance.
REFERENCES
- 1.Alexeyev, M. F., R. A. Roberts, R. M. Daugherty, J. P. Audia, and H. H. Winkler. 2004. Cysteine-scanning mutagenesis and thiol modification of the Rickettsia prowazekii ATP/ADP translocase: evidence that transmembrane regions I and II, but not III, are structural components of the aqueous translocation channel. Biochemistry 43:6995-7002. [DOI] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F., and H. H. Winkler. 2002. Complete replacement of basic amino acid residues with cysteines in Rickettsia prowazekii ATP/ADP translocase. Biochim. Biophys. Acta 1565:136-142. [DOI] [PubMed] [Google Scholar]
- 3.Alexeyev, M. F., and H. H. Winkler. 1999. Gene synthesis, bacterial expression and purification of the Rickettsia prowazekii ATP/ADP translocase. Biochim. Biophys. Acta 1419:299-306. [DOI] [PubMed] [Google Scholar]
- 4.Alexeyev, M. F., and H. H. Winkler. 1999. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase reported by novel dual pho-lac reporters. J. Mol. Biol. 285:1503-1513. [DOI] [PubMed] [Google Scholar]
- 5.Alexeyev, M. F., and H. H. Winkler. 2002. Transposable dual reporters for studying the structure-function relationships in membrane proteins: permissive sites in R. prowazekii ATP/ADP translocase. Biochemistry 41:406-414. [DOI] [PubMed] [Google Scholar]
- 6.Amiri, H., O. Karlberg, and S. G. Andersson. 2003. Deep origin of plastid/parasite ATP/ADP translocases. J. Mol. Evol. 56:137-150. [DOI] [PubMed] [Google Scholar]
- 7.Andersson, S. G. E., and C. G. Kurland. 1995. Genomic evolution drives the evolution of the translation system. Biochem. Cell Biol. 73:775-787. [DOI] [PubMed] [Google Scholar]
- 8.Andersson, S. G. E., and C. G. Kurland. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263-268. [DOI] [PubMed] [Google Scholar]
- 9.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. M. Alsmark, R. M. Podowdki, A. K. Naslund, A.-S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-143. [DOI] [PubMed] [Google Scholar]
- 10.Andersson, S. G. E., A. Zomorodipour, H. H. Winkler, and C. G. Kurland. 1995. Unusual organization of the rRNA genes in Rickettsia prowazekii. J. Bacteriol. 177:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkinson, W. H., and H. H. Winkler. 1989. Permeability of Rickettsia prowazekii to NAD. J. Bacteriol. 171:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkinson, W. H., and H. H. Winkler. 1985. Transport of AMP by Rickettsia prowazekii. J. Bacteriol. 161:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Audia, J. P., R. A. W. Roberts, and H. H. Winkler. 2006. Cysteine-scanning mutagenesis and thiol modification of the Rickettsia prowazekii ATP/ADP translocase: characterization of TMs IV-VII and IX-XII and their accessibility to the aqueous translocation pathway. Biochemistry 45:2648-2656. [DOI] [PubMed] [Google Scholar]
- 14.Cai, J., and H. H. Winkler. 1997. Transcriptional regulation of the GLTA and TLC genes in Rickettsia prowazekii growing in a respiration-deficient host cell. Acta Virol. 41:285-288. [PubMed] [Google Scholar]
- 15.Collingro, A., E. R. Toenshoff, M. W. Taylor, T. R. Fritsche, M. Wagner, and M. Horn. 2005. ‘Candidatus Protochlamydia amoebophila’, an endosymbiont of Acanthamoeba spp. Int. J. Syst. Evol. Microbiol. 55:1863-1866. [DOI] [PubMed] [Google Scholar]
- 16.Daugherty, R. M., N. Linka, J. P. Audia, C. Urbany, H. E. Neuhaus, and H. H. Winkler. 2004. The nucleotide transporter of Caedibacter caryophilus exhibits an extended substrate spectrum compared to the analogous ATP/ADP translocase of Rickettsia prowazekii. J. Bacteriol. 186:3262-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driskell, L. O., A. M. Tucker, H. H. Winkler, and D. O. Wood. 2005. Rickettsial metK-encoded methionine adenosyltransferase expression in an Escherichia coli metK deletion strain. J. Bacteriol. 187:5719-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunbar, S. A., and H. H. Winkler. 1997. Increased and controlled expression of the Rickettsia prowazekii ATP/ADP translocase and analysis of cysteine-less mutant translocase. Microbiology 143:3661-3669. [DOI] [PubMed] [Google Scholar]
- 19.Fox, C. F., and E. P. Kennedy. 1965. Specific labeling and partial purification of the M protein, a component of the β-galactoside transport system of Escherichia coli. Proc. Natl. Acad. Sci. USA 54:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gimenez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 21.Greub, G., and D. Raoult. 2003. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl. Environ. Microbiol. 69:5530-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haferkamp, I., S. Schmitz-Esser, N. Linka, C. Urbany, A. Collingro, M. Wagner, M. Horn, and H. E. Neuhaus. 2004. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature 432:622-625. [DOI] [PubMed] [Google Scholar]
- 23.Hatch, T. P., E. Al-Hossainy, and J. A. Silverman. 1982. Adenine nucleotide and lysine transport in Chlamydia psittaci. J. Bacteriol. 150:662-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horn, M., A. Collingro, S. Schmitz-Esser, C. L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G. J. Nyakatura, M. Droege, D. Frishman, T. Rattei, H. W. Mewes, and M. Wagner. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728-730. [DOI] [PubMed] [Google Scholar]
- 25.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 26.Kampfenkel, K., T. Möhlmann, O. Batz, M. Van Montagu, D. Inzé, and H. E. Neuhaus. 1995. Molecular characterization of an Arabidopsis thaliana cDNA encoding a novel putative adenylate translocator of higher plants. FEBS Lett. 374:351-355. [DOI] [PubMed] [Google Scholar]
- 27.Krause, D. C., H. H. Winkler, and D. O. Wood. 1985. Cloning and expression of the Rickettsia prowazekii ADP/ATP translocator in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 29.Linka, N., H. Hurka, B. F. Lang, G. Burger, H. H. Winkler, C. Stamme, C. Urbany, I. Seil, J. Kusch, and H. E. Neuhaus. 2003. Phylogenetic relationships of non-mitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene 306:27-35. [DOI] [PubMed] [Google Scholar]
- 30.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 31.Möhlmann, T., J. Tjaden, C. Schwöppe, H. H. Winkler, K. Kampfenkel, and H. E. Neuhaus. 1998. Occurrence of two plastidic ATP/ADP transporters in Arabidopsis thaliana L—molecular characterisation and comparative structural analysis of homologous ATP/ADP translocators from plastids and Rickettsia prowazekii. Eur. J. Biochem. 252:353-359. [DOI] [PubMed] [Google Scholar]
- 32.Neuhaus, H. E., G. Henrichs, and R. Scheibe. 1993. Characterization of glucose-6-phosphate incorporation into starch by isolated intact cauliflower-bud plastids. Plant Physiol. 101:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang, H., and H. H. Winkler. 1993. Copy number of the 16S rRNA gene in Rickettsia prowazekii. J. Bacteriol. 175:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 36.Plano, G. V., and H. H. Winkler. 1991. Identification and initial topological analysis of the Rickettsia prowazekii ATP/ADP translocase. J. Bacteriol. 173:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randerath, K., and E. Randerath. 1967. Thin-layer separation methods for nucleic acid derivatives. Methods Enzymol. 12:323-347. [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning; a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schmitz-Esser, S., N. Linka, A. Collingro, C. L. Beier, H. E. Neuhaus, M. Wagner, and M. Horn. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to chlamydiae and rickettsiae. J. Bacteriol. 186:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schünemann, D., S. Borchert, U. I. Flügge, and H. W. Heldt. 1993. ADP/ATP translocator from pea root plastids (comparison with translocators from spinach chloroplasts and pea leaf mitochondria). Plant Physiol. 103:131-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sicheritz-Pontén, T., C. G. Kurland, and S. G. E. Andersson. 1998. A phylogenetic analysis of the cytochrome b and cytochrome c oxidase I genes supports an origin of mitochondria from within the Rickettsiaceae. Biochim. Biophys. Acta 1365:545-551. [DOI] [PubMed] [Google Scholar]
- 42.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 43.Syvänen, A.-C., H. Amiri, A. Jamal, S. G. E. Andersson, and C. G. Kurland. 1996. A chimeric disposition of the elongation factor genes in Rickettsia prowazekii. J. Bacteriol. 178:6192-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tjaden, J., H. H. Winkler, C. Schwöppe, M. van der Laan, T. Möhlmann, and H. E. Neuhaus. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181:1196-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tucker, A. M., H. H. Winkler, L. O. Driskell, and D. O. Wood. 2003. S-Adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185:3031-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 47.Viale, A., and A. K. Arakaki. 1994. The chaperone connection to the origins of the eukaryotic organelles. FEBS Lett. 341:146-151. [DOI] [PubMed] [Google Scholar]
- 48.Weinglass, A. B., and H. R. Kaback. 2000. The central cytoplasmic loop of the major facilitator superfamily of transport proteins governs efficient membrane insertion. Proc. Natl. Acad. Sci. USA 97:8938-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weisburg, W. G., M. E. Dobson, J. E. Samuel, G. A. Dasch, L. P. Mallavia, O. Baca, L. Mandelco, J. E. Sechrest, E. Weiss, and C. R. Woese. 1989. Phylogenetic diversity of the rickettsiae. J. Bacteriol. 171:4202-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williamson, L. R., G. V. Plano, H. H. Winkler, D. C. Krause, and D. O. Wood. 1989. Nucleotide sequence of the Rickettsia prowazekii ATP/ADP translocase-encoding gene. Gene 80:269-278. [DOI] [PubMed] [Google Scholar]
- 51.Winkler, H. H. 1986. Membrane transport in rickettsiae. Methods Enzymol. 125:253-259. [DOI] [PubMed] [Google Scholar]
- 52.Winkler, H. H. 1990. Rickettsia species (as organisms). Annu. Rev. Microbiol. 44:131-153. [DOI] [PubMed] [Google Scholar]
- 53.Winkler, H. H. 1976. Rickettsial permeability: an ADP-ATP transport system. J. Biol. Chem. 251:389-396. [PubMed] [Google Scholar]
- 54.Winkler, H. H., R. Daugherty, and F. Hu. 1999. Rickettsia prowazekii transports UMP and GMP, but not CMP, as building blocks for RNA synthesis. J. Bacteriol. 181:3238-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkler, H. H., and R. M. Daugherty. 1986. Acquisition of glucose by Rickettsia prowazekii through the nucleotide intermediate uridine 5′-diphosphoglucose. J. Bacteriol. 167:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkler, H. H., R. M. Daugherty, and J. P. Audia. 2003. Cysteine-scanning mutagenesis and thiol modification of the Rickettsia prowazekii ATP/ADP translocase: evidence that TM VIII faces an aqueous channel. Biochemistry 42:12562-12569. [DOI] [PubMed] [Google Scholar]
- 57.Winkler, H. H., and H. E. Neuhaus. 1999. Non-mitochondrial adenylate transport: a plant plastid to obligate intracellular bacterium connection. Trends Biochem. Sci. 277:64-68. [Google Scholar]
- 58.Wright, J. K., R. M. Feather, and P. Overath. 1979. Lactose carrier protein of Escherichia coli: the riddle of the two binding sites, p. 239-248. In M. Klinkenberg, F. Palmieri, S. Papa, and E. Quagliariello (ed.), Function and molecular aspects of biomembrane transport. Elsevier/North Holland Biomedical Press, Amsterdam, The Netherlands.