Abstract
In Streptomyces coelicolor, replication is initiated by the DnaA protein in the centrally located oriC region and proceeds bidirectionally until the replication forks reach the ends of the linear chromosome. We identified three clusters of DnaA boxes (H69, H24, and D78) which are in a relatively short segment of the chromosome centered on the oriC region. Of the clusters analyzed, D78 exhibited the highest affinity for the DnaA protein; the affinity of DnaA for the D78 cluster was about eightfold higher than the affinity for oriC. The high-affinity DnaA boxes appear to be involved in the control of chromosome replication. Deletion of D78 resulted in more frequent chromosome replication (an elevated ratio of origins to chromosome ends was observed) and activated aerial mycelium formation, leading to earlier colony maturation. In contrast, extra copies of D78 (delivered on a plasmid) caused slow colony growth, presumably because of a reduction in the frequency of initiation of chromosome replication. This suggests that the number of high-affinity DnaA boxes is relatively constant in hyphal compartments and that deletion of D78 therefore permits an increased copy number of either the chromosomal origin region or a plasmid harboring the D78 cluster. This system conceivably influences the timing of decisions to initiate aerial mycelial formation and sporulation.
The genus Streptomyces comprises gram-positive soil bacteria that are known for their ability to produce many valuable antibiotics and other secondary metabolites. Unusual for bacteria, they undergo complex morphological differentiation (7, 8, 12). Germination of a spore leads to the formation of a vegetative mycelium consisting of branching hyphae. During further development, new branches grow into the air, forming a layer of aerial mycelium. The compartments of vegetative hyphae contain several copies of uncondensed chromosomes between occasional cross walls. The syncytial aerial hyphal tips may contain more than 50 copies of the chromosome. After cessation of growth in aerial hyphae, the chromosomes are condensed and regular ladders of sporulation septa are laid down, forming prespore compartments that differentiate into chains of uninucleoid exospores. Streptomycetes possess a large linear chromosome (8 to 9 Mbp) with a high G+C content (70 to 75%) (2, 16). A central core region, comprising approximately one-half of the chromosome, contains all the genes likely to be unconditionally essential, while the arms contain many “contingency” loci coding for nonessential functions.
In eubacteria, in eukaryotes, and very likely in archaea, replication is controlled at the initiation stage (1, 13, 25). Bacterial chromosome replication is initiated at a single origin, oriC, by the initiator protein DnaA (10), which specifically interacts with 9-bp nonpalindromic sequences (DnaA boxes) at oriC (for reviews, see references 38, 39, and 50). In Streptomyces coelicolor, replication proceeds bidirectionally from the centrally located oriC region toward the ends of the chromosome (41). In Escherichia coli and S. coelicolor the replication origins are different sizes (250 bp and 1,000 bp, respectively) and have different numbers of DnaA boxes (5 and 19, respectively) (58). The S. coelicolor DnaA protein exhibits the highest affinity for the consensus sequence TT(A/G)TCCACA, which is designated the “strong” DnaA box (36). Like all other DnaA proteins, the Streptomyces DnaA protein consists of four domains; domain III and the carboxy-terminal part (domain IV) are responsible for binding of ATP and DNA, respectively, and the N-terminal part (domain I) and domain III contain oligomerization sites. The presence of a long flexible domain II in S. coelicolor DnaA allows it to bind widely spaced DnaA boxes within the extended oriC region (18).
Replication initiation has to occur at the correct time in the cell cycle, and any one origin must initiate once and only once per cell cycle. Control of initiation relies on a reduction in the availability and/or activity of the two key elements, DnaA and the oriC region. Among bacteria, the initiation of replication and its regulation are best understood in E. coli, in which the following three mechanisms prevent reinitiation from the newly replicated origins: (i) sequestration of oriC (5, 31), (ii) conversion of active DnaA protein into an inactive form (21-24), and (iii) reduction in the level of DnaA (27, 28). E. coli oriC contains a high number of GATC sequences, which are the recognition sites for the Dam methyltransferase (3). The newly replicated, and therefore hemimethylated, GATC sequences are bound by the SeqA protein, and oriC is “arrested” (i.e., bound to the membrane). Conversion of the active ATP form of DnaA to the inactive ADP form occurs by the RIDA mechanism (regulatory inactivation of DnaA) (23). This mechanism is dependent on the DnaN sliding clamp of DNA polymerase III and the Hda protein, which together activate the intrinsic ATPase activity of the DnaA protein (4). The availability of DnaA at oriC is reduced by DnaA boxes distributed over the chromosome, particularly by a cluster of five DnaA boxes (datA [DnaA titration]) that titrates a large number of DnaA molecules (43).
In most studies of the regulation of chromosome replication the workers have focused on unicellular, rod-shaped bacteria, particularly E. coli, which divide by binary fission and have a single circular chromosome. The obvious differences between these bacteria and filamentous Streptomyces strains containing elongated compartments with multiple copies of a linear chromosome implies that there may be differences in the regulation of chromosome replication. Cells of the fast-growing organism E. coli divide every 20 min, while chromosome replication requires about 45 min; thus, rounds of replication overlap, and two, four, or even eight origins may coexist. However, the newly replicated oriC regions are temporarily arrested (sequestered) to prevent untimely reinitiation and asynchronous initiation until oriC is fully methylated, which occurs about one-third of a cell cycle after initiation has taken place. This may explain why E. coli can harbor large numbers (more than 10) of oriC-containing minichromosomes (which are also sequestered) without incompatibility problems (40). In contrast to E. coli minichromosomes, Streptomyces minichromosomes are unstable, and only low copy numbers occur (53, 56). Very little is known about chromosome replication, particularly the synchronization of this process in multinucleoid compartments, in Streptomyces (54). It should be noted that at least the Streptomyces species that have been studied do not have a methylation system comparable to the Dam system of E. coli and, presumably, are not able to sequester their oriC regions. On the other hand, inactivation of ATP-DnaA by ATP hydrolysis is likely to take place in Streptomyces, since Streptomyces DnaA has an ATPase activity (33) similar to that of E. coli DnaA. Sequence analysis of the entire S. coelicolor chromosome revealed the presence of clusters of “strong” DnaA boxes which may contribute to the regulation of initiation of S. coelicolor chromosome replication. In this study, we addressed for the first time the question of the importance of a DnaA box cluster in the regulation of replication in a species other than E. coli. Below we describe the in silico and in vitro identification and characterization of the high-affinity binding sites for the DnaA protein. In addition, we examined the influence of deletion of or the presence of extra copies of DnaA-binding sites on chromosome replication, growth, and colony differentiation of S. coelicolor.
MATERIALS AND METHODS
DNA manipulation and bacterial growth conditions.
DNA manipulations were carried out by using standard protocols (47). Enzymes were supplied by Roche, New England BioLabs, or Fermentas; isotopes were obtained from Amersham-Pharmacia-Biotech; and oligonucleotides were obtained from Invitrogen or the Institute of Biochemistry and Biophysics (Warsaw, Poland). The S. coelicolor and E. coli strains used are listed in Table 1. For culture, transformation, and conjugation we used general procedures described previously for E. coli (47) and Streptomyces (26). S. coelicolor was cultivated in tryptic soy broth-yeast extract-malt extract (YEME) (1:1) complex liquid medium or on minimal medium or soy flour-mannitol (SFM) agar plates. To reduce clumping in liquid cultures, S. coelicolor cells were cultivated with vigorous shaking in flasks containing springs. Apramycin (50 μg/ml), ampicillin (100 μg/ml), and thiostrepton (10 μg/ml) were added when they were required for selection. Spore suspensions were prepared as described previously (26). Briefly, 2 × 106 spores were cultured on SFM agar, and after 44 and 64 h the spores were harvested and counted with a hemocytometer (Buerker's chamber). Each experiment was performed at least in triplicate.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| AG115 | lacX74 galU galK araD139 strA hsdR17/F′ lacIqlacZ::Tn10 | 37 |
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab stock |
| WM2121 | araΔ(lac-pro) fis::Km recA56 rpsL srlC300::Tn10 thi | 29 |
| WM1905 | dam-13::Tn9 dcm hsdR mcrA mcrB | 52 |
| ET12567/pUZ8002 | dam-13::Tn9 dcm cat tet hsdM hsdR zjj-201::Tn10/tra neoRP4 | 44 |
| S. coelicolor strains | ||
| M145 | SCP1− SCP2− | 2 |
| J3338 | M145 ΔdnaAboxesD78::aac(3)IV | This study |
| J3339 | M145 ΔdnaAboxesH24::aac(3)IV | This study |
| J3341 | M145 ΔdnaAboxesD78::Swa | This study |
| J3342 | M145 ΔdnaAboxesH24::Swa | This study |
| M145/pWHM3 D78 | M145 with multiple copies of D78 DnaA boxes | This study |
| M145/pWHM3 H24 | M145 with multiple copies of H24 DnaA boxes | This study |
| M145/pWHM3 H69 | M145 with multiple copies of H69 DnaA boxes | This study |
| J3338/pWHM3 D78 | M145 ΔdnaAboxesD78::aac(3)IV with multiple copies of D78 DnaA boxes | This study |
| J3337 | M145 dnaN egfp aac(3)IV | Ruban-Ośmiałowska et al., in preparation |
| DJ510 | J3341 dnaN egfp aac(3)IV | This study |
Southern blot hybridization.
Southern hybridization was performed as described previously (47). Total cellular DNA digested with the SalI enzyme was separated on a 1.0% agarose gel, transferred to Roti Nylon Plus (Roth), and probed with labeled DNA fragments derived from the oriC region (PCR-amplified fragment of gyrB gene; 500 bp) (Table 2), a chromosome end (PCR-amplified fragment of argG; 550 bp) (Table 2), or the pWHM3 plasmid (∼500-bp SalI-XhoI fragment of pWHM3). Probes were labeled with digoxigenin-11-dUTP using a Random Primed DNA labeling kit (Roche). The signals were detected by chemiluminescence with the CSPD reagent (a substrate for alkaline phosphatase; Roche). The blots were exposed to Kodak BioMax film, which was later scanned with a Typhoon 8600 variable-mode imager. Signals were quantified using the ImageQuant software. Each Southern hybridization was performed in triplicate.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a |
|---|---|
| pdnaNfw | GGCCTACAAGTACCTGATCATGCCGGTGCGGCTGAGCGGCCTGCCGGGCCCGGAGCTG |
| pdnaNrv | GGACAGCCGCCGACGGCGCCACCGGCCGGCGGGCGACGGTTGTAGGCTGGAGCTGCTTC |
| H24-fw | GGATCCTGCGCGCGGGTGCCTGTGTC |
| H24-rv | AAGCTTGGCCTGCGGGCACCGTCAGC |
| H69-fw | GGATCCTGCTGAGATCGGCGCAGCTC |
| H69-rv | AAGCTTCCCCGGCGGTGGCTGCCC |
| D78-fw | GGATCCGCACGGGCGCCGGGTTGAC |
| D78-3rv | AAGCTTCTGACCCTCGGGCCTATCTG |
| ΔD78-fw | GGCGCCGCACGGGCGCCGGGTTGACCGGATTTCCTTGTGATTTAAATTCCGGGGATCCGTCGA |
| ΔD78-rv | ACGGCCGGTTCTGACCCTCGGGCCTATCTGCTGATTTTCGATTTAAATGTAGGCTGGAGCTGCTTC |
| ΔH24-fw | CGGGTGCCCGCGGGTGCCTGTGTCATGAGCACCAATCTAATTTAAATGTAGGCTGGAGCTGCTTC |
| ΔH24-rv | CGGATAACCGTAGGCCTGCGGGCACCGTCAGCCAATCTGGATTTAAATTCCGGGGATCCGTCGACC |
| arg-fw | CGGATCATCGAGGCCAAGAGC |
| arg-rv | GTCGGCGGAGACCTCGCCGC |
| gyr-fw | GGTACTGCGGGTTCCGGCCGG |
| gyr-rv | CGCACGAGGGCGGCAGCGACG |
Restriction sites are indicated by boldface type.
Isolation of fragments harboring DnaA boxes using affinity chromatography.
Affinity chromatography was performed as described previously (17, 34). Briefly, the DNA-binding domain (BD) of the S. coelicolor DnaA protein fused to the C terminus of glutathione S-transferase (GST) was bound to glutathione-Sepharose beads and then was used as an affinity reagent to evaluate the binding of DNA fragments containing DnaA boxes. The DNA fragments specifically interacting with the DnaA BD were eluted with high-salt buffer, and this was followed by isopropanol precipitation. The DNA was resuspended in 10 mM Tris-HCl-1 mM EDTA (pH 8) and analyzed on a 1% agarose gel (stained with SYBR).
Purification of the S. coelicolor DnaA protein.
The DnaA protein of S. coelicolor was overexpressed in E. coli WM2121 as a His-tagged protein and then purified on a Ni2+-nitrilotriacetic acid-agarose column (QIAGEN) as described previously (33, 35).
Electrophoretic mobility shift assay.
For binding assays, 32P-labeled DNA (5 fmol) was incubated with DnaA protein in the presence of the nonspecific competitor poly(dA-dC)(dT-dG) (100 ng) at 20°C for 20 min in binding buffer (20 mM HEPES/KOH [pH 7.6], 5 mM magnesium acetate, 1 mM EDTA, 4 mM dithiothreitol, 0.2% Triton X-100, 3 mM ATP, 50 μg/ml bovine serum albumin) (45). The bound complexes were separated by electrophoresis in 4% polyacrylamide gels (0.25× Tris-borate-EDTA, 4 V/cm, 4°C). The gels were dried and analyzed with a Typhoon 8600 variable-mode imager. The apparent equilibrium dissociation constant [KD(app)] was determined as described previously (3, 35, 36). The reaction mixtures contained a fixed amount of DNA and various concentrations of DnaA protein. The DNA concentration used was much lower than the protein concentration required for half-maximal binding, so the protein concentration at half-maximal binding was very close to KD(app). The KD(app) was deduced from a curve (percentage of unbound DNA versus DnaA concentration [nM]), based on the equation KD = [S] · [P] · [SP]−1, where [S] is the DNA concentration, [P] is the protein concentration, and [SP] is the DNA-protein complex concentration. When [S] was ≪KD, then [P]free ≅ [P]total, so KD = [P]total · [S] · [SP]−1.
Construction of strains carrying an extra copy or deletion of clusters of DnaA boxes.
DNA fragments carrying clusters of DnaA boxes were PCR amplified using appropriate primers (Table 2) and then cloned into shuttle vector pWHM3 (Table 3). The S. coelicolor M145 protoplasts were transformed with the pWHM3 derivatives. A knockout strategy (14, 15) was used for construction of deletions of the H24 and D78 DnaA clusters. Deletions were created by introducing the apramycin resistance cassette (apra) flanked by SwaI restriction sites amplified with oligonucleotides pH24fw and pH24rv for cosmid H24 (positions 9635 to 9885) and with oligonucleotides pD78fw and pD784rv for cosmid D78 (positions 33910 to 34170) (for details, see Tables 1, 2, and 3). The resulting constructs, H24ΔDnaAbox::apra and D78ΔDnaAbox::apra, were used to transform ET12567/pUZ8002, from which they were mobilized into S. coelicolor M145, resulting in strains J3339 and J3338, respectively. Clean knockout constructs were created by restriction digestion of cosmids H24ΔDnaAbox::apra and D78ΔDnaAbox::apra with SwaI and religation. Subsequently, the kan gene in the SuperCos part of the resulting cosmids was exchanged for a vio-oriT cassette, and the resulting cosmids were then used for conjugation into J3339 and J3338. Vior exconjugants were subcultured on antibiotic-free medium and screened for the loss of both Vior and Aprar, which indicated that there was double-crossover allelic exchange in J3338 and J3339, to obtain strains J3341 (ΔD78) and J3342 (ΔH24). A knockin strategy (14, 15) was used to construct S. coelicolor ΔD78 dnaN-egfp, which expressed chromosomally encoded enhanced green fluorescent protein (EGFP)-tagged DnaN instead of the wild-type protein (Ruban-Ośmiałowska et al., manuscript in preparation). Chromosomal DNA of all of the strains constructed was checked by PCR and/or by Southern hybridization, and the presence of the DnaN-EGFP fusion protein was examined by phosphorimager scanning of the sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel.
TABLE 3.
Cosmids and plasmids used in this study
| Plasmid or cosmid | Relevant characteristics | Source or reference |
|---|---|---|
| pGEM-T Easy | bla, T vector for cloning PCR-amplified fragments | Promega |
| pGEM-T Easy D78 | pGEM-T Easy derivative containing cluster of D78 | This study |
| pGEM-T Easy H24 | pGEM-T Easy derivative containing cluster of H24 | This study |
| pGEM-T Easy H69 | pGEM-T Easy derivative containing cluster of H69 | This study |
| pWHM3 | tsr bla lacZα, bifunctional vector derivative of pIJ486 | 51 |
| pWHM3 D78 | pWHM3 derivative containing additional cluster of D78 | This study |
| pWHM3 H24 | pWHM3 derivative containing additional cluster of H24 | This study |
| pWHM3 H69 | pWHM3 derivative containing additional cluster of H69 | This study |
| pGEXDnaA(BD) | Expression vector, BD of S. coelicolor DnaA protein fused to glutathione S-transferase | 34 |
| pLEXDnaAx6His | Expression vector, isolation of S. coelicolor DnaA protein | 33 |
| H24ΔDnaAbox::apra | Cosmid H24 ΔDnaAboxes32620<>32955::aac(3)IV | This study |
| D78ΔDnaAbox::apra | Cosmid D78 ΔDnaAboxes9635<>9885::aac(3)IV | This study |
| H24ΔDnaAbox::Swa_relig | Cosmid H24 ΔDnaAboxes32620<>32955::SwaI religation, kan::vph | This study |
| D78ΔDnaAbox::Swa_relig | Cosmid D78 ΔDnaAboxes9635<>9885::SwaI religation, kan::vph | This study |
Microscopy.
Strains used for fluorescence microscopic observations were inoculated in the acute-angle junction of coverslips inserted at an angle of 45° in minimal medium agar containing 1% mannitol (26). The staining procedures were the procedures described previously (20, 48). Briefly, mycelium was fixed for 10 min with a paraformaldehyde-glutaraldehyde mixture, digested for 2 min with 2 mg ml−1 lysozyme, and incubated for 1 h with 10 μg ml−1 wheat germ agglutinin-tetramethylrhodamine conjugate (Molecular Probes) for cell wall visualization. After five washes with phosphate-buffered saline, the coverslips were mounted in Vectashield (Vector Laboratories) antifade reagent. Fluorescence microscopy was carried out using a Nikon Eclipse 600 or Zeiss Axio Imager Z1 microscope equipped with a ×100 objective.
Scanning electron microscopy of the mycelium of S. coelicolor M145 and the deletion mutant ΔD78 was preformed as described previously (11).
RESULTS
Clusters of high-affinity DnaA boxes are located around the oriC region in the center of the S. coelicolor linear chromosome.
The S. coelicolor DnaA protein exhibits the highest affinity for the 5′-TT(A/G)TCCACA-3′ “strong” DnaA box sequence defined by Majka et al. (35). In the oriC region, only 1 of 19 DnaA boxes is a strong box. To identify potential DnaA boxes outside the oriC region, we looked for all 28 possible nonamers that differed at no more than one position from the strong DnaA box (32). We found 631 weak DnaA boxes with a single mismatch and 51 strong DnaA boxes. The weak DnaA boxes are randomly distributed along the chromosome. In contrast, all but 3 of the 51 strong DnaA boxes are located in the core region of the chromosome, particularly around the oriC region (Fig. 1). Interestingly, most of the strong DnaA boxes (42 of 51) lie in noncoding DNA regions upstream of genes. In many cases a single strong DnaA box is present in a putative promoter region, and sometimes a strong box is accompanied by a weak one, as illustrated by the promoter region of dnaA itself (19, 36). Thus, the DnaA boxes might serve as regulatory elements. Indeed, the two boxes in the dnaA promoter are bound by DnaA, and the expression of dnaA is autoregulated (19). In addition, we identified clusters of DnaA boxes containing five or six DnaA boxes within a relatively short distance (213 to 329 bp) (Fig. 1). These clusters were named after the cosmids, H24, H69, and D78, in which they are located. Cluster H24 (nucleotides 4306411 to 4306674) contains one strong box, while clusters H69 and D78 (nucleotides 4239631 to 4239844 and 4361979 to 4362308, respectively) both contain four strong boxes. The clusters all are near the oriC region (Fig. 1).
FIG. 1.
Distribution of “strong” DnaA boxes in the S. coelicolor A3(2) chromosome and localization of clusters of DnaA boxes.
DnaA protein exhibits higher affinity for the D78 cluster than for the oriC region.
To examine whether the clusters are bound by DnaA, a DNA-binding assay for isolation of specific sequences directly from cosmid DNA was performed. In this assay, GST-DnaA BD fusion protein bound to glutathione-Sepharose beads was used as an affinity reagent to evaluate binding of DNA fragments containing DnaA boxes. In our experiments, cosmids H24, H69, and D78 digested with SalI were incubated with the immobilized binding domain of DnaA (Fig. 2). Under low-salt conditions, all DNA fragments were nonspecifically bound, while under medium-salt conditions the fragments containing DnaA boxes were selectively retained (Fig. 2). With each cosmid, fragments containing the cluster of DnaA boxes were bound efficiently (H24, 2,560 bp; H69, 2,900 bp; D78, 4,670 bp, 3,080 bp, 1,430 bp, and 500 bp plus the cosmid backbone containing three DnaA boxes [∼10,000 bp] [Fig. 2]). Interactions between individual clusters of DnaA boxes and DnaA protein were confirmed by gel retardation assays. In our experiments, three clusters of DnaA boxes were amplified by PCR, using corresponding cosmids as templates and the pairs of primers listed in Table 2 (Fig. 2). The labeled fragments were incubated with increasing amounts of purified entire DnaA protein (all four domains), and then nucleoprotein complexes were analyzed in a 4% native polyacrylamide gel. The multiple nucleoprotein complexes were formed in a manner that depended on the protein concentration (Fig. 3A). The apparent dissociation constants (Fig. 3A) were calculated from the gel retardation assay results, as previously described in detail (6, 35, 36). Our results show that the affinities of the clusters of DnaA boxes analyzed for DnaA protein vary significantly. The DnaA protein exhibits the highest affinity for the D78 cluster (50-fold higher than the affinity for the H24 fragment).
FIG. 2.
In vitro analysis of the interactions between DnaA protein and multiple DnaA sequences. DnaA box-containing DNA fragments of cosmid DNA (H24, H69, or D78; 3 μg) digested with the SalI restriction enzyme were selectively bound to GST-DnaA BD beads. Fragments were analyzed by agarose gel electrophoresis. Lane 0, DNA before affinity chromatography; lane L, DNA incubated with the fusion protein beads and eluted with low-salt buffer; lane M, DNA nonspecifically bound to the fusion protein and eluted with medium-salt buffer; lane H, DNA specifically bound to the fusion protein and released from the beads by washing with high-salt buffer. Lane λ/PstI contained a standard size marker. Localization of DnaA boxes on cosmid inserts is shown on the right (the distances between adjacent DnaA boxes are indicated above and below the cosmids). The clusters of DnaA sequences are enclosed in boxes. Uppercase letters (to the right and above the cosmids) indicate the retained fragments of the cosmid insert.
FIG. 3.
Interactions of DnaA with clusters of DnaA boxes: gel retardation assay. The assay was performed using 32P-labeled DNA fragments containing clusters of DnaA boxes (∼2 fmol; H24 H69, D78; the sizes of the fragments analyzed are shown in Fig. 1). The DNA fragments were incubated with increasing amounts of DnaA protein. The DNA-protein complexes were separated on a 4% polyacrylamide gel and subjected to autoradiography. (A) Interaction of the DnaA protein with individual clusters of DnaA boxes. Lane 1, no DnaA; lane 2, 0.28 nM DnaA; lane 3, 1.4 nM DnaA; lane 4, 6.9 nM DnaA; lane 5, 20.7 nM DnaA; lane 6, 27.6 nM DnaA; lane 7, 41.4 nM DnaA. (B) Competition gel retardation assay. All four 32P-labeled fragments, H24, H69, D78, and oriC (∼2 fmol of each fragment), were incubated together with increasing amounts of the Dna A protein. Lane 1, no DnaA; lane 2, 0.28 nM DnaA; lane 3, 0.56 nM DnaA; lane 4, 1.4 nM DnaA; lane 5, 2.8 nM DnaA; lane 6, 6.9 nM DnaA; lane 7, 13.8 nM DnaA; lane 8, 20.7 nM DnaA; lane 9, 27.6 nM DnaA; lane 10, 34.7 nM DnaA; lane 11, 41.4 nM DnaA; lane 12, 48.6 nM DnaA; lane 13, 55.2 nM DnaA; lane 14, 69.4 nM DnaA.
To compare DnaA protein-binding specificities for the clusters and oriC, a competition gel retardation assay was performed. A mixture of the H24, H69, and D78 clusters and oriC was incubated with increasing amounts of DnaA (Fig. 3B). DnaA protein exhibited the highest affinity for the D78 fragment, which was completely bound at the lowest protein concentration (Fig. 3B). The H69 fragment was bound by the DnaA protein with approximately the same affinity as the oriC region, while the H24 fragment was retarded only at elevated protein concentrations. The affinity of DnaA for the D78 cluster was eightfold higher than that for oriC. This suggests that the DnaA box clusters, especially D78, may act as a reservoir for DnaA molecules.
Relative copy number of the D78 DnaA box cluster affects the timing of sporulation.
To find out whether the number of DnaA boxes affects Streptomyces growth, the H24, H69, and D78 DnaA box clusters were delivered into wild-type S. coelicolor on plasmid pWHM3, which is based on the high-copy-number pIJ101 replicon. In each case, transformants were viable, but they grew markedly more slowly than the control strain transformed with the empty pWHM3 vector grew. This effect was especially strong in transformants with extra copies of the D78 cluster. We noticed a significant delay in sporulation on minimal and rich solid agar media (S. coelicolor sporulates only on solid media) (Fig. 4A). Note that colonies growing on selective medium (with thiostrepton), including colonies harboring empty plasmid pWHM3, grew more slowly than the wild-type strain grew.
FIG. 4.
Relative copy number of the D78 DnaA box cluster affects growth and the timing of sporulation. (A) Growth (48 h) of S. coelicolor/pWHM3+D78, S. coelicolor ΔD78, and appropriate control strains on minimal medium (MM) or rich medium (SFM) (26). (B) Scanning electron microscopy of mycelium of S. coelicolor M145 and the ΔD78 mutant. Specimens were taken from cultures grown for 48 h. An increased frequency of coiled aerial hyphae in the mutant and a more advanced developmental stage (more mature spore chains) were observed. WT, wild type.
To investigate the effect of a deficiency of DnaA boxes on Streptomyces growth, we constructed mutant strains ΔH24 and ΔD78 harboring deletions of the H24 and D78 clusters, respectively (see Materials and Methods). In liquid media, it was rather difficult to observe any differences among the strains analyzed; the growth curves of the deletion mutants compared with the growth curve the wild type did not change significantly. S. coelicolor tends to grow as clumps of mycelium. Despite the use of various conditions that reduced clumping we were not able to completely eliminate aggregation. Thus, the growth of the strains was probably influenced by clumping of the mycelium, and this might have affected the growth rates. In contrast, on solid media the ΔD78 mutant started to sporulate earlier than the wild type started to sporulate. Scanning electron microscopy revealed that after 2 days, aerial hyphae of the deletion mutant had already differentiated into long spore chains, while spores appeared only sporadically in the wild-type control (Fig. 4B). To examine this phenomenon further, we counted the number of spores that could be scraped from the solid medium (see Materials and Methods). After 44 to 64 h, the ΔD78 deletion mutant produced approximately 10 times more spores than the wild-type strain produced (Table 4). At later times (>120 h), the two strains, ΔD78 and the wild type, produced approximately the same number of spores (data not shown). The viability (heat and lysozyme resistance) of ΔD78 spores was similar to that of wild-type spores (data not shown).
TABLE 4.
Sporulation of wild-type strain M145 and deletion mutant ΔD78 of S. coelicolor
| Strain | No. of sporesa
|
|
|---|---|---|
| 44 h | 64 h | |
| Wild type | 1.5 × 107 | 2.1 × 1011 |
| ΔD78 | 1.3 × 108 | 1.4 × 1012 |
Each experiment was repeated in triplicate (see Materials and Methods).
Chromosome replication is more frequent in the D78 deletion mutant than in the wild-type strain.
Since the DnaA protein exhibits high affinity for the D78 cluster, we speculated that particularly this cluster might be involved in the regulation of chromosome replication. In order to observe ongoing replication directly in individual hyphal compartments, we used a functional DnaN-EGFP fusion that facilitated visualization of replication forks (Ruban-Ośmiałowska et al., in preparation). We introduced the fusion into the ΔD78 mutant and compared the fluorescence with that of the wild-type S. coelicolor dnaN-egfp strain (strain J3337) (Ruban-Ośmiałowska et al., in preparation). Fluorescence microscopy showed that the hyphal compartments contained several visible green foci whose sizes, intensities, and distributions were variable (Fig. 5A). In neither strain was there any spatial correlation between the foci and tips or septa. Examination of substantial numbers of hyphal compartments revealed that in the deletion mutant foci were more densely packed along hyphae than they were in the wild type (Fig. 5B, left panel). ΔD78 contained more compartments with bright and compact foci than the wild type contained (Fig. 5B, left panel); the faint, diffuse foci were more predominant in compartments of the wild-type control (Fig. 5A).
FIG. 5.
Distribution of DnaN-EGFP foci in mycelium of S. coelicolor M145 and the ΔD78 mutant. (A) S. coelicolor strains M145 and ΔD78 carrying dnaN-egfp were grown for 26 and 42 h. The images show overlays of two fluorescence signals: DnaN-EGFP foci and cell wall stained with wheat germ agglutinin-tetramethylrhodamine conjugate. Bars, 5 μm. The arrows indicate types of fluorescence (green arrows, compact foci; lilac arrows, diffuse foci; yellow arrows, dispersed foci), and the red arrowheads indicate boundaries of compartments. (B) Fractions of compartments of S. coelicolor strains M145 and ΔD78 harboring compact foci (580 hyphal compartments of each strain analyzed were examined). WT, wild type.
Number of DnaA boxes affects the timing of chromosome replication in S. coelicolor.
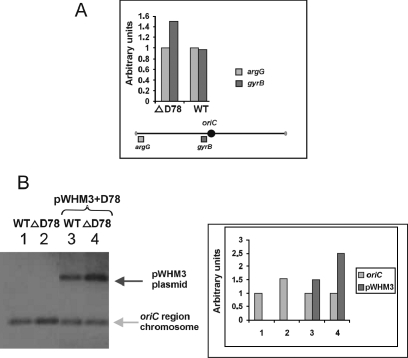
To further investigate the effect of deletion of the high-affinity cluster D78 on chromosome replication, we determined the relative copy numbers of the oriC region and chromosome end by quantification of Southern blot hybridization signals. Southern blots with the same amounts of SalI-digested DNA were hybridized with probes specific for the origin region and one of the chromosomal ends (Fig. 6A). The ratio of origin regions to chromosome ends was about 50% higher in the deletion mutant than in the wild type (Fig. 6A). This was presumably due to an increased copy number of the chromosomal origin in the ΔD78 mutant (Fig. 6B, left panel, lanes 1 and 2). In an additional Southern blot experiment, the relative copy numbers of oriC and plasmid pWHM3+D78 in the wild type and the ΔD78 mutant were compared. Interestingly, the copy number of pWHM3+D78 was about 70% higher in the deletion mutant than in the wild type (Fig. 6B), suggesting that the total number of high-affinity boxes may remain constant.
FIG. 6.
Deletion of the D78 cluster increases the ratio of the oriC region to a terminal region and the copy number of the pWHM+D78 plasmid. Total DNA from S. coelicolor M145 and ΔD78, with and without pWHM+D78, was isolated from cultures grown in YEME-tryptic soy broth (26) for 24 h. Southern blots of DNA (3 μg) digested with SalI were hybridized with the labeled probes indicated. The probes consisted of sequences specific for the oriC region (gyrB gene; 500 bp), a chromosome end (argG; 550 bp) (41), and the pWHM3 plasmid (∼500-bp SalI-XhoI fragment of the pWHM3 plasmid). The hybridization signals were quantified by using the ImageQuant program. (A) Quantification of hybridization signals. The graph shows the relative copy numbers of the oriC regions (gyrB) and the chromosomal ends (argG). The locations of the probes on the chromosome are indicated below the graph. (B) (Left panel) Southern blot hybridization. Lane 1, S. coelicolor M145; lane 2, S. coelicolor ΔD78; lane 3, S. coelicolor M145/pWHM3+D78; lane 4, S. coelicolor ΔD78/pWHM3+D78. (Right panel) Quantification of hybridization signals. The graph shows the relative copy numbers of the oriC region (gyrB) and the pWHM3 plasmid. WT, wild type.
DISCUSSION
S. coelicolor, like many other bacteria, including Bacillus subtilis, possesses no Dam-like methylation system, SeqA homologue, or Hda homologue of the kind involved in the regulation of initiation of chromosome replication in E. coli (4, 5, 22-24, 39). In contrast to E. coli cells, growing vegetative hyphal compartments of Streptomyces contain several chromosomes, while the copy number of minichromosomes is only one copy per chromosome, compared with the multiple copies of E. coli minichromosomes (56). So far, the regulation of chromosome number in Streptomyces hyphal compartments has not been studied. Thus, it is interesting to study the mechanism(s) of replication regulation in these filamentous organisms. Here we characterized a cluster of high-affinity DnaA boxes, a putative counterpart of the E. coli datA locus, and its role in the regulation of S. coelicolor linear chromosome replication in multinucleoid compartments.
Streptomyces origin of replication is flanked by high-affinity DnaA-binding sites.
We identified three clusters of high-affinity DnaA boxes, each containing five or six DnaA boxes (Fig. 1, 2, and 3). In E. coli, the datA site consists of five DnaA boxes, two of which are “strong” (27, 43). Interestingly, the chromosomal clusters of DnaA boxes identified in S. coelicolor are in a ca. 120-kb short segment centered on the oriC region. Streptomyces avermitilis, a second Streptomyces species whose genome sequence is known (16), has the same three clusters of high-affinity DnaA boxes, despite the fact that the two species are not very closely related. This suggests that the clusters play a biologically significant role. Of the clusters analyzed, D78 exhibits the highest affinity for the DnaA protein, and the affinity is approximately eightfold higher than the affinity of DnaA for oriC (Fig. 3B). The S. coelicolor DnaA protein oligomerizes via two domains (domains I and III) (36) and presumably therefore forms complexes in which there are far more protein molecules than DnaA boxes. Thus, the high-affinity binding sites, particularly the D78 cluster, may influence the level of free DnaA protein. Indeed, introduction of extra copies of DnaA boxes from the D78 cluster (delivered on a plasmid) resulted in an increase in the level of DnaA protein (data not shown).
Number of DnaA boxes influences regulation of replication and colony maturation.
Little is known about Streptomyces chromosome organization within hyphal compartments, apart from the findings of previous studies which showed that replicating activity did not appear to be confined to particular regions of hyphal compartments (30; Ruban-Ośmiałowska et al., in preparation). Deletion of the D78 cluster of DnaA boxes resulted in an elevated ratio of origins to chromosome ends (Fig. 6) that could have been a result of more frequent initiation or slower replication progression. The latter possibility seems unlikely; recently, Simmons et al. (49) demonstrated that in E. coli the high-affinity cluster of DnaA boxes (datA locus) does not influence progression of the replication fork. Colonies of the ΔD78 mutant sporulated earlier than colonies of the wild-type strain sporulated. Disturbed expression of genes localized near the D78 cluster, including the genes involved in DNA metabolism and DNA replication, has been ruled out by microarray analysis of RNA from the D78 deletion mutant (data not shown). It therefore seems possible that some aspect of the chromosome or origin number per compartment or the origin/terminus ratio may contribute checkpoint information to the decision-making that precedes aerial growth (8). In particular, overreplication could present particular problems in subapical hyphal compartments, in which there is no cell wall growth until the emergence of a new branch. We speculate that in these circumstances, overreplication might often result in more frequent branching, which might be manifested by earlier growth of an aerial mycelium and subsequent sporulation. Chromosome number could also be critical for triggering sporulation in aerial hyphal apical compartments, which may contain many chromosomes (up to 50 or so) before sporulation occurs. It is not known whether there is any upper limit for the number of chromosomes per aerial hyphal compartment, although two developmental regulatory genes (whiA and whiB) have been identified as possible participants in such a checkpoint (11).
Visualization of ongoing replication in vegetative hyphae, using a DnaN-EGFP fusion protein, allowed us to compare the wild type and the ΔD78 mutant. Analysis of DnaN-EGFP revealed that bright, compact DnaN-EGFP foci result from ongoing replication, while diffuse fluorescence results from disassembly of the replication machinery; DnaN-EGFP foci disappear when DNA replication is inhibited by addition of novobiocin (Ruban-Ośmiałowska, in preparation) (Fig. 5). Hyphal compartments of the ΔD78 mutant contained more bright foci than hyphal compartments of the wild type contained, in which fluorescence was sometimes diffuse. We speculate that the earlier maturation and hypersporulation of aerial hyphae of the mutant may result from this more frequent replication, which could cause the system for indicating chromosome numbers to be activated early; consequently, each round of replication goes to completion earlier, shortly before it is possible to initiate sporulation (Table 4 and Fig. 4). In contrast, delivery of extra copies of D78 resulted in slow colony growth, presumably as a consequence of a reduction in the frequency of initiation of chromosome replication.
A cluster of high-affinity DnaA boxes is involved in the control of chromosome copy number.
We suggest that there is a system to keep the number of high-affinity DnaA boxes within hyphal compartments relatively constant. Therefore, deletion of high-affinity DnaA cluster D78 permitted an increased copy number of either the chromosomal origin region or a plasmid harboring the D78 cluster (Fig. 6). Our previous data corroborate this observation; in contrast to E. coli minichromosomes, Streptomyces minichromosomes are unstable and occur at low copy numbers (56, 57). In B. subtilis, DnaA boxes are probably also involved in the regulation of chromosome replication; the oriC region is also flanked by several clusters of DnaA boxes (32, 42), and strong incompatibility was observed between plasmid and chromosomal oriC regions when an additional cluster of DnaA boxes was delivered on a plasmid (55). Chromosomal replication control in Streptomyces and B. subtilis has similarities to replication control of low-copy-number plasmids, such as P1, RK2, pSC101 P1, RK2, pSC101, and F, which harbor binding sites for initiator protein (iterons) which are found not only in the origin but also outside ori (9, 46). When deleted and cloned in trans, these sites increase and reduce plasmid copy number, respectively. The origins of these low-copy-number plasmids can pair via the bound initiator protein. This “handcuffing” mechanism has been suggested to cause steric hindrance for initiation and thereby control the copy number. Our results suggest that handcuffing may also apply to the chromosomal replication control in Streptomyces; pairing of the oriC fragments in the presence of DnaA protein enhanced the kinetics of intermolecular DNA ligation (observed as a ladder of oriC fragments [see Fig. S1 in the supplemental material]).
In summary, high-affinity DnaA boxes appear to be involved in the control of Streptomyces chromosome replication. Deletion of high-affinity DnaA boxes results in more “intensive” replication, so the upper limits of the chromosome number occur earlier than they occur in the wild type. This causes early aerial growth, perhaps because of increased branching of subapical compartments, and earlier sporulation of aerial hyphae in a model linking origin number to the initiation of sporulation. Delivery of extra copies of high-affinity DnaA boxes has the opposite effect: sporulation is significantly delayed. The extra DnaA boxes may increase the handcuffing probability and thereby sterically inhibit replication.
Supplementary Material
Acknowledgments
We thank Kim Findlay for performing scanning electron microscopy. We are grateful to Miroslaw Dudek and Pawel Mackiewicz for help with searching for DnaA boxes.
This work was supported by the Ministry of Scientific Research and Information Research (grant 2P04A 054 29). D.J. was supported by Marie Curie Reintegration Grant MERG-6-CT-2005-014851. Some of this work was done by A.S.-K. as a Marie Curie Fellow under grant QLK2-CT-2001-60081 from the European Commission.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Boye, E., A. Lobner-Olesen, and K. Skarstad. 2000. Limiting DNA replication to once and only once. EMBO Rev. 1:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camara, J. E., A. M. Breier, T. Brendler, S. Austin, N. R. Cozzarelli, and E. Crooke. 2005. Hda inactivation of DnaA is the predominant mechanism preventing hyperinitiation of Escherichia coli DNA replication. EMBO J. 6:736-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, J. L., and L. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from Dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 6.Carey, J. 1991. Gel retardation. Methods Enzymol. 208:103-117. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465-1478. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 9.Das, N., M. Valjavec-Gratian, A. N. Basuray, R. A. Fekete, P. P. Papp, J. Paulsson, and D. K. Chattoraj. 2005. Multiple homeostatic mechanisms in the control of P1 plasmid replication. Proc. Natl. Acad. Sci. USA 102:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erzberger, J. P., M. M. Pirruccello, and J. M. Berger. 2002. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21:4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flardh, K., K. C. Findlay, and K. F. Chater. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145:2229-2243. [DOI] [PubMed] [Google Scholar]
- 12.Flardh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564-571. [DOI] [PubMed] [Google Scholar]
- 13.Giraldo, R. 2003. Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev. 26:533-554. [DOI] [PubMed] [Google Scholar]
- 14.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gust, B., G. Chandra, D. Jakimowicz, T. Yuqing, C. J. Bruton, and K. F. Chater. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54:107-128. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M., Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 17.Jakimowicz, D., J. Majka, W. Messer, C. Speck, M. Fernandez, M. C. Martin, J. Sanchez, F. Schauwecker, U. Keller, H. Schrempf, and J. Zakrzewska-Czerwinska. 1998. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology 144:1281-1290. [DOI] [PubMed] [Google Scholar]
- 18.Jakimowicz, D., J. Majka, G. Konopa, G. Węgrzyn, W. Messer, H. Schrempf, and J. Zakrzewska-Czerwińska. 2000. Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 298:351-364. [DOI] [PubMed] [Google Scholar]
- 19.Jakimowicz, D., J. Majka, B. Lis, G. Konopa, G. Węgrzyn, H. Messer, H. Schrempf, and J. Zakrzewska-Czerwinska. 2000. Structure and regulation of the dnaA promoter region in three Streptomyces species. Mol. Gen. Genet. 262:1093-1102. [DOI] [PubMed] [Google Scholar]
- 20.Jakimowicz, D., B. Gust, J. Zakrzewska-Czerwińska, and K. F. Chater. 2005. Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 187:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 22.Katayama, T. 2001. Feedback controls restrain the initiation of Escherichia coli chromosomal replication. Mol. Microbiol. 41:9-17. [DOI] [PubMed] [Google Scholar]
- 23.Kato, J. 2005. Regulatory network of the initiation of chromosomal replication in Escherichia coli. Crit. Rev. Biochem. Mol. Biol. 40:331-342. [DOI] [PubMed] [Google Scholar]
- 24.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelman, L. M., and Z. Kelman. 2003. Archaea: an archetype for replication initiation studies? Mol. Microbiol. 48:605-615. [DOI] [PubMed] [Google Scholar]
- 26.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 27.Kitagawa, R., H. Mitsuki, T. Okazaki, and T. Ogawa. 1996. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19:1137-1147. [DOI] [PubMed] [Google Scholar]
- 28.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch, C., J. Vandekerckhove, and R. Kahmann. 1988. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc. Natl. Acad. Sci. USA 85:4237-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kummer, C., and S. Kretschmer. 1986. DNA replication is not restricted to specific regions in young vegetative Streptomyces mycelia. J. Basic Microbiol. 26:27-31. [DOI] [PubMed] [Google Scholar]
- 31.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1997. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 32.Mackiewicz, P., J. Zakrzewska-Czerwińska, A. Zawilak, M. R. Dudek, and S. Cebrat. 2004. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 32:3781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majka, J., W. Messer, H. Schrempf, and J. Zakrzewska-Czerwińska. 1997. Purification and characterization of the Streptomyces lividans initiator protein DnaA. J. Bacteriol. 179:2426-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majka, J., D. Jakimowicz, M. Zarko-Postawka, and J. Zakrzewska-Czerwińska. 1997. Glutathione S-transferase fusion proteins as an affinity reagent for rapid isolation of specific sequence directly from genomic DNA. Nucleic Acids Res. 25:2537-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majka, J., D. Jakimowicz, W. Messer, H. Schrempf, M. Lisowski, and J. Zakrzewska-Czerwińska. 1999. Interactions of the Streptomyces lividans initiator protein DnaA with its target. Eur. J. Biochem. 260:325-335. [DOI] [PubMed] [Google Scholar]
- 36.Majka, J., J. Zakrzewska-Czerwińska, and W. Messer. 2001. Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces. J. Biol. Chem. 276:6243-6252. [DOI] [PubMed] [Google Scholar]
- 37.Mattern, S. 1992. Regulation of the galactose operon of Streptomyces lividans. Ph.D. thesis. Universität Osnabrück, Osnabrück, Germany.
- 38.Messer, W., F. Blaesing, D. Jakimowicz, M. Krause, J. Majka, J. Nardmann, S. Schaper, H. Seitz, C. Speck, C. Weigel, G. Węgrzyn, M. Welzeck, and J. Zakrzewska-Czerwinska. 2001. Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading. Biochimie 83:5-12. [DOI] [PubMed] [Google Scholar]
- 39.Messer, W. 2002. The bacterial replication initiator DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26:355-374. [DOI] [PubMed] [Google Scholar]
- 40.Morigen, F. Molina, K. and Skarstad. 2005. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 187:3913-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musialowski, M. S., F. Flett, G. B. Scott, G. Hobbs, C. P. Smith, and S. G. Oliver. 1994. Functional evidence that the principal DNA replication origin of the Streptomyces coelicolor chromosome is close to the dnaA-gyrB region. J. Bacteriol. 17:5123-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogasawara, N., S. Moriya, and H. Yoshikawa. 1991. Initiation of chromosome replication: structure and function of oriC and DnaA protein in eubacteria. Res. Microbiol. 142:851-859. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa, T., Y. Yamada, T. Kuroda, T. Kishi, and S. Moriya. 2002. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 44:1367-1375. [DOI] [PubMed] [Google Scholar]
- 44.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parada, C., and K. J. Marinas. 1991. Mechanism of DNA A protein-dependent pBR322 DNA replication. DNA A protein-mediated trans-strand loading of the DNA B protein at the origin of pBR322 DNA. J. Biol. Chem. 266:18895-18906. [PubMed] [Google Scholar]
- 46.Park, K., E. Han, J. Paulsson, and D. K. Chattoraj. 2001. Origin pairing (′handcuffing') as a mode of negative control of P1 plasmid copy number. EMBO J. 24:7323-7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25:847-858. [DOI] [PubMed] [Google Scholar]
- 49.Simmons, L. A., A. M. Breier, N. R. Cozzarelli, and J. M. Kaguni. 2004. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 51:349-358. [DOI] [PubMed] [Google Scholar]
- 50.Speck, C., and W. Messer. 2001. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vara, J., M. Lewandowska-Skarbek, Y.-G. Wang, S. Donadio, and C. R. Hutchinson. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthetic pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171:5872-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weigel, C., A. Schmidt, B. Ruckert, R. Lurz, and W. Messer. 1997. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 16:6574-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenner, T., V. Roth, G. Fischer, C. Fourrier, B. Aigle, B. Decaris, and P. Leblond. 2003. End-to-end fusion of linear deleted chromosomes initiates a cycle of genome instability in Streptomyces ambofaciens. Mol. Microbiol. 50:411-425. [DOI] [PubMed] [Google Scholar]
- 54.Yang, M. C., and R. Losick. 2001. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J. Bacteriol. 183:5180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshikawa, H., and R. G. Wake. 1993. Initiation and termination of chromosome replication, p. 507-528. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 56.Zakrzewska-Czerwińska, J., D. Jakimowicz, J. Majka, W. Messer, and H. Schrempf. 2000. Initiation of the Streptomyces chromosome replication. Antonie Leeuwenhoek 78:211-221. [DOI] [PubMed] [Google Scholar]
- 57.Zakrzewska-Czerwińska, J., J. Majka, and H. Schrempf. 1995. Minimal requirements of the Streptomyces lividans 66 oriC region and its transcriptional and translational activities. J. Bacteriol. 177:4765-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zawilak-Pawlik, A., A. Kois, J. Majka, D. Jakimowicz, A. Smulczyk-Krawczyszyn, W. Messer, and J. Zakrzewska-Czerwińska. 2005. Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria. Biochem. J. 389:471-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.