Abstract
Molecular modeling and mutational analysis (site-directed mutagenesis and saturation mutagenesis) were used to probe the molecular determinants of the substrate specificity of aminopyrrolnitrin oxygenase (PrnD) from Pseudomonas fluorescens Pf-5. There are 17 putative substrate-contacting residues, and mutations at two of the positions, positions 312 and 277, could modulate the enzyme substrate specificity separately or in combination. Interestingly, several of the mutants obtained exhibited higher catalytic efficiency (approximately two- to sevenfold higher) with the physiological substrate aminopyrrolnitrin than the wild-type enzyme exhibited.
Rieske oxygenases are widely distributed in bacteria and are used in a variety of biosynthetic and metabolic reactions, including cis-dihydroxylation, monohydroxylation, desaturation, sulfoxidation, and O- and N-dealkylation (4, 20). Recently, we described biochemical characterization of a novel Rieske N-oxygenase, aminopyrrolnitrin oxygenase (PrnD) from Pseudomonas fluorescens Pf-5, that catalyzes arylamine oxidation (15). PrnD is involved in the biosynthesis of the antibiotic pyrrolnitrin, which is produced by many pseudomonads and has broad-spectrum antifungal activity (1, 5, 12, 18). In the proposed biosynthetic pathway of pyrrolnitrin, PrnD catalyzes the oxidation of the amino group of aminopyrrolnitrin (Aprn) to a nitro group, forming pyrrolnitrin (22). Arylamine oxygenases seem to be widespread and are used in diverse metabolic reactions (6, 11, 13, 16, 17, 19). However, PrnD is the only biochemically characterized example of an arylamine N-oxygenase involved in arylnitro group formation. Characterization of the other known arylamine-oxygenating enzyme, AurF, was partial or less informative, since it was based on whole-cell experiments rather than in vitro experiments using purified enzyme (23). More recently, we determined the catalytic mechanism of PrnD, which involves two monooxygenation steps and one dehydrogenation step via hydroxylamine and nitroso compounds as intermediates (14). Here we used molecular modeling and mutational analysis via site-directed mutagenesis and saturation mutagenesis to probe the substrate specificity of PrnD, with the goal of understanding the molecular determinants of this enzyme and facilitating future protein engineering of new substrate specificity for biotechnological applications.
MATERIALS AND METHODS
Materials.
The pMal-c2x expression vector, malE primer, factor Xa, amylose resin, Taq DNA polymerase, T4 DNA ligase, DNase I, and restriction endonucleases were purchased from New England Biolabs (Beverly, MA). Plasmid pQE-80L was obtained from QIAGEN (Valencia, CA). A BCA protein assay kit and pyrrolnitrin were obtained from Sigma-Aldrich (St. Louis, MO). Materials for PCR product purification, gel extraction, and plasmid preparation were obtained from QIAGEN. Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA). The physiological substrate aminopyrrolnitrin was a kind gift from J. W. Frost of Michigan State University (East Lansing, MI).
Bacterial strains and growth conditions.
Escherichia coli BL21(DE3) and DH5α were obtained from Novagen (Madison, WI) and the University of Illinois Biochemistry Department's Media Preparation Facility (Urbana, IL), respectively. Pseudomonas fluorescens Pf-5 (ATCC BAA-447) was purchased from the American Type Culture Collection (Manassas, VA). E. coli strains DH5α and BL21(DE3) were grown aerobically at 37°C or 30°C in Luria-Bertani medium with constant shaking (220 rpm). When necessary, kanamycin was added at a concentration of 50 μg/ml and ampicillin was added at a concentration of 100 μg/ml.
Construction of the pTKXb-PrnD plasmid.
The PCR product obtained by using oligonucleotide primers 5′-GGCATATGAACGACATTCAATTGGATCAAG-3′ (NdeI restriction site underlined) and 5′-GGCTCGAGTCACCGCTCACTTGCGACGCG-3′ (XhoI restriction site underlined) was cleaved by NdeI and XhoI and was purified using a QIAEX II gel purification kit (QIAGEN). The purified product was cloned into the NdeI- and XhoI-digested expression vector p6xHTKXb119 (10) to obtain pTKXb-PrnD, which was transformed into E. coli BL21(DE3).
Site-directed mutagenesis and saturation mutagenesis of pTKXb-PrnD.
Site-directed mutagenesis was carried out by using a QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA). The pTKXb-PrnD plasmid was used as the DNA template. All 14 residues in the substrate binding pocket (SBP) except two Ala residues were mutated to Ala or Val individually. The plasmids containing the correct mutant genes were then used to transform E. coli BL21(DE3), and colonies selected by kanamycin resistance were used for protein expression. The pTKXb-PrnD mutants were expressed by using the procedure used for the wild-type (WT) enzyme, as described above. Saturation mutagenesis at Phe-312 and Leu-277 was carried out by using the overlap extension PCR-based method described elsewhere (3).
Expression and purification of the fusion protein.
Overnight cultures of BL21(DE3) cells transformed with the pMal-c2x-PrnD vectors were diluted 1:200 in LB medium supplemented with ampicillin (100 μg/ml) and grown at 30°C until the absorbance at 600 nm was ∼0.6. Then protein expression was induced by addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM, and incubation was continued for 3 h. Cells were harvested by centrifugation at 4°C for 20 min at 6,000 × g, rinsed with phosphate-buffered saline, and frozen and stored at −20°C. The yield was approximately 3 g (wet weight) of bacteria/liter of culture. A bacterial lysate was prepared by thawing and resuspending cells from a 1-liter culture in 40 ml of buffer containing 20 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol (DTT), 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 0.1 mM EDTA (starting buffer); the resulting suspension was treated with 10 mg of lysozyme for 30 min. The suspension was sonicated on ice with a Branson sonicator for 1 min five times with 1-min intervals between treatments. After centrifugation for 20 min at 10,000 × g, the concentration of NaCl in the supernatant was adjusted to 0.5 M and the supernatant was loaded onto an amylose resin column (3 mg of fusion protein/ml of amylose). The column was washed with 10 volumes of 0.5 M NaCl in starting buffer and with 20 volumes 20 mM Tris-HCl (pH 7.8)-100 mM NaCl-1 mM DTT before elution with 10 mM maltose in the latter buffer. For cleavage with factor Xa, the dialyzed maltose binding protein-PrnD fusion protein was incubated with factor Xa (1 μg/200 μg of fusion protein) for 16 h at 4°C. Maltose was removed with hydroxyapatite resin, and maltose binding protein was removed after cleavage by a second adsorption to the amylose resin. Fractions from the amylose column were then concentrated with Centricon-10 ultrafiltration units (Amicon), and the concentrations of NaCl, Tris-HCI (pH 7.8), and DTT were adjusted to 100 mM, 20 mM, and l mM, respectively.
In vitro reconstitution of the iron-sulfur clusters in the PrnD proteins.
The reconstitution procedures were carried out in an anaerobic chamber (Jacomex, Dagneux, France). All the solutions were incubated anaerobically for 2 h before the beginning of each experiment. The purified PrnD was diluted to obtain concentrations of 10 to 20 μM with N2-sparged 20 mM Tris-HCl buffer (pH 7.8) containing 10 mM DTT, 200 mM NaCl, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 10% (vol/vol) glycerol. β-Mercaptoethanol was added to the protein solution at a concentration of 1.0% (vol/vol), and the solution was gently mixed and left for 90 min. Fe(NH4)2(SO4)2 and Na2S were added to the solution at final concentrations of ∼1.0 mM. The reconstitution reaction proceeded for 2 to 3 h. The solution was diluted eightfold by addition of N2-sparged 20 mM Tris-HCl buffer (pH 7.8), diluted twofold by addition of 20 mM Tris-HCl buffer (pH 7.8), equilibrated in air, and then dialyzed against the same buffer. Excess Fe2+ and S2− were removed with a desalting column (10-DG; Bio-Rad). The reconstituted PrnD protein was concentrated with Centricon-30 ultrafiltration units (Amicon, Beverly, MA) to a final volume of 0.2 ml and stored at 4°C or (for longer times) at −80°C. The iron-sulfur cluster incorporation was analyzed by electron paramagnetic resonance spectroscopy.
Enzyme kinetics of PrnD and mutants.
Enzyme activity was routinely assayed by high-performance liquid chromatography (HPLC). Each assay mixture (final volume, 0.5 ml) contained 500 μM NADPH, 3 μM flavin mononucleotide, 500 μM substrate, and SsuE reductase and PrnD at an SsuE/PrnD molar ratio of 4.0 in 20 mM Tris-HCl (pH 7.8), and it was stirred at 30°C. The reactions were started by addition of PrnD to the reaction mixtures, and the reaction mixtures were analyzed by HPLC. One unit of activity was defined as the amount of enzyme that formed 1 μmol of product per min at 30°C under the standard assay conditions. Kinetic parameters determined in atmospheric oxygen were obtained by fitting the data to the Michaelis-Menten equation. SsuE was prepared as described elsewhere (15).
Protein electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed with a Mini-PROTEAN II system (Bio-Rad, Richmond, CA) with 12% polyacrylamide gels (30% acrylamide/0.8% bisacrylamide stock) under denaturing conditions. Protein concentrations were determined using the BCA method (Sigma) according to the manufacturer's instructions, with bovine serum albumin as a standard. The samples used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis were incubated, typically for 5 min at 90 to 100°C, with an equal volume of denaturing sample buffer. Broad-range molecular weight standards were purchased from Bio-Rad.
Homology modeling.
A homology model of PrnD was constructed using the Insight II software (Insight II, version 2000; Accelrys Inc., San Diego, CA). The crystal structure of the α-subunit of naphthalene dioxygenases (NDO) (2, 8, 9) was used as a template. The sequence of PrnD was aligned with that of NDO using CLUSTALW (http://workbench.sdsc.edu/) and was adjusted to ensure that critical residues, such as the catalytic iron coordinating the facial triad of PrnD (H186, H191, and D323), were aligned with critical residues of NDO (H208, H213, and D362). Gaps in regions of secondary structures were avoided when the sequences were aligned. Three loop optimization models were generated for each model constructed with Insight II. All the models were checked with the PROSTAT and Profiles-3D functions in Insight II. The model with the highest overall score was chosen.
Aminopyrrolnitrin and para-aminobenzyl amine (pABA) were docked in the homology models of wild-type PrnD and the F312S and L277V mutants by using the Molecular Operating Environment (MOE) software (Chemical Computing Group Inc., Montreal, Canada). Mutations were introduced into the PrnD model by using the Rotamer Explorer function, and the rotamer with the lowest free energy was chosen. The approximate initial position of the substrate was determined based on the possible binding sites identified by the Site Finder function in MOE, as well as the relative position of indole in the crystal structure of naphthalene dioxygenase (PDB code 1O7N). Each docking run consisted of 25 independent docks with six iteration cycles, and a random start was used to generate substrate positions within the docking box. From the results, the substrate orientation which gave the lowest interaction energy was chosen for another round of docking. A nonrandom start was used in this case. This process was repeated two times or until there was no significant decrease in the interaction energy of the substrate. The Conolly surface of the substrate binding pocket was generated using the Molecular Surface function in MOE.
Analytical methods.
Optical spectra were recorded with a Varian Cary 100 Bio UV-Vis spectrophotometer (Varian, Palo Alto, CA). Enzyme reaction products were analyzed with an Agilent series 1100 HPLC system (Agilent Technologies, Palo Alto, CA). A sample was eluted on a ZORBAX SB-C8 column (4.6 by 150 mm; Agilent). The HPLC parameters were as follows: 25°C; solvent A, 1% acetic acid in water; solvent B, methanol; gradient, 5% solvent B for 2 min, followed by an increase to 100% solvent B in 18 min and finally 100% solvent B for 2 min; flow rate, 1.0 ml/min; detection by UV spectroscopy at 254 nm.
Protein database search.
The amino acid sequences deduced from the prnD gene sequences of P. fluorescens Pf-5 were compared with the sequences of related enzymes from other sources using the BLAST Network at the National Center for Biotechnology Information. Multiple-sequence alignment of P. fluorescens PrnD and related enzymes was performed with CLUSTAL W.
RESULTS
Homology modeling.
A homology model of PrnD was constructed on the basis of the crystal structure of the α-subunit of NDO (2, 8, 9) by using the Insight II software. The level of sequence identity between PrnD and NDO is relatively low (∼19%); however, our previous work confirmed that PrnD belongs to the Rieske oxygenase class of enzymes, in which the Rieske 2Fe-2S center and the mononuclear iron site are evolutionarily conserved (15), suggesting that the overall fold of the enzymes should be preserved. Thus, the Rieske 2Fe-2S center coordinating residues, C69, H71, C88, and H91, as well as the nonheme iron coordinating facial triad residues, H186, H191, and D323, of PrnD (corresponding to C81, H83, C101, and H104 and to H186, H191, and D323, respectively, in NDO), were used as constraints during threading of the sequences. The physiological substrate Aprn and its analog pABA were docked into the homology model using the MOE software. Fourteen residues plus the three highly conserved residues involved in the 2-His-1-carboxylate facial triad were identified to be within 4.5 Å of the substrates, forming the SBP. These fourteen residues are Ala-184, Ala-187, Ile-195, Leu-202, Trp-209, Thr-229, Arg-232, Pro-236, Asp-269, Leu-277, Gln-278, Val-310, Phe-312, and Gln-318.
Site-directed mutagenesis of the residues in the substrate binding pocket.
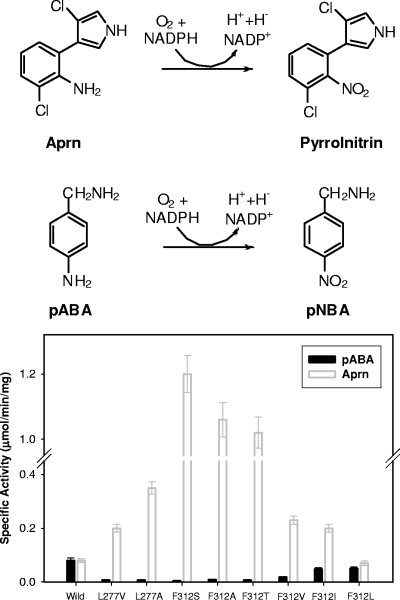
To probe their functional roles in substrate specificity, all 14 residues except two alanine residues in the SBP were replaced with a smaller residue, alanine or valine, using the QuikChange site-directed mutagenesis kit. The catalytic activities of the PrnD mutants with Aprn and pABA were determined and compared with those of WT PrnD. Most mutants exhibited significantly decreased activity with both Aprn and pABA. This is consistent with the notion that these residues are important for the substrate specificity and/or catalytic activity of PrnD. Interestingly, the F312A and L277A mutants exhibited significantly increased conversion of the bulky hydrophobic substrate Aprn but significantly decreased activity with pABA (Fig. 1). The specific activities of the F312A mutant were determined to be 0.009 μmol/min/mg protein for pABA and 1.06 μmol/min/mg protein for Aprn, which are 11% and 1,330% of the specific activities of the WT enzyme, respectively. The specific activities of the L277A mutant were determined to be 0.007 μmol/min/mg protein for pABA and 0.35 μmol/min/mg protein for Aprn, which are 9% and 438% of the specific activities of the WT enzyme, respectively. Even though replacement of the remaining 10 residues by smaller amino acids had no positive effect on the activity with the bulky substrate Aprn, our mutational study of the Phe-312 and Leu-277 residues indicated that enlargement of the binding pocket near the substrate significantly modulated the substrate specificity with the large hydrophobic Aprn molecule. Thus, these two positions are considered to be crucial determinants of the substrate specificity of PrnD, and their importance for activity and substrate specificity was investigated further by saturation mutagenesis.
FIG. 1.
Specific activities of wild-type PrnD and mutants generated by saturation mutagenesis at positions 277 and 312. In a mutant library, the enzymes with low activity for pABA and high activity for Aprn were selected, and specific activities with pABA and Aprn were determined. pNBA, p-nitrobenzyl amine.
Saturation mutagenesis at Leu-277 and Phe-312.
Saturation mutagenesis was performed at positions 277 and 312 using an overlap extension PCR-based method (3). Mutants exhibiting lower activity for pABA and higher activity for Aprn were identified using Gibbs reagent-based activity assays (7, 21) and were confirmed by determining whole-cell activity with Aprn. The positive mutants that exhibited higher activity for Aprn were found to be the F312A, F312S, F312V, F312L, F312I, L277A, and L277V mutants. The mutant genes were introduced into the pMal-c2x plasmid and overexpressed, and each mutant enzyme was purified and reconstituted for kinetic studies as described elsewhere (15). Interestingly, as shown in Fig. 1, as the hydrophobic side chain at position 312 became smaller in the order leucine-isoleucine-valine-threonine-alanine, the specific activity with the bulky hydrophobic substrate Aprn increased compared to the specific activity of the WT enzyme. With the small and less hydrophobic substrate pABA, the opposite trend was observed; the activity decreased as the size of the hydrophobic side chain of the amino acid decreased. The F312S and F312T mutants had slightly different characteristics, likely due to the presence of the hydroxyl group of their side chains. The F312S mutant exhibited a slightly lower kcat/Km than the F312T mutant containing an extra CH group for Aprn, but the change in the substrate specificity was more dramatic than that for the F312T mutant due to the significantly low activity for pABA (Table 1).
TABLE 1.
Kinetic constants of wild-type PrnD and mutantsa
| Enzyme | Aprn
|
pABA
|
(kcat/Km)Aprn/(kcat/Km)pABA | ||||
|---|---|---|---|---|---|---|---|
| kcat (min−1) | Km (μM) | kcat/Km (min−1 μM−1) | kcat (min−1) | Km (μM) | kcat/Km (min−1 μM−1) | ||
| WT | 6.8 | 191 | 0.036 | 6.5 | 379 | 0.0172 | 2.09 |
| F312L | 5.6 | 84.2 | 0.066 | 4.3 | 343 | 0.0125 | 5.28 |
| F312I | 17.4 | 124 | 0.140 | 4.1 | 734 | 0.0056 | 25.0 |
| F312V | 19.8 | 122 | 0.162 | 1.5 | 782 | 0.0019 | 85.3 |
| F312T | 86.5 | 590 | 0.147 | 0.61 | 893 | 0.0007 | 210 |
| F312S | 102 | 771 | 0.132 | 0.33 | 1,420 | 0.0002 | 660 |
| F312A | 90.3 | 512 | 0.176 | 0.78 | 925 | 0.0008 | 220 |
| L277V | 17.2 | 134 | 0.128 | 0.66 | 940 | 0.0007 | 183 |
| L277A | 30.1 | 256 | 0.118 | 0.62 | 962 | 0.0006 | 197 |
| F312A/L277V | 123 | 505 | 0.243 | 0.60 | 1,115 | 0.0005 | 486 |
Each value is the mean of triplicate measurements and varied from the mean by no more than 15%.
Kinetic analysis of the PrnD variants.
For further evaluation, the kinetic constants of the mutant enzymes for pABA and Aprn were determined (Table 1). While the kcat for Aprn increased with a decrease in the size of the amino acid side chain, the kcat and the affinity for pABA decreased as the side chain became smaller. For the F312S mutant, the low activity for pABA seems to have resulted from both a decreased kcat (0.33 min−1) and a decreased affinity (Km, 1,420 μM). In contrast, the high activity for Aprn seems to have resulted mainly from an increased kcat (102 min−1). Consequently, the F312S mutant displayed a remarkably altered substrate preference from pABA to Aprn, which was supported by the fact that the ratio of kcat/Km values for Aprn to the ratio of kcat/Km values for pABA (selectivity ratio) changed from 2.09 to 660. The F312S and F312A mutants showed the highest substrate specificity and catalytic efficiency with Aprn, with a selectivity ratio of 660 and a kcat/Km value of 0.176.
In the case of L277 mutants, the L277V and L277A mutants exhibited higher activity with Aprn than WT PrnD exhibited. For the L277V mutant, the high activity with Aprn seems to have resulted from both an increased kcat (17.2 min−1) and an increased affinity (Km, 134 μM). The residue corresponding to L277 in PrnD (L305 in NDO) has not been linked previously to the catalytic activity of any Rieske nonheme iron oxygenase. When the two mutations were combined, the F312A/L277V mutant showed a cumulative effect, and there was a further increase in the activity with Aprn. Thus, mutations that result in smaller amino acids at these two residues are expected to enlarge the SBP, which might result in loose binding of substrates and alleviate the steric hindrance when the bulky substrate binds to the active site.
DISCUSSION
In the present study, we constructed a homology model of PrnD and used mutational analysis, including site-directed mutagenesis and saturation mutagenesis, to probe the substrate specificity of PrnD. There were putative substrate-contacting residues, and mutations at positions 312 and 277 were found to modulate the enzyme substrate specificity separately or in combination.
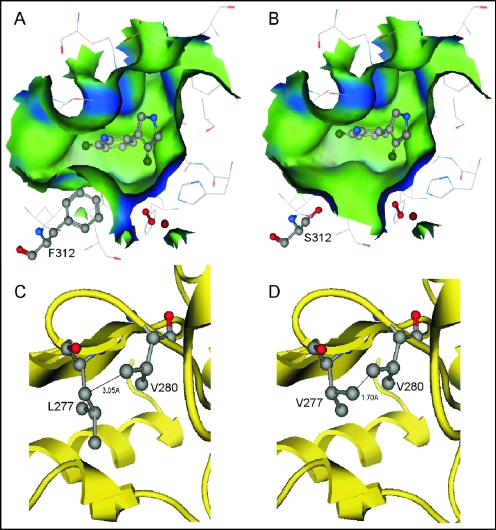
According to the docking results for Aprn in the PrnD homology model, residue 312 lies in the deepest part of the SBP. The F312S mutation resulted in a binding pocket in PrnD that was much deeper, wider, and slightly more polar than the WT binding pocket (Fig. 2A and B). In the WT molecule, the orientation of Aprn is not favorable for efficient turnover due to the steric hindrance between the hydrophobic group of the substrate and the bulky side chain of F312. Removal of the large hydrophobic phenyl group of F312 allows the bulky Aprn substrate to approach closer to the active site iron. Introduction of serine into the binding pocket did not result in formation of any new hydrogen bonds between the residue and the substrate, as the amine group of Aprn is located more than 4 Å from the oxygen in the serine side chain. The residues within 4 Å of the S312 side chain are V280, M292, V310, L311, Q318, and A319. These residues are the residues most likely to interact with S312, but no hydrogen bond was observed between S312 and these residues. In the WT PrnD model, the side chain of F312 interacts with the same residues that serine interacts with in the F312S mutant. This further indicates that the effect of the F312S mutation on the enzyme activity is primarily due to enlargement of the SBP. The L277V mutation did not cause a significant change in the topography of the binding pocket. L277 in the WT and V277 in the mutant have the same neighboring residues within 4 Å of their side chains. However, in the L277V mutant, the methyl group of V277 comes within 1.7 Å of another methyl group from V280. This may cause a steric clash that can alter the position of V280, which is in the secondary shell of the binding pocket, resulting in more subtle changes to the SBP and the enzyme activity than the F312S mutation. An X-ray crystal structure of PrnD is needed to confirm the homology model and the structural interpretation discussed above.
FIG. 2.
(A and B) Molecular surfaces in the substrate binding pockets of wild-type PrnD (A) and the F312S mutant (B). Blue and green indicate hydrophilic and hydrophobic surfaces, respectively. (C and D) The intermolecular distances between V280 and L277 in wild-type PrnD (C) and V277 in the L277V mutant (D) are 3.50 and 1.70 Å, respectively. The molecular surfaces and intermolecular distances are the result of modeling.
In summary, we showed that the substrate specificity of PrnD, an arylamine N-oxygenase, can be modulated by manipulating residues 312 and 277 in the SBP. In addition, several resulting mutants exhibited significantly higher activity with the physiological substrate Aprn. Our results should improve our understanding of arylamine oxidation in biological processes and set the stage for more practical investigations of PrnD as a biocatalyst for synthesis of industrially important nonnatural arylnitro compounds.
Acknowledgments
This research was supported by grant N00014-02-1-0725 from the Office of Naval Research. J.-K.L. was also partially supported by a Shen Fellowship from the Department of Chemical and Biomoelcular Engineering at the University of Illinois at Urbana-Champaign.
REFERENCES
- 1.Burkhead, K. D., D. A. Schisler, and P. J. Slininger. 1994. Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl. Environ. Microbiol. 60:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carredano, E., A. Karlsson, B. Kauppi, D. Choudhury, R. E. Parales, J. V. Parales, K. Lee, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2000. Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J. Mol. Biol. 296:701-712. [DOI] [PubMed] [Google Scholar]
- 3.Chockalingam, K., Z. Chen, J. A. Katzenellenbogen, and H. Zhao. 2005. Directed evolution of specific receptor-ligand pairs for use in the creation of gene switches. Proc. Nat. Acad. Sci. USA 102:5691-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costas, M., M. P. Mehn, M. P. Jensen, and L. Que. 2004. Dioxygen activation at mononuclear nonheme iron active sites: enzymes, models, and intermediates. Chem. Rev. 104:939-986. [DOI] [PubMed] [Google Scholar]
- 5.Hammer, P. E., D. S. Hill, S. T. Lam, K. H. VanPee, and J. M. Ligon. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63:2147-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He, J., and C. Hertweck. 2004. Biosynthetic origin of the rare nitroaryl moiety of the polyketide antibiotic aureothin: involvement of an unprecedented N-oxygenase. J. Am. Chem. Soc. 126:3694-3695. [DOI] [PubMed] [Google Scholar]
- 7.Joern, J. M., T. Sakamoto, A. A. Arisawa, and F. H. Arnold. 2001. A versatile high throughput screen for dioxygenase activity using solid-phase digital imaging. J. Biomol. Screen. 6:219-223. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson, A., J. V. Parales, R. E. Parales, D. T. Gibson, H Eklund, and S. Ramaswamy. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039-1042. [DOI] [PubMed]
- 9.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 10.Kim, Y. W., J. H. Choi, J. W. Kim, C. Park, J. W. Kim, H. Cha, S. B. Lee, B. H. Oh, T. W. Moon, and K. H. Park. 2003. Directed evolution of Thermus maltogenic amylase toward enhanced thermal resistance. Appl. Environ. Microbiol. 69:4866-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King, R. R., C. H. Lawrence, and L. A. Calhoun. 1998. Unusual production of 5-nitroanthranilic acid by Streptomyces scabies. Phytochemistry 49:1265-1267. [Google Scholar]
- 12.Lambert, B., F. Leyns, L. Vanrooyen, F. Gossele, Y. Papon, and J. Swings. 1987. Rhizobacteria of maize and their antifungal activities. Appl. Environ. Microbiol. 53:1866-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lancini, G. C., D. Kluepfel, E. Lazzari, and G. Sartori. 1966. Origin of the nitro group of azomycin. Biochim. Biophy. Acta 130:37-44. [Google Scholar]
- 14.Lee, J. K., and H. Zhao. 2006. Mechanistic studies on the conversion of arylamine to arylnitro compounds by aminopyrrolnitrin oxygenase: identification of intermediates and kinetic studies. Angew. Chem. Int. Ed. Engl. 45:622-625. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. K., M. Simurdiak, and H. Zhao. 2005. Reconstitution and characterization of aminopyrrolnitrin oxygenase, a Rieske N-oxygenase that catalyzes unusual arylamine oxidation. J. Biol. Chem. 280:36719-36728. [DOI] [PubMed] [Google Scholar]
- 16.Meunier, B., S. P. de Visser, and S. Shaik. 2004. Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem. Rev. 104:3947-3980. [DOI] [PubMed] [Google Scholar]
- 17.Morris, M., G. Pagan, and H. Warmke. 1954. Hiptagenic acid, a toxic component of Indigofera endecaphylla. Science 119:322-323. [DOI] [PubMed] [Google Scholar]
- 18.Pfender, W. F., J. Kraus, and J. E. Loper. 1993. A genomic region from Pseudomonas fluorescens Pf-5 required for pyrrolnitrin production and inhibition of Pyrenophora tritici repentis in wheat straw. Phytopathology 83:1223-1228. [Google Scholar]
- 19.Rebstock, C., H. J. Crooks, J. Controulis, and Q. Bartz. 1949. Chloramphenicol (Chloromycetin). IV. Chemical studies. J. Am. Chem. Soc. 71:2458-2462. [Google Scholar]
- 20.Resnick, S. M., K. Lee, and D. T. Gibson. 1996. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J. Ind. Microbiol. Biotechnol. 17:438-457. [Google Scholar]
- 21.Sakamoto, T., J. M. Joern, A. Arisawa, and F. H. Arnold. 2001. Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl. Environ. Microbiol. 67:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanpee, K. H., O. Salcher, and F. Lingens. 1980. Formation of pyrrolnitrin and 3-(2-amino-3-chlorophenyl)pyrrole from 7-chlorotryptophan. Angew. Chem. Int. Ed. Engl. 19:828-829. [Google Scholar]
- 23.Winkler, R., and C. Hertweck. 2005. Sequential enzymatic oxidation of aminoarenes to nitroarenes via hydroxylamines. Angew. Chem. Int. Ed. Engl. 44:4083-4087. [DOI] [PubMed] [Google Scholar]