Abstract
Bacteria commonly exchange genetic information by the horizontal transfer of conjugative plasmids. In gram-negative conjugation, a relaxase enzyme is absolutely required to prepare plasmid DNA for transit into the recipient via a type IV secretion system. Here we report a mutagenesis of the F plasmid relaxase gene traI using in-frame, 31-codon insertions. Phenotypic analysis of our mutant library revealed that several mutant proteins are functional in conjugation, highlighting regions of TraI that can tolerate insertions of a moderate size. We also demonstrate that wild-type TraI, when overexpressed, plays a dominant-negative regulatory role in conjugation, repressing plasmid transfer frequencies ∼100-fold. Mutant TraI proteins with insertions in a region of approximately 400 residues between the consensus relaxase and helicase sequences did not cause conjugative repression. These unrestrictive TraI variants have normal relaxase activity in vivo, and several have wild-type conjugative functions when expressed at normal levels. We postulate that TraI negatively regulates conjugation by interacting with and sequestering some component of the conjugative apparatus. Our data indicate that the domain responsible for conjugative repression resides in the central region of TraI between the protein's catalytic domains.
Much of the lateral gene transfer between bacteria occurs through the action of conjugative plasmids that encode all of the functions necessary for their hosts to transmit them to recipient cells. Plasmid transfer is achieved through direct cell contact and active transport of DNA by the donor. In gram-negative conjugation systems, typified by the F plasmid of Escherichia coli, only one strand of DNA is translocated, so single-strand cleavage and unwinding of the substrate DNA must occur prior to transfer (9). Strand scission is performed by plasmid-encoded “relaxases” that cleave their cognate plasmid at a specific site called nic within the origin-of-transfer region (oriT). In the case of the F and related plasmids, the accessory proteins TraM, TraY, and integration host factor also bind at oriT as part of the relaxosome complex (15, 17, 19, 32). DNA unwinding is usually performed by a separate helicase, though in some systems, such as the F and R388 plasmids of E. coli, relaxase and helicase activities are both present in the relaxase (11, 27, 42). After making a single-stranded break at nic, the relaxase is thought to deliver the DNA to a type IV secretory apparatus that can translocate it across the recipient cell membrane. The relaxase remains covalently bound to the nic site and religates the scission once DNA transfer is complete. The replication of single-stranded plasmid DNA in the donor and recipient regenerates the double-stranded DNA plasmid in both cells.
The F plasmid relaxase TraI is a well-studied model of structure and function for relaxases. The single-stranded DNA-cleaving activity of TraI is present in the first ∼310 residues of this 1,756-residue protein, while the helicase motifs are located towards the C terminus (residues ∼990 to 1450) (Fig. 1) (4, 28, 41). The C-terminal region of TraI following the helicase domain appears to play a role in conjugation and may function in protein-protein interactions (28). TraI binds nic with exquisite sequence specificity (40), and some structural determinants of this specificity have been revealed by site-directed mutagenesis and crystallographic analyses (8, 13, 14, 20).
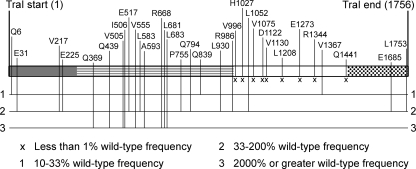
FIG. 1.
Diagram of i31 library and conjugation phenotypes of traI::i31 mutants. The bar shows the length of the TraI protein, with four regions highlighted by different patterns: gray, relaxase domain; lined, region of unknown function; white, helicase domain; spotted, putative protein-protein interaction domain. Each allele (except the Q794 allele) was introduced on a pTrc-based vector to a CAW400 pOXΔtraI donor, and the resulting strains were assayed for their abilities to transfer pOXΔtraI to an F− recipient. For the Q794 allele, the complementation phenotype was assayed in an XK1502 F′ΔI donor. Mutants are listed by position of i31 insert, and their phenotypes relative to the ΔtraI/p99I+ control donor are shown by the labeled vertical lines. The pOXΔtraI/p99I+ control donor had a conjugation frequency of 8.0 × 10−3 transconjugants/donor.
Despite the extensive biochemical analysis of TraI, a general mutagenesis of the traI gene has not been previously described. We mutagenized F traI using the λTnlacZ/in system (25, 26) to generate mutants with a defined 31-codon insertion (the “i31”) at different positions. i31 libraries are useful tools for studying structure-function relationships and mapping functional regions within a protein because i31 lesions are vicinal in their effects. The i31 disrupts nearby protein folding, which can be deleterious to the function of the disrupted region. In some cases, however, the insert does not affect function, and such inserts allow us to identify “permissive sites” in the protein that can tolerate insertions of a moderate size. Prior applications of i31 libraries include defining the topology of transmembrane proteins, studying protein-protein interactions, mapping functional domains, and investigating the interactions of bacterial proteins with eukaryotic cells (18, 21, 22, 38, 39, 44, 45). In one study focusing on the E. coli LacI repressor, which is, like TraI, a cytoplasmic DNA binding protein, the phenotypes resulting from i31 mutations and previously characterized missense mutations at nearby positions were explicitly compared and found to agree quite well (31).
We tested the phenotypes of TraI::i31 mutants in conjugation and in vivo oriT cleavage assays. Several mutations render TraI nonfunctional in conjugation, while others mark permissive sites in the protein. An analysis of mutant with insertions between the consensus relaxase and helicase domains of the protein allowed us to identify a novel role for the intervening region, which functions as a negative regulator of F plasmid conjugation. The mechanism of negative regulation is unclear, though it may represent a titration or sequestration of some vital conjugative component by TraI.
MATERIALS AND METHODS
Growth media, strains, and plasmids.
E. coli strains and plasmids used in this study are summarized in Table 1. The rich (LB) and minimal (M63) media used in these experiments have been described previously (30). Tetracycline sensitivity selection was performed on minimal plates as described previously (3), supplemented with fusaric acid (6 mg/liter), lactose (0.2% wt/vol), thiamine (100 μg/ml), and hydrophobic amino acids (isoleucine, phenylalanine, tryptophan, and tyrosine at 50 μg/ml). Medium supplements were used at the following concentrations: 100 μg/ml, ampicillin; 30 μg/ml, chloramphenicol; 15 μg/ml, tetracycline; 100 μg/ml, streptomycin; 40 μg/ml, kanamycin; 5 or 10 μg/ml, gentamicin; 40 μg/ml, 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal); 1 mM, isopropyl-β-d-galactopyranoside (IPTG).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference/source |
|---|---|---|
| E. coli strains | ||
| CAW400 | MC4100 valR recA56 | 35, 46 |
| CC191 | MC1000 F′128(lacIqZΔM15) phoAΔ20 rpoB argE(Am) recA′ | 37, 38 |
| CC1254 | MC1000 F42 ΔrecBCD::red-kan phoAΔ20 rpoB argE(Am) | C. Manoil |
| HS3169 | MC4100 malKΔ16 zjb-729::Tn10 | 33 |
| XK1502 | F− ΔlacU169 nalA | 34 |
| AG1 | F−endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 | Stratagene |
| Plasmids | ||
| F′42 | F′lac JCFL0 | 1 |
| F′ΔI | F′42 with traI deleted and replaced by Tn10 tetRA sequence | This study |
| p99D+ | pTrc99A with wild-type traD cloned into the MCSa | This study |
| p99M+ | pTrc99A with wild-type traM cloned into the MCS | This study |
| p99I+ | pTrc99A with wild-type traI cloned into the MCS | This study |
| pACYC184 | Cmr cloning vector with p19A rep functions | 5 |
| pACYCM+ | pACYC184 with wild-type traM | This study |
| pEG100 | pACYC184-derived vector with MCS from pTrc99A | This study |
| pEG103 | pEG103 with traD cloned into the MCS | This study |
| pGK111M0 | 1.2-kb BglII-Pst fragment of plasmid R1 containing oriT, traM, and finP in pUC119; traM not expressed due to G-to-C substitution in start codon ATG | 36 |
| pLOW2 | Kmr vector with p15A rep functions and MCS from pUC18 NotI | 12 |
| pLOW2traM0oriT | EcoRI-Pst oriT-traM0 fragment of pGK111M0 in pLOW2 | This study |
| pNLK5 | Apr vector carrying traD under araBAD promoter | 21 |
| pKD46 | Apr plasmid with λ Red genes gam, bet, and exo | 7 |
| pOXΔtraI | Gmr pOX38 derivative with dhfr gene in place of traI | 46 |
| pPD1 | pHUB2-derived plasmid carrying traD and traI | 43 |
| pRFM2T75 | Cloning vector carrying traM under control of a T7 promoter | L. S. Frost |
| pTrc99A | Apr cloning vector | 2 |
MCS, multiple cloning site.
The traI gene was cloned into pTrc99A as follows. The coding region of traI was amplified from pPD1 (43) using primers that added an Acc65I site to the 5′ end and an XbaI site to the 3′ end. These primers have the following sequences (the traI coding sequence is in upper case): cgggggtacccaaagggatatacgtttATGA (forward) and gtcctaagcgtttgtctagatgtatcaGTCTCCA (reverse). The PCR product was digested with Acc65I and XbaI and ligated into the Acc65I and XbaI sites of pTrc99A to generate p99I+. The construct was tested for its ability to complement a traI-deficient donor for conjugation, and stable production of TraI protein was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
A derivative of the F plasmid lacking traI was generated as follows. The traI gene in F′42 (Table 1) was replaced with a PCR-amplified tetracycline resistance cassette from Tn10 by λ Red recombination (7) in strain CC1254 to generate F′42ΔtraI::tetRA (“F′ΔI”). The following primers were used for tetRA PCR amplification (homology to tetRA is shown in upper case): gggatatacgttatgatgagtattgcgcaggtcagatcgTTAAGACCCACTTTCACATT (forward) and tcagtctccacccagggttttctctttctgcaggtcgcggCTAAGCACTTGTCTCCTG (reverse). Only the first 27 nucleotides and the final 40 nucleotides of traI remained intact. This particular arrangement was chosen because the traI termini provided sufficient homology to the traI::i31 alleles carried on pTrc vectors (44 base pairs on either side) to allow λ Red recombination. The F′ΔI construct was verified by PCR and by its ability to be complemented for conjugative DNA transfer by traI in trans.
To insert mutant traI alleles into the F′42, individual alleles were cut from their pTrc-based vectors using Acc65I and HindIII and recombined into the F′ΔI in strain XK1502 using the λ Red system present on pKD46 (7). Putative recombinants were selected on lactose minimal plates containing fusaric acid (see above) and purified on lactose minimal plates. Candidates were screened by conjugative proficiency and/or PCR, and production of TraI::i31 proteins was verified by Western blotting with an i31-specific antibody (25).
For increased expression of traM and traD, these genes were cloned into pACYC184-based vectors. traM was amplified from pRFM2T75 by PCR and cloned into the pTrc99A vector using BamHI and SalI to generate p99M+. The SalI/BamHI fragment of p99M+ was cloned into those same sites in pACYC184 to generate pACYCM+. traD was cloned from pNLK5 (21) into pTrc99A (generating p99D+) using Acc65I and HindIII and then with the same restriction enzymes into pEG100 to generate pEG103.
To create low-copy recombinant oriT substrates for activity measurements of TraI-catalyzed nic cleavage in vivo, we used the pLOW2 vector described by Hansen et al. (12). A subcloned 1.2-kb BglII/PstI fragment of the IncFII R1 oriT region was excised from pGK111M0 (36) and ligated to the polylinker of pLOW2. To avoid transcription into the oriT region from the lac promoter immediately adjacent to the polylinker, a subsequent step was necessary to reverse the orientation of the insert. The polylinker of pLOW2 is flanked by dual NotI sites. NotI digestion, followed by religation of the products, generated the desired recombinant, designated pLOW2traM0oriT, with the lac promoter 1.2 kb distant from oriT.
Mutagenesis of traI.
λTnlacZ/in mutagenesis (25) of traI was carried out in strain CC191 p99I+. From approximately 100,000 screened colonies, we isolated 37 plasmids with independent in-frame traI-lacZ fusions and located the insertions by sequencing outwards from the transposon. Among this set of fusions, there were 33 different insertion locations, with insertions at the Q839 and V506 codons isolated twice and the insertion at the Q6 codon isolated three times. Transposon sequences were converted to i31 inserts as described previously (25) to generate “p99traI::i31” plasmids. The resulting traI alleles contain in-frame insertions of 31 codons with a defined sequence, with 27 of the 31 codons being identical in all mutants. Mutant alleles and proteins are referred to by the codon into which the insertion occurred (e.g., E1685 or TraIiE1685). The production of TraI::i31 proteins was assayed by Coomassie staining of gels after SDS-PAGE and by Western blot detection using an i31-specific antibody (25).
Internal deletions within traI were generated using the unique BamHI site present in the i31 sequence of the p99traI::i31 plasmids as previously described (26). To generate the deletion from codons 6 to 225, the plasmids p99traI::Q6 and p99traI::E225 were digested with BamHI and PvuI, mixed, and ligated. To generate the deletion from codons 996 to 1753, p99traI::V996 and p99traI::L1753 were cut with BamHI and EcoRI and then mixed and ligated. Mutants were assayed for their abilities to produce protein by Western blotting with an anti-i31 antibody in the same manner as that used to detect full-length i31 mutant derivatives.
Quantitative conjugation.
A total of 0.1 ml of donor and HS3169 recipient overnight cultures were added to 0.8 ml LB, and the mixture was rotated slowly at 37°C for 45 min. For the initial characterization of conjugative proficiency of the traI::i31 mutations on pTrc99A, CAW400 pOXΔtraI was the donor strain; for all subsequent experiments, the donor was XK1502 carrying the F′42 or F′ΔI plasmid. pOXΔtraI transconjugants were selected on rich plates containing gentamicin and streptomycin, while F′lac transconjugants were selected on lactose minimal agar supplemented with leucine, isoleucine, and streptomycin. The trc promoter was not induced when donors carried traI alleles on pTrc-based plasmids since the amount of TraI present without induction was higher than that in strains carrying the wild-type F′42 (see Results).
TraI expression analysis.
For the quantitation of radiolabeled proteins, cells were diluted 1:100 from overnight cultures into M63-glycerol and grown to an optical density at 600 nm (OD600) of ∼0.37 with shaking at 37°C. A total of 750 μl of these cultures was labeled with 30 μCi of 35S-Met/Cys (NEG-072 EXPRE35S35S [35S] protein labeling mix from PerkinElmer Life Sciences) for 1 min and then treated with 0.05% unlabeled methionine and immediately put on ice. TraI::i31 protein was immunoprecipitated with i31-specific antiserum (25). Precipitated proteins were separated using SDS-PAGE, and TraI was quantified using a PhosphorImager (Molecular Dynamics).
For analysis by Western blotting, cells were grown to mid-log phase in rich media without IPTG induction and then harvested. Cell extracts were separated on polyacrylamide gels, and TraI derivatives were detected by Western blotting using a polyclonal anti-i31 antibody (25) or a commercial polyclonal anti-LacZ antibody (5′→3′, Inc.) as the primary antibody and an alkaline-phosphatase-conjugated secondary antibody. Bound antibodies were visualized by exposure to nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (XP). When appropriate, blots were then scanned and bands were quantified using ImageQuant software (Molecular Dynamics).
Intracellular oriT cleavage assays.
In vivo nic cleavage catalyzed by TraI and various i31 mutants was assayed with a runoff DNA synthesis technique (47) using primer 8 as previously described (17). Briefly, E. coli AG1 strains carrying a recombinant IncF oriT traM-null cleavage substrate, pLOW2traM0oriT, and an expression construct for the traI allele were grown in the presence of ampicillin, kanamycin, and 0.1 mM IPTG. Cells were harvested in exponential growth phase, and an equivalent cell mass (OD600) was used as a template for in vitro unidirectional DNA synthesis in the presence of α32P-labeled deoxynucleoside triphosphate precursors (Amersham). The reaction mixtures also contained 2 ng of a KpnI-linearized fragment of control DNA. DNA synthesis on this standard DNA fragment generates a specific product of 72 nucleotides in length, which can be easily distinguished from the 121-nucleotide product synthesized on the oriT plasmids cleaved intracellularly at nic. The control product is used to normalize the yield of each reaction. Reaction products were applied immediately to denaturing gels and resolved with a polynucleotide size marker as described previously (17). Products were visualized by autoradiography of the dried gels.
RESULTS
Mutagenesis of traI-generated alleles that outperformed the wild type in conjugative complementation.
We isolated a library of 33 distinct traI::i31 mutants in the vector pTrc99A. Given the large size of TraI, an examination of the protein in cell extracts by SDS-PAGE gives a reasonable indication of protein production and stability. Coomassie staining of gels with cell extracts from cultures induced with 1 mM IPTG for 30 min revealed that all of these mutants produce similar amounts of stable, full-length protein except the V1130 mutant, which makes a truncated product (data not shown).
The 33 alleles (in pTrc-derived vectors) were assayed for the ability to complement transfer of the F-derived, traI-deficient plasmid pOXΔtraI; the results are summarized in Fig. 1. Compared to that of the pOXΔtraI/p99I+ control, mutant conjugation frequencies fell into four broad categories corresponding to the groups defined in Fig. 1: extremely defective (x), partially defective (class 1), wild type (class 2), and higher than wild type (class 3). Twelve alleles complemented conjugation in a manner similar to that of the wild type, revealing permissive sites scattered across the length of the protein. Ten of the 12 extremely and partially defective alleles had insertions in the helicase domain (residues ∼990 to 1450). Of the remaining two class 1 alleles, the reduced function of the Q6 allele is not surprising given the proximity of its insertion to active-site residues in the relaxase domain (8), but the insertion in the Q839 allele is not in a characterized functional motif. Strikingly, all mutants with i31 lesions between residues 369 and 683 were functional in conjugation and most allowed frequencies of conjugation higher those observed with wild-type traI (Fig. 1). An important corollary to this observation was that the pOXΔtraI/p99I+ donor had a conjugative efficiency of 8.0 × 10−3 transconjugants/donor, which is relatively low (approximately 1% of the conjugative efficiency of the Tra+ F plasmid under similar conditions). Some alleles, such as the A593 and L681 alleles, complemented pOXΔtraI 100 times better than did wild-type traI in this assay, achieving normal F transfer rates. The surprising phenotype prompted further investigation of these class 3 alleles.
nic cleavage activity is unchanged in class 3 TraI mutants.
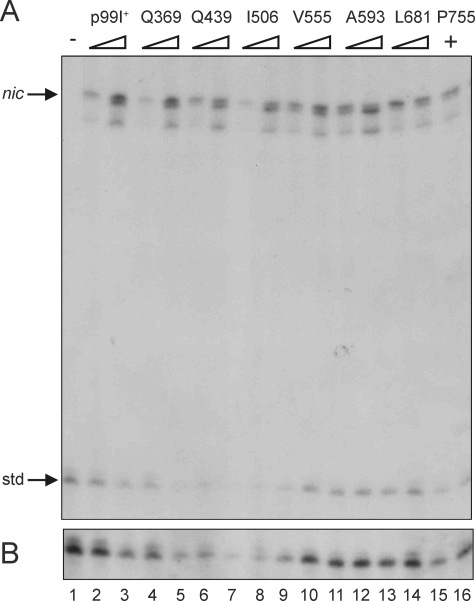
One possible explanation for the increased conjugation frequencies observed with class 3 mutants is that the site- and strand-specific cleavage catalyzed by TraI in preparation for DNA transfer could be strongly enhanced in these insertion mutants. This hypothesis was tested by measuring the level of cleavage at nic catalyzed by TraI in vivo, which was determined by using an in vitro runoff DNA synthesis assay (17). Intracellular cleavage of a recombinant oriT substrate in hosts expressing a selection of traI::i31 alleles (six class 3 and one class 2) was compared to cleavage of the same substrate in cells carrying wild-type traI (Fig. 2). One- and twofold equivalents of cell mass (as indicated by OD600) from each strain were included in the reaction mixtures; the number of viable cells in each culture was determined at cell harvest and indicated similar levels of CFU per reaction mixture. The efficiency of the runoff DNA synthesis reaction can be normalized using the specific product produced on a known amount of linear control DNA added to each reaction mixture (Fig. 2B). While this is not a strictly quantitative assay, a twofold increase in the amount of cleaved product used for in vitro DNA synthesis results in a visible difference in output (compare the one- and twofold equivalent lanes for each allele). Cells with increased TraI cleavage activity would have more cleaved product per cell and therefore result in more runoff synthesis product per viable cell input to the reaction. Despite variations in conjugative efficiency between the TraI mutants tested here, no notable difference in oriT cleavage activity was detected between the mutants (lanes 4 to 16) and wild-type TraI (lanes 2 and 3). The observation that class 3 alleles have wild-type conjugation phenotypes when present on the F′42 (see below) is further evidence that they have normal relaxase activity.
FIG. 2.
Intracellular oriT cleavage by TraI and TraI::i31 mutants. (A) Autoradiogram of runoff DNA synthesis products measuring TraI-catalyzed cleavage activity. E. coli AG1 strains harboring pLOW2traM0oriT and TraI-expressing plasmids as shown above were harvested after subculture in fresh medium and IPTG induction. All reaction mixtures contained equivalent cell mass and 2 ng of a purified DNA standard (std). The numbers of viable cells in the reaction mixtures resolved in lanes 1 to 16 were 2, 0.27, 0.54, 0.16, 0.32, 0.25, 0.5, 0.27, 0.54, 0.3, 0.6, 1.5, 3, 0.7, 1.4, and 0.81 (107 CFU), respectively. The source of products that are terminated within the A stretch proximal to nic is not known, but these and shorter extension products are consistently observed when high concentrations of cells are present in reaction mixtures. Also typical for this assay is the apparent double band at nic, which is consistent with the terminal transferase activity of the polymerase used in vitro. (B) Standard DNA product after a threefold- longer exposure time. +, present; −, negative control reaction.
Expression of traI derivatives in trans can affect conjugation phenotype.
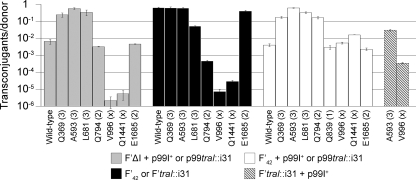
To ensure that the phenotype of class 3 mutants was not somehow related to the complementation of pOXΔtraI conjugation in particular, we used p99I+ and several p99traI::i31 plasmids to complement conjugative transfer of the plasmid F′ΔI, a derivative of the wild-type F′42 in which the traI gene was replaced with a tetracycline resistance marker (Fig. 3). In this system, we again found that p99I+ complemented plasmid transfer at an ∼100-fold-reduced frequency relative to that of F′42 (6.6 × 10−3 transconjugants/donor). All other plasmids also complemented conjugation as they had for pOXΔtraI: the V996 and Q1441 i31 mutants were still defective, class 2 mutants (the Q794 and E1685 mutants) still performed equivalently to the wild type, and class 3 mutants (the Q369, A593, and L681 mutants) again outperformed the wild type.
FIG. 3.
Complementation and dominance phenotypes of traI and traI::i31 alleles in conjugation. XK1502 donors carrying different combinations of F′42- and pTrc-derived plasmids (as indicated) were assayed for their ability to transfer an F′lac plasmid into an F− recipient. Each bar represents the conjugative efficiency of one donor strain. Similarly, colored bars represent the four types of experiments: gray, complementation of F′ΔI in trans; black, phenotype of traI::i31 mutants in cis; white, dominance of p99I+ or p99traI::i31s over the F′42; hatched, dominance of p99I+ over F′traI::i31s. Alleles are listed by position of insert (and functional class); “wild type” denotes wild-type traI. An F′42/pTrc99A donor had the same conjugation frequency as did a donor carrying the F′42 alone. Error bars represent ±standard errors of the means of results of two to four independent biological replicates.
One trivial explanation for the phenotype of class 3 mutants is that strains carrying these alleles on pTrc-based vectors might grow substantially better than do strains with pTrc-based plasmids bearing other alleles. We followed the growth in rich media of XK1502-derived strains carrying the traI+ and traI::A593 alleles on pTrc-based vectors and found less than a 10% difference between their doubling times. We concluded that the class 3 phenotype could not be explained by differential effects of traI alleles on growth rate.
We next set out to test the phenotype of i31 mutants when TraI was expressed at physiological levels. While we did not induce TraI expression in our prior conjugation experiments, leaky transcription from the trc promoter on the pTrc-derived plasmids resulted in increased TraI expression relative to F′lac (see below). For complementation experiments in cis, we replaced the wild-type traI locus of F′42 with a variety of traI::i31 alleles using λ Red recombination to make markerless replacements and avoid upstream or downstream lesions that might affect expression. We chose three class 3 alleles (the Q369, A593, and L681 alleles), two class 2 alleles (the Q794 and E1685 alleles), and two extremely defective alleles (the V996 and Q1441 alleles). The conjugation frequencies of the resulting F′lac plasmids were determined and compared to the F′ΔI/p99traI complementation data in Fig. 3. The two defective alleles tested remained defective on F′lac. The Q794 allele appeared similar to the wild type in high-copy complementations but had a severe defect when present on F′lac, while the E1685 allele was indistinguishable from the wild type in both conjugation assays. F′lac plasmids carrying any of the three class 3 alleles transferred in a manner similar to that of the wild-type F′42 (though F′traI::L681 seems somewhat defective). In summary, we observed that class 3 alleles supported robust transfer efficiencies regardless of copy number, while the wild-type traI and the functional E1685 mutation repressed conjugation when expressed from pTrc-derived plasmids. We therefore designated the class 3 alleles as “unrestrictive” to describe their inability to repress conjugative efficiency.
The inability of unrestrictive alleles to suppress conjugation at high copy suggested that the region mutated might be a negative regulatory domain. To test this conjecture, we performed conjugative dominance experiments in which wild-type traI was present on unmodified F′42 and a second allele was introduced on a pTrc derivative (Fig. 3). Conjugation frequencies ranged widely, depending upon the pTrc-expressed allele. p99I+ and p99traI::E1685 displayed negative dominance over F′42, reducing the frequency of conjugation by 2 orders of magnitude (compare F′42 or traI::i31 and F′42 + p99I+ or p99traI::i31s in Fig. 3). The V996 and Q1441 alleles, two defective alleles with lesions well outside the domain associated with unrestrictive mutations, reduced F′42 conjugation frequency in a manner similar to that of wild-type traI when present on the pTrc vector but did not show further dominance beyond that noted for p99I+. In contrast, plasmid-borne class 3 alleles (the Q369, A593, and L681 alleles) did not exhibit conjugative repression of coresident F′42. While the Q794 allele did not display negative dominance for conjugation, the Q839 allele did, suggesting that the C-terminal boundary of the “restrictive” domain lies between residues 794 and 839.
We continued our conjugative dominance experiments by determining the conjugation frequencies of two strains carrying p99I+ and F′traI::i31 plasmids (Fig. 3). The V996 allele was recessive to wild-type traI on the high-copy plasmid, while the A593 allele showed partial sensitivity to the high-copy repression. We concluded that the expression of TraI from the pTrc-derived multicopy plasmid led to a significant and dominant decrease in conjugative efficiency for coresident F′lac plasmids, which could be rescued by insertions in the region between residues 369 and 794 of TraI.
To determine whether the restrictive domain could function in the absence of the other functional domains of TraI, we used the i31 library to generate two internal deletions: an N-terminal deletion from residues 6 to 225 (deleting the majority of the relaxase domain) and a C-terminal deletion from residues 996 to 1753 (deleting the helicase and putative protein-protein interaction domains). When expressed from the pTrc vector, neither of these mutants had an effect on the conjugation frequency of a coresident F′42 plasmid in conjugative dominance experiments, but the levels of mutant proteins produced by donor cells were determined by Western blotting to be quite low relative to the levels of full-length TraI::i31 proteins (data not shown). Because the overexpression of TraI is probably a critical factor in conjugative repression (see below), we do not find it surprising that the deletion mutants failed to exert dominant effects on conjugation.
To address the question of domain sufficiency in a different way, we assayed the conjugative dominance phenotypes of a number of TraI-LacZ fusions. These fusions are generated as part of the i31 mutagenesis, so we have a TraI-LacZ translational fusion at the same position as each i31 allele in the library (see Materials and Methods). At the point of fusion, native TraI sequence ends and the sequence of LacZ follows, resulting in a C-terminally truncated TraI derivative. Various TraI-LacZ fusions were expressed from the pTrc vector in the presence of wild-type F′42; the conjugative frequencies associated with each tested donor are shown in Table 2, which also summarizes which domains are thought to be present in each mutant. Most fusion proteins had little, if any, effect on conjugative efficiency. The only fusion protein which had a considerable dominant-negative effect on conjugation was the protein with a fusion at position 1753, and this fusion caused only a fivefold decrease in conjugative efficiency (relative to ∼100-fold for wild-type TraI). Western blot analysis showed that all of the fusion proteins were expressed at levels comparable to those of full-length TraI::i31 proteins (data not shown). These results suggest that the restrictive domain may require the C-terminal region of the protein to function properly. We cannot, however, rule out the possibility that the tetramerization of LacZ interferes with the function of the TraI restrictive domain by sequestering it or decreasing its effective concentration within the cell.
TABLE 2.
Conjugative dominance of TraI-LacZ fusionsa
| Position of fusion | Relaxase domain | Restrictive domain | Helicase domain | Interaction domain | Fraction of positive control transfer frequencyb |
|---|---|---|---|---|---|
| Q6 | − | − | − | − | 0.77 |
| Q369 | + | − | − | − | 0.99 |
| L930 | + | + | − | − | 0.79 |
| V996 | + | + | − | − | 0.78 |
| V1130 | + | + | − | − | 0.84 |
| L1208 | + | + | − | − | 0.81 |
| E1685 | + | + | + | − | 0.59 |
| L1753 | + | + | + | ? | 0.18 |
+, domain is present; −, domain is absent or disrupted; ?, domain may be disrupted.
Positive control strain was XK1502 F′42 pTrc99A. Values shown are means of two to four independent biological replicates.
TraI expression is higher from pTrc vectors than from F′lac.
We predicted that pTrc-derived plasmids might allow higher expression of TraI than would F′lac, which could potentially cause the observed repression of conjugation. To assess TraI production, we radiolabeled cells carrying traI::L681 on F′lac or on a pTrc vector and quantified the amount of TraIiL681 present in these strains after immunoprecipitation with i31-specific antibodies and SDS-PAGE. To ensure that expression levels were comparable to expression during conjugation experiments, we did not induce the trc promoter on p99traI::L681. We found that cells carrying p99traI::L681 expressed ∼7.5 times as much TraI as did those harboring F′traI::L681, and quantitative analysis of protein stability showed no degradation of TraIiL681 over 15 min (data not shown).
We also used Western blotting to compare the expression levels of TraIiQ369, TraIiL681, and TraIiE1685 from F′lac and pTrc-derived plasmids (without inducing the trc promoter). Qualitatively, the ratios of TraI expression from pTrc to that from F′lac looked consistently similar; quantitation of TraI bands gave mean pTrc:F′lac expression ratios in the range of 10:1 for all three mutant proteins. We infer that cells carrying a traI allele on a pTrc-based vector, without induction of the trc promoter, express on the order of 10-fold-more TraI protein than do cells carrying the same allele on F′lac.
Increasing traD or traM copy number did not reverse repression by TraI.
The conjugative repression caused by elevated levels of TraI might be the result of TraI binding to and sequestering of another protein. We considered two possible candidates for sequestration: TraM, a protein that associates with TraI and stimulates nic cleavage in the related R1 plasmid system (17, 19), and TraD, the coupling protein of the F Tra system that connects the relaxosome to the membrane-spanning DNA transfer complex (23, 38). Introduction of traM or traD on a pACYC184-based plasmid had little effect on conjugation whether traI was present at low or high copy, suggesting that neither is uniquely limiting for conjugation during traI-mediated repression (Table 3).
TABLE 3.
TraI repression rescue experiments
| Relevant donor genotype
|
Transfer frequencya (transconjugants/donor) | ||
|---|---|---|---|
| F′lac allele (single copy) | pTrc allele (15 to 30 copies) | pACYC allele (10 to 12 copies) | |
| traI+ | Vector only | Vector only | 0.09 ± 0.01 |
| traI+ | Vector only | traD | 0.14 ± 0.03 |
| traI+ | Vector only | traM | 0.18 ± 0.07 |
| ΔtraI | traI+ | Vector only | 0.0011 ± 0.0001 |
| ΔtraI | traI+ | traD | 0.0009 ± 0.0001 |
| ΔtraI | traI+ | traM | 0.0008 ± 0.0003 |
Values are shown as means ± standard error of the means of two to four independent biological replicates.
DISCUSSION
To aid in the structure-function characterization of the F plasmid relaxase TraI, we generated a mutant library in which each mutant has a 31-residue insertion at a defined location in the protein. In conjugation and strand cleavage assays, the phenotypes of our mutants were consistent with the prevailing model that the core biochemical activities of DNA cleavage and unwinding lie in conserved regions between amino acid residues ∼1 to 310 and ∼990 to 1450 (4, 13, 14, 16, 28, 29). Furthermore, we identified numerous “permissive sites” at which the 31-residue insertion was functionally tolerated. We also found that the expression of wild-type TraI from our multicopy plasmid led to a dominant ∼100-fold reduction in the frequency of conjugation. Several of the mutations did not cause a dominant decrease in conjugation frequency, though they appeared otherwise normal in conjugation and oriT cleavage assays. Using three functional alleles, we found that the high-copy pTrc vector expressed ∼10-fold more TraI during growth under noninducing conditions than did F′lac. We hypothesize that increased amounts of TraI protein cause the repression of conjugation and that the unrestrictive mutant proteins have a defect in the normal repressive function.
Others have previously reported that complementing a ΔtraI strain with traI on a multicopy plasmid led to a reduction in conjugation frequency relative to the parental F-derived plasmid and speculated that the decrease in conjugation efficiency occurred because recipients were traI deficient and therefore unable to facilitate plasmid retransfer (28, 29). To test the hypothesis that a lack of plasmid retransfer could explain our data, we performed conjugation experiments in which the donors carried p99I+ in the presence of intact F′lac instead of F′ΔI. F transfer frequencies using these donors were reduced 100-fold relative to those of donors carrying the empty vector in place of p99I+, even though recipients were fully capable of plasmid retransfer. p99traI::Q839, p99traI::V996, and p99traI::Q1441 also displayed negative dominance in the presence of intact F′lac. p99traI::Q369, p99traI::A593, p99traI::L681, and p99traI::Q794 showed either no dominance or very modest negative dominance for conjugation, and F′traI::A593 exhibited partial resistance to conjugative repression by p99I+. Based on these observations, we rejected the retransfer hypothesis as an explanation for our results.
Taken together, our data suggest that TraI contains a negative regulatory domain that limits conjugation as TraI increases in abundance. This “restrictive” domain appears to span much of the region between the consensus relaxase and helicase sequences, encompassing at least residues 369 to 794. The presence of a negative regulatory domain in this region of TraI explains why defective mutants with lesions outside the restrictive domain (the V996 and Q1441 mutants) complemented conjugation better when expressed at single copy than at high copy, while the defective Q794 mutant complemented conjugation better at high copy than at single copy (Fig. 3). Such results highlight the complications inherent in performing complementation experiments in trans in this system.
TraIiQ839, which contains the i31 upstream of the conserved helicase domain, retains the restrictive activity, and we therefore believe that the negative regulatory domain does not overlap the consensus helicase domain. We infer that the restrictive domain does not require helicase function from the negative dominance of the V996 mutant, whose defect in conjugation is almost certainly due to a loss of helicase activity. However, we were unable to recapitulate the dominant-negative conjugation phenotype of overexpressed, wild-type TraI using internal deletions or successive C-terminal LacZ fusions. Internal deletion mutants, consistent with their lack of conjugative dominance, did not express levels of protein comparable to those of the full-length alleles. The failure of TraI-LacZ fusions to repress conjugation in trans suggests that the C-terminal region of TraI may be important for proper folding or function of the restrictive domain. Work is ongoing to assess the requirement for an intact C terminus in restrictive activity.
We observed that a 10-fold increase in TraI levels could have a large effect on the conjugative efficiency of a strain. Tenfold changes in TraI expression could conceivably occur during the lifetime of a given cell, depending on the conditions; for example, Frost and Manchak observed differential expression of TraI during different stages of growth in batch culture (10). The mechanism by which TraI's negative regulatory domain restricts conjugation remains unclear, however. This region of TraI is not similar to that of other proteins except closely related relaxases, so we cannot derive clues about its function by analogy to homologous sequences. We have ruled out the hypothesis that unrestrictive mutant proteins have an intrinsically higher single-stranded DNA cleavage activity in vivo, but it is possible that the negative regulatory domain could regulate TraI relaxase or helicase activity at some point during conjugation. However, the observation that increased TraI expression drives down conjugation frequency in trans and that this effect can be abrogated by specific mutations suggests instead that the protein may titrate out some vital component of the conjugative apparatus. Our data suggest that neither TraD nor TraM is limiting under conditions of TraI-mediated repression, but we do not rule out the sequestration of a binding partner(s) by TraI.
In conjugative dominance assays, neither of the tested helicase domain insertion mutations lowered conjugation frequencies more than wild-type TraI did. One dominant-negative helicase mutation has been described for the related relaxase TrwC, and it was postulated that the mutant protein cleaved plasmid DNA and then interfered with unwinding (24). Other helicase-defective TrwC mutants did not display negative dominance, however. Further work on the dominance of helicase mutants may shed light on the cooperativity of multiple relaxase molecules in processing plasmid DNA.
In addition to broadening our understanding of the functional domains of TraI, our mutant library allowed us to identify a number of permissive sites in the protein. These are sites at which insertions of a moderate size are tolerated, allowing experimenters to engineer in a tag or functional moiety with relative impunity. In this vein, one of our mutants has been used to construct a cuprous oxide binding derivative of TraI that enables the assembly of cuprous oxide nanoparticles under conditions in which Cu2O precipitation would normally be thermodynamically unfavorable (6). The development of other TraI derivatives with novel characteristics is ongoing.
In summary, we have generated a mutant library for assessing structure-function relationships of the F plasmid relaxase TraI. We have demonstrated the utility of this library by identifying and mapping a previously undiscovered regulatory domain with no known homologs outside of closely related relaxases. The library was generated on a cloning vector, but we recombined several different alleles onto the F plasmid in a markerless manner for complementation experiments in cis. The traI::i31 library should facilitate further characterization of the role of TraI in F plasmid conjugation, shedding light on relaxases, helicases, and the mechanisms of conjugation in general.
Acknowledgments
The work in the Traxler lab was supported by the National Science Foundation (MCB-0345018) and the U.S. Army Research Office DURINT Program (DAAD19-01-1-04999). Work performed in the Zechner lab was supported by FWF grants P16722-B12 and p18607-B12. G. Palacios was supported in part by the University of Washington STAR program (NHLBI 5T35 HL07763). R. Haft was supported in part by the National Institute of General Medical Sciences (NRSA T32 GM07270).
We are grateful to John Hodges and Lisa Winterroth for assistance in the construction of some strains. We thank Laura Frost, Colin Manoil, and Kelly Hughes for discussions, advice, and strains that facilitated this work.
REFERENCES
- 1.Achtman, M., N. Willetts, and A. J. Clark. 1971. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J. Bacteriol. 106:529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 3.Bochner, B. R., H. C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd, D. R., J. K. Sampson, H. M. Ragonese, and S. W. Matson. 2002. Structure-function analysis of Escherichia coli DNA helicase I reveals non-overlapping transesterase and helicase domains. J. Biol. Chem. 277:42645-42653. [DOI] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai, H., W. S. Choe, C. K. Thai, M. Sarikaya, B. A. Traxler, F. Baneyx, and D. T. Schwartz. 2005. Nonequilibrium synthesis and assembly of hybrid inorganic-protein nanostructures using an engineered DNA binding protein. J. Am. Chem. Soc. 127:15637-15643. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta, S., C. Larkin, and J. F. Schildbach. 2003. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure 11:1369-1379. [DOI] [PubMed] [Google Scholar]
- 9.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F-factor and mechanisms of conjugation, p. 2377-2382. In F. C. Neidhardt, R. Curtiss III, C. A. Gross, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 10.Frost, L. S., and J. Manchak. 1998. F− phenocopies: characterization of expression of the F transfer region in stationary phase. Microbiology 144:2579-2587. [DOI] [PubMed] [Google Scholar]
- 11.Grandoso, G., M. Llosa, J. C. Zabala, and F. de la Cruz. 1994. Purification and biochemical characterization of TrwC, the helicase involved in plasmid R388 conjugal DNA transfer. Eur. J. Biochem. 226:403-412. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, L. H., S. J. Sorensen, and L. B. Jensen. 1997. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene 186:167-173. [DOI] [PubMed] [Google Scholar]
- 13.Harley, M. J., and J. F. Schildbach. 2003. Swapping single-stranded DNA sequence specificities of relaxases from conjugative plasmids F and R100. Proc. Natl. Acad. Sci. USA 100:11243-11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harley, M. J., D. Toptygin, T. Troxler, and J. F. Schildbach. 2002. R150A mutant of F TraI relaxase domain: reduced affinity and specificity for single-stranded DNA and altered fluorescence anisotropy of a bound labeled oligonucleotide. Biochemistry 41:6460-6468. [DOI] [PubMed] [Google Scholar]
- 15.Howard, M. T., W. C. Nelson, and S. W. Matson. 1995. Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J. Biol. Chem. 270:28381-28386. [PubMed] [Google Scholar]
- 16.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karl, W., M. Bamberger, and E. L. Zechner. 2001. Transfer protein TraY of plasmid R1 stimulates TraI-catalyzed oriT cleavage in vivo. J. Bacteriol. 183:909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy, K. A., E. G. Gachelet, and B. Traxler. 2004. Evidence for multiple pathways in the assembly of the Escherichia coli maltose transport complex. J. Biol. Chem. 279:33290-33297. [DOI] [PubMed] [Google Scholar]
- 19.Kupelwieser, G., M. Schwab, G. Hogenauer, G. Koraimann, and E. L. Zechner. 1998. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 275:81-94. [DOI] [PubMed] [Google Scholar]
- 20.Larkin, C., S. Datta, M. J. Harley, B. J. Anderson, A. Ebie, V. Hargreaves, and J. F. Schildbach. 2005. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure 13:1533-1544. [DOI] [PubMed] [Google Scholar]
- 21.Lee, M. H., N. Kosuk, J. Bailey, B. Traxler, and C. Manoil. 1999. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J. Bacteriol. 181:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lippincott, J., and B. Traxler. 1997. MalFGK complex assembly and transport and regulatory characteristics of MalK insertion mutants. J. Bacteriol. 179:1337-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Llosa, M., G. Grandoso, M. A. Hernando, and F. de la Cruz. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264:56-67. [DOI] [PubMed] [Google Scholar]
- 25.Manoil, C., and J. Bailey. 1997. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J. Mol. Biol. 267:250-263. [DOI] [PubMed] [Google Scholar]
- 26.Manoil, C., and B. Traxler. 2000. Insertion of in-frame sequence tags into proteins using transposons. Methods 20:55-61. [DOI] [PubMed] [Google Scholar]
- 27.Matson, S., and B. Morton. 1991. Escherichia coli DNA helicase I catalyzes a site- and strand-specific nicking reaction at the F plasmid oriT. J. Biol. Chem. 266:16232-16237. [PubMed] [Google Scholar]
- 28.Matson, S. W., and H. Ragonese. 2005. The F-plasmid TraI protein contains three functional domains required for conjugative DNA strand transfer. J. Bacteriol. 187:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matson, S. W., J. K. Sampson, and D. R. Byrd. 2001. F plasmid conjugative DNA transfer: the TraI helicase activity is essential for DNA strand transfer. J. Biol. Chem. 276:2372-2379. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Nelson, B., C. Manoil, and B. Traxler. 1997. Insertion mutagenesis of the lac repressor and its implications for structure-function analysis. J. Bacteriol. 179:3721-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, W. C., M. T. Howard, J. A. Sherman, and S. W. Matson. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 270:28374-28380. [PubMed] [Google Scholar]
- 33.Panagiotidis, C., M. Reyes, A. Sievertsen, W. Boos, and H. Shuman. 1993. Characterization of the structural requirements for assembly and nucleotide binding of an ATP-binding cassette transporter: the maltose transport system of Escherichia coli. J. Biol. Chem. 268:23685-23696. [PubMed] [Google Scholar]
- 34.Panicker, M. M., and E. G. Minkley, Jr. 1985. DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. J. Bacteriol. 162:584-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters, J. E., T. E. Thate, and N. L. Craig. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pölzleitner, E., E. L. Zechner, W. Renner, R. Fratte, B. Jauk, G. Hogenauer, and G. Koraimann. 1997. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol. Microbiol. 25:495-507. [DOI] [PubMed] [Google Scholar]
- 37.Rietsch, A., D. Belin, N. Martin, and J. Beckwith. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, K. D., E. Andersen-Nissen, F. Hayashi, K. Strobe, M. A. Bergman, S. L. R. Barrett, B. T. Cookson, and A. Aderem. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4:1247-1253. [DOI] [PubMed] [Google Scholar]
- 40.Stern, J. C., and J. F. Schildbach. 2001. DNA recognition by F factor TraI36: highly sequence-specific binding of single-stranded DNA. Biochemistry 40:11586-11595. [DOI] [PubMed] [Google Scholar]
- 41.Street, L. M., M. J. Harley, J. C. Stern, C. Larkin, S. L. Williams, D. L. Miller, J. A. Dohm, M. E. Rodgers, and J. F. Schildbach. 2003. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim. Biophys. Acta 1646:86-99. [DOI] [PubMed] [Google Scholar]
- 42.Traxler, B. A., and E. G. Minkley, Jr. 1988. Evidence that DNA helicase I and oriT site-specific nicking are both functions of the F TraI protein. J. Mol. Biol. 204:205-209. [DOI] [PubMed] [Google Scholar]
- 43.Traxler, B. A., and E. G. Minkley, Jr. 1987. Revised genetic map of the distal end of the F transfer operon: implications for DNA helicase I, nicking at oriT, and conjugal DNA transport. J. Bacteriol. 169:3251-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters, C. M., and G. M. Dunny. 2001. Analysis of functional domains of the Enterococcus faecalis pheromone-induced surface protein aggregation substance. J. Bacteriol. 183:5659-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters, C. M., H. Hirt, J. K. McCormick, P. M. Schlievert, C. L. Wells, and G. M. Dunny. 2004. An amino-terminal domain of Enterococcus faecalis aggregation substance is required for aggregation, bacterial internalization by epithelial cells and binding to lipoteichoic acid. Mol. Microbiol. 52:1159-1171. [DOI] [PubMed] [Google Scholar]
- 46.Wolkow, C. A., R. T. DeBoy, and N. L. Craig. 1996. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 10:2145-2157. [DOI] [PubMed] [Google Scholar]
- 47.Zechner, E. L., H. Pruger, E. Grohmann, M. Espinosa, and G. Hogenauer. 1997. Specific cleavage of chromosomal and plasmid DNA strands in gram-positive and gram-negative bacteria can be detected with nucleotide resolution. Proc. Natl. Acad. Sci. USA 94:7435-7440. [DOI] [PMC free article] [PubMed] [Google Scholar]