Abstract
Negative regulation of epr in Bacillus subtilis 168 is mediated jointly by both ScoC and SinR, which bind to their respective target sites 62 bp apart. Increasing the distance between the two sites abolishes repression, indicating that the two proteins interact, thereby suggesting a mechanism of corepression.
The expression of many of the genes coding for enzymes such as proteases, amylases, etc. (28), is repressed in the exponential phase by a group of regulatory proteins called “transition state regulators” (15, 27, 34). AbrB, ScoC, and SinR are among the best-characterized transition state regulators (33, 34, 36). Both ScoC and SinR are known to be negative regulators of protease production that bind to a DNA sequence whose consensus sequences appear to be 5′-RATANTATY-3′ (14, 16, 27, 35) and 5′-GNCNCGAAATACA-3′, respectively (12, 31). The active state of SinR is in the tetrameric form, and its activity is antagonized by SinI (3, 6, 21).
We had previously shown that Epr, a minor extracellular protease in Bacillus subtilis (5, 32), is transcribed by a σD-dependent RNA polymerase and that it is involved in swarm activity (8, 25). We were thus interested in determining how this gene is regulated. In this study, we show that negative regulation of epr requires both ScoC and SinR and that their mode of action appears to be through a mechanism of corepression.
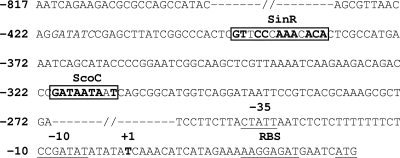
Inspection of the sequence upstream of the epr promoter revealed the presence of putative ScoC and SinR binding sites, 5′-GATAATAAT-3′ and 5′-GTTCCCAAACACA-3′, respectively (Fig. 1), that display an 8/9 match and a 10/13 match with the consensus binding sites for ScoC and SinR. To determine whether these two sites conferred negative regulation on epr, two DNA fragments of 457 bp (−424 to +33) and 343 bp (−310 to +33), with and without the two sites, respectively, and containing the epr promoter, ribosome binding site (RBS), and ATG were PCR amplified from pPZ (Table 1) with primers KKR28/KKR36 and KKR103/KKR36 (Table 2). The amplified products were digested with HindIII/BamHI and PstI/BamHI, respectively, fused in the translational frame to the lacZ gene in pRB381, a replicative multicopy plasmid (4), to give pSZ and pHZ (Table 1) and then were transformed into B. subtilis 168 to give 168-SZ and 168-HZ, respectively. Both strains were grown at 37°C in Penassay broth to the stationary phase (optical density at 600 nm [OD600] of ∼2.0), and the β-galactosidase activities were determined (26). The activity in 168-HZ was 3,500 Miller units, as compared to 200 Miller units in 168-SZ, indicating that the region between −422 and −308, containing the putative binding sites for ScoC and SinR, negatively regulates epr expression. Further deletions from −308 to −70 did not show any significant change in activity compared to 168-HZ (data not shown).
FIG. 1.
Sequence of epr promoter region (−817 to +33) (5, 8, 30). ScoC and SinR binding sites are indicated by the boxed nucleotides. Nucleotides marked in bold within these sites are identical to the consensus recognition sequence (16, 29).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Description or genotypea | Source or referenceb |
|---|---|---|
| Plasmids | ||
| pRB381 | E. coli-Bacillus shuttle vector for translational fusion with β-galactosidase gene; ′lacZ Kmr Apr | BGSC (4) |
| pPZ | pRB381 bearing 850-bp insert (−817 to +33) containing ScoC and SinR binding sites, epr promoter, RBS, and ATG in translation fusion with lacZ gene | Laboratory stock |
| pSZ | pRB381 bearing 455-bp insert (−422 to +33) containing ScoC and SinR binding sites, epr promoter, RBS, and ATG in translation fusion with lacZ gene | This study |
| pHZ | pRB381 bearing 341-bp insert (−312 to +33) containing epr promoter, RBS, and ATG in translation fusion with lacZ gene | This study |
| pSHZ | pSZ with mutated ScoC binding site | This study |
| pSRZ | pSZ with mutated SinR binding site | This study |
| pS200Z | pSZ with 200-bp DNA insertion between ScoC and SinR binding sites | This study |
| Strains | ||
| 168 | trpC2 | BGSC |
| 168-SZ | B. subtilis 168 bearing pSZ | This study |
| 168-HZ | B. subtilis 168 bearing pHZ | This study |
| 168ΔH | trpC2 scoC::Spr | Laboratory stock |
| 168ΔR | trpC2 sinR::Cmr | Laboratory stock |
| 168ΔH-SZ | B. subtilis 168ΔH bearing pSZ | This study |
| 168ΔR-SZ | B. subtilis 168ΔR bearing pSZ | This study |
| 168-SHZ | B. subtilis 168 bearing pSHZ | This study |
| 168-SRZ | B. subtilis 168 bearing pSRZ | This study |
| 168-S200Z | B. subtilis 168 bearing pS200Z | This study |
Kmr, kanamycin resistance; Apr, ampicillin resistance; Spr, spectinomycin resistance; Cmr, chloramphenicol resistance.
BGSC, Bacillus Genetic Stock Center.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Restriction site(s)b |
|---|---|---|
| KKR28 | GGTAAAGCTTAGGATATCCGAGC | HindIII then EcoRV |
| KKR36 | CTTAGGATCCATGATTCATCTCC | BamHI |
| KKR67 | CGTAAGCTTAATCAGAAGACGCGC | HindIII |
| KKR103 | CGACTGCAGCGGCATGGTCAGGAT | PstI |
| KKR127 | CGCTTTGCGTGACGGATTATC | |
| KKR215 | GCACCCGGGCAGCGGCATGGTCA | SmaI |
| KKR216 | GCACCCGGGATCGGGTCTGTCTTC | SmaI |
| KKR217 | ATTTATAACTGCCCATGAAATCAGC | SspI |
| KKR218 | ATTAAAAGTGGGCCGATAAGGTC | SspI |
| KKR253 | GCAGCTAGCAGCTCGTTAAAATCAAG | NheI |
| KKR254 | CGAGCTAGCTTCCGGGGTATGCTG | NheI |
Nucleotides in boldface are complementary to the genome sequence.
Underlined in the corresponding sequence.
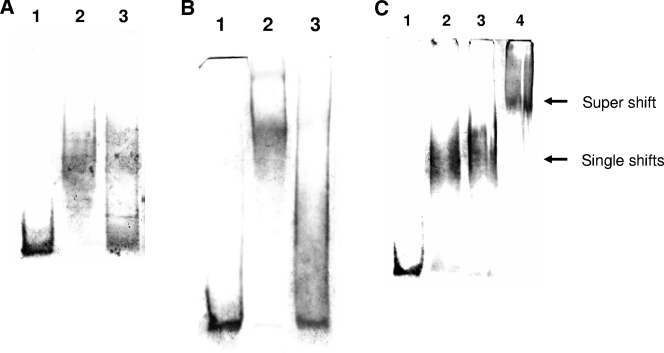
We then assessed whether ScoC and SinR bind to their respective sites by the electrophoretic mobility shift assay (EMSA). The genes coding for ScoC (27) and SinR (11) were cloned into pET28a and pET43.1b, respectively, expressed in Escherichia coli BL21(DE3), and the two proteins were purified on Ni-nitrilotriacetic acid columns. Binding reactions were carried out in a 20-μl reaction mixture at 37°C for 15 min, with either 1 μM ScoC or 12 μM SinR, and a 150-bp DNA containing both binding sites was obtained by PCR amplification from pPZ with primers KKR28 and KKR127 (Table 2) and end labeled with digoxigenin (DIG). (Procedures for DNA labeling, DNA binding, and detection were performed as per Roche Applied Science, catalog no. 3353591.) The reaction mixture was electrophoresed on a 5% polyacrylamide gel and electroblotted onto Nylon membrane, and DNA was detected with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP). Figure 2A shows that the DNA probe is significantly retarded in the presence of ScoC (lane 2) compared to in its absence (lane 1). The amount of DNA probe showing retardation is significantly reduced, with a 100× molar excess of unlabeled probe (lane 3) showing the specificity of binding. Similarly in Fig. 2B, the probe is retarded in the presence of SinR (lane 2) compared to in its absence (lane 1). The retardation is abolished, with a 100× molar excess of unlabeled probe (lane 3) showing the specificity of binding. Thirty-base-pair oligonucleotides containing either the putative ScoC or SinR binding site also were able to compete with ScoC or SinR binding to the labeled probe, respectively, but not reciprocally (data not shown). We thus conclude that both ScoC and SinR bind to a specific site within the epr promoter.
FIG. 2.
(A) EMSA with ScoC. Binding reactions were carried out with 10 nM DIG-labeled epr probe (∼150 bp) and ScoC (1 μM) in a 20-μl reaction buffer containing 20 mM HEPES, pH 7.6, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol, Tween 20 (0.2% [wt/vol]), 30 mM KCl, 1 μg poly(dI-dC), and 0.1 μg poly-l-lysine at 37°C for 15 min. The bound product was electrophoresed on a 5% polyacrylamide gel in 0.25× Tris-borate-EDTA buffer at 4°C and electroblotted onto Nylon membrane, and DNA was detected with NBT/BCIP (Roche Applied Science; www.roche-applied-science.com/pack-insert/3353591a.pdf). Lane 1, DIG-labeled epr probe; lane 2, epr probe plus ScoC; and lane 3, epr probe plus ScoC plus 100× molar excess unlabeled probe. (B) EMSA with SinR (12 μM). (The binding conditions, electrophoresis, and detection method are as described for panel A. Lane 1, DIG-labeled epr probe (∼150 bp, 10 nM); lane 2, epr probe plus SinR; and lane 3, epr probe plus SinR plus 100× molar excess unlabeled probe. (C) EMSA with ScoC (1 μM) and SinR (12 μM). (The binding conditions, electrophoresis, and detection method are as described for panel A). Lane 1, DIG-labeled epr probe (∼150 bp, 10 nM); lane 2, epr probe plus ScoC; lane 3, epr probe plus SinR; and lane 4, epr probe plus ScoC plus SinR.
To determine whether ScoC and SinR were involved in the repression of epr, we transformed pSZ into 168ΔH and 168ΔR (Table 1), scoC and sinR disruptants, respectively, to give 168ΔH-SZ and 168ΔR-SZ. The β-galactosidase activities in 168ΔH-SZ, 168ΔR-SZ, and 168-SZ were 3,400, 2,000, and 200 Miller units, respectively, showing that negative regulation of epr was dependent on both ScoC and SinR and that neither, individually, can fully repress epr. This observation thus suggests a mechanism of corepression that may involve interaction of the two proteins. Since SinI is also known to interact with SinR (3), it may regulate epr expression by preventing corepression. The reduced level of derepression in 168ΔR-SZ compared to 168ΔH-SZ may be due to a reduction in σD levels in the sinR disruptant since SinR appears to positively regulate σD levels via FlgM (9).
In further support of the idea that both ScoC and SinR were required for repression, we made two constructs, pSHZ and pSRZ (Table 1), carrying either a mutated ScoC (5′-GATCCCGGG-3′) or mutated SinR (5′-TTAATATTTATAA-3′) binding site (mutations underlined), respectively, but identical in all other respects to pSZ and assessed whether they independently lead to relief from repression. To create the mutated ScoC binding site, two PCR products were obtained from pPZ with primers KKR67/KKR216 and KKR215/KKR36 (Table 2). The first product was restricted with HindIII and SmaI and cloned into pBluescript SK+. The second PCR product was restricted with SmaI and BamHI and cloned downstream of the first PCR product in pBluescript SK+. The epr promoter containing the mutated ScoC binding site was reamplified with primers KKR28/KKR36 (Table 2) and cloned in pRB381 to give pSHZ. pSRZ was similarly constructed from two PCR products obtained from pPZ with primers KKR67/KKR218 and KKR217/KKR36 (Table 2) and sequentially cloned into pBR322 at HindIII/SspI and SspI/BamHI, respectively. The fused product was reamplified with primers KKR28/KKR36 and cloned in pRB381 to give pSRZ. Mutations of ScoC and SinR binding sites were confirmed by digestions with SmaI and SspI, respectively, as these sites were introduced in the primers to create the mutations. pSHZ and pSRZ were introduced into B. subtilis 168 to give 168-SHZ and 168-SRZ, and the promoter activity was compared with those of 168-SZ and 168-HZ. The β-galactosidase activities in the three strains 168-SHZ, 168-SRZ, and 168-HZ were very similar (3,450, 3,550, and 3,500 Miller units, respectively), as compared to 200 Miller units in 168-SZ. The results show that the ScoC and SinR binding sites are important for negative regulation of epr and that mutation of either of them completely relieves repression. The observation of an “all-or-none” repression once again emphasizes the requirement of both proteins for repression. Figure 2C shows that both proteins are capable of binding to the epr promoter, as evidenced by a supershift in retardation in the EMSA (lane 4) compared to ScoC or SinR alone (lanes 2 and 3, respectively), indicating that they do not affect each other's binding. Furthermore, mutation of either the ScoC or SinR binding site eliminated the binding of their specific repressor but not the other (data not shown). Taken together, our results suggest that binding of the two repressors to their respective sites could result in a synergistic interaction between the two proteins and that the distance between the two sites could be critical for their interaction. The distance between the ScoC and SinR binding sites is 62 bp. If the distance between the two sites were increased, then repression by ScoC and SinR might be abolished. To determine if this was the case, the promoter activity in a construct, pS200Z (Table 1), in which the distance between the two sites was separated by an additional 200 bp was compared with that of pSZ in B. subtilis 168. pS200Z was constructed by PCR amplification of two products obtained from pPZ with primers KKR67/KKR253 and KKR254/KKR36 (Table 2) and cloned sequentially into pBR322 at HindIII/NheI and NheI/BamHI, respectively. A 200-bp DNA fragment was derived from plasmid pET3a by EcoRV digestion and inserted between the ScoC and SinR binding sites within the pBR322 recombinant that was restricted with NheI and filled in with Klenow enzyme. The epr promoter segment was reamplified with primers KKR28/KKR36 and cloned in pRB381 to give pS200Z, which was introduced into B. subtilis 168 to give 168-S200Z. Whereas the β-galactosidase activity in 168-SZ was 200 Miller units, the activity in 168-S200Z was 3,300 Miller units, comparable to that of 168-HZ (3,500 Miller units), showing the dependence on distance for repression by ScoC and SinR and thus suggesting the interaction of the two proteins for corepression. Insertion of the 200-bp DNA does not, however, affect the binding of the two proteins, as evidenced by the observation of a supershift in the presence of the two proteins (data not shown).
Corepression by ScoC and SinR has not been previously reported in B. subtilis, although the capability of these two proteins to interact has been demonstrated in a LexA-based bacterial two-hybrid system (30). In fact, there appear to be only a few examples of corepression described in both prokaryotes and eukaryotes (7, 23, 24, 38). Several mechanisms have been described for corepression. They may involve direct contacts between proteins that bind DNA, as observed with CytR and cyclic AMP (cAMP)-cAMP receptor protein (CRP) in E. coli (17, 18, 29) and with MecI and BlaI in Staphylococcus aureus (24). The interaction between two DNA binding proteins may either require an additional factor to link the two proteins, as in the case of the nuclear protein CBP that links the basal transcription factor TFIIB with CREB (19), or may assist in the bending of DNA, thereby facilitating the interaction of the two proteins. In E. coli, bending of DNA by integration host factor (IHF) and HU facilitates interaction between flanking DNA-bound dimers of ParB (10, 13) and GalR (1, 2, 22), respectively. In some instances, one protein may regulate another DNA binding protein, as in the case of bacteriophage P1, in which the Bof protein affects the conformation of C1 and stimulates its binding with DNA (20, 37). In another instance, in phage P1, Doc and Phd autoregulate their own transcription by corepression (23). When only Phd was expressed, partial repression of the operon was observed. However, when both Phd and Doc were coexpressed, there was a dramatic enhancement in repression. In contrast, partial repression of epr is not observed with ScoC or SinR alone. Only when both are present does repression of epr occurs. It is possible that this system could be used to screen a library of genes whose products interfere with corepression, allowing one to identify proteins that interact with ScoC, SinR, or both proteins.
Acknowledgments
P. Kodgire acknowledges the Council for Scientific and Industrial Research, India, for the Ph.D. research fellowship [9/87(328)/2003-EMR-I].
REFERENCES
- 1.Aki, T., and S. Adhya. 1997. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 16:3666-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aki, T., H. E. Choy, and S. Adhya. 1996. Histone-like protein HU as a specific transcriptional regulator: co-factor role in repression of gal transcription by GAL repressor. Genes Cells 1:179-188. [DOI] [PubMed] [Google Scholar]
- 3.Bai, U., I. Mandic-Mulec, and I. Smith. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 7:139-148. [DOI] [PubMed] [Google Scholar]
- 4.Bruckner, R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187-192. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner, R., O. Shosheyov, and R. H. Doi. 1990. Multiple active forms of a novel serine protease from Bacillus subtilis. Mol. Gen. Genet. 221:486-490. [DOI] [PubMed] [Google Scholar]
- 6.Cervin, M. A., R. J. Lewis, J. A. Brannigan, and G. B. Spiegelman. 1998. The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase. Nucleic Acids Res. 26:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeSilva, H., K. Lee, and M. A. Osley. 1998. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics 148:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixit, M., C. S. Murudkar, and K. K. Rao. 2002. epr is transcribed from a σD promoter and is involved in swarming of Bacillus subtilis. J. Bacteriol. 184:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fredrick, K., and J. D. Helmann. 1996. FlgM is a primary regulator of σD activity, and its absence restores motility to a sinR mutant. J. Bacteriol. 178:7010-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funnell, B. E. 1991. The P1 plasmid partition complex at parS. The influence of Escherichia coli integration host factor and of substrate topology. J. Biol. Chem. 266:14328-14337. [PubMed] [Google Scholar]
- 11.Gaur, N. K., E. Dubnau, and I. Smith. 1986. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J. Bacteriol. 168:860-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur, N. K., J. Oppenheim, and I. Smith. 1991. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173:678-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes, F., and S. Austin. 1994. Topological scanning of the P1 plasmid partition site. J. Mol. Biol. 243:190-198. [DOI] [PubMed] [Google Scholar]
- 14.Henner, D. J., E. Ferrari, M. Perego, and J. A. Hoch. 1988. Location of the targets of the hpr-97, sacU32(Hy), and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J. Bacteriol. 170:296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoch, J. A. 1993. spo0 genes, the phosphorelay, and the initiation of sporulation, p. 747-756. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 16.Kallio, P. T., J. E. Fagelson, J. A. Hoch, and M. A. Strauch. 1991. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266:13411-13417. [PubMed] [Google Scholar]
- 17.Kallipolitis, B. H., M. Norregaard-Madsen, and P. Valentin-Hansen. 1997. Protein-protein communication: structural model of the repression complex formed by CytR and the global regulator CRP. Cell 89:1101-1109. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen, H. H., P. Valentin-Hansen, and L. Sogaard-Andersen. 1996. CytR/cAMP-CRP nucleoprotein formation in E. coli: the CytR repressor binds its operator as a stable dimer in a ternary complex with cAMP-CRP. J. Mol. Biol. 260:113-119. [DOI] [PubMed] [Google Scholar]
- 19.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223-226. [DOI] [PubMed] [Google Scholar]
- 20.Lehnherr, H., M. Velleman, A. Guidolin, and W. Arber. 1992. Bacteriophage P1 gene 10 is expressed from a promoter-operator sequence controlled by C1 and Bof proteins. J. Bacteriol. 174:6138-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, R. J., J. A. Brannigan, W. A. Offen, I. Smith, and A. J. Wilkinson. 1998. An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J. Mol. Biol. 283:907-912. [DOI] [PubMed] [Google Scholar]
- 22.Lyubchenko, Y. L., L. S. Shlyakhtenko, T. Aki, and S. Adhya. 1997. Atomic force microscopic demonstration of DNA looping by GalR and HU. Nucleic Acids Res. 25:873-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnuson, R., and M. B. Yarmolinsky. 1998. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 180:6342-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and β-lactamase regulators. J. Bacteriol. 183:6862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murudkar, C. S., P. Kodgire, and K. K. Rao. 2006. The carboxy terminal domain of Epr, a minor extracellular serine protease, is essential for the swarming motility of Bacillus subtilis 168. FEMS Microbiol. Lett. 257:24-31. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 442-443. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., New York, N.Y.
- 27.Perego, M., and J. A. Hoch. 1988. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J. Bacteriol. 170:2560-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priest, F. G. 1977. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol. Rev. 41:711-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen, P. B., B. Holst, and P. Valentin-Hansen. 1996. Dual-function regulators: the cAMP receptor protein and the CytR regulator can act either to repress or to activate transcription depending on the context. Proc. Natl. Acad. Sci. USA 93:10151-10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez, A., and J. Olmos. 2004. Bacillus subtilis transcriptional regulators interaction. Biotechnol. Lett. 26:403-407. [DOI] [PubMed] [Google Scholar]
- 31.Shafikhani, S. H., I. Mandic-Mulec, M. A. Strauch, I. Smith, and T. Leighton. 2002. Postexponential regulation of sin operon expression in Bacillus subtilis. J. Bacteriol. 184:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sloma, A., A. Ally, D. Ally, and J. Pero. 1988. Gene encoding a minor extracellular protease in Bacillus subtilis. J. Bacteriol. 170:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, I. 1993. Regulatory proteins that control late-growth developement, p. 785-800. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 34.Strauch, M. A. 1993. AbrB, a transition state regulator, p. 757-764. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 35.Strauch, M. A., and J. A. Hoch. 1992. Control of postexponential gene expression by transition state regulators, p. 105-121. In R. H. Doi and M. McGloughlin (ed.), Biology of bacilli: application to industry. Butterworth-Heinemann, Stoneham, Mass. [PubMed]
- 36.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velleman, M., T. Heinzel, and H. Schuster. 1992. The Bof protein of bacteriophage P1 exerts its modulating function by formation of a ternary complex with operator DNA and C1 repressor. J. Biol. Chem. 267:12174-12181. [PubMed] [Google Scholar]
- 38.Wade, P. A., and J. A. Jaehning. 1996. Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol. Cell. Biol. 16:1641-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]