Abstract
The wall-less prokaryote Mycoplasma pneumoniae, a common cause of chronic respiratory tract infections in humans, is considered to be among the smallest and simplest known cells capable of self-replication, yet it has a complex architecture with a novel cytoskeleton and a differentiated terminal organelle that function in adherence, cell division, and gliding motility. Recent findings have begun to elucidate the hierarchy of protein interactions required for terminal organelle assembly, but the engineering of its gliding machinery is largely unknown. In the current study, we assessed gliding in cytadherence mutants lacking terminal organelle proteins B, C, P1, and HMW1. Furthermore, we screened over 3,500 M. pneumoniae transposon mutants individually to identify genes associated with gliding but dispensable for cytadherence. Forty-seven transformants having motility defects were characterized further, with transposon insertions mapping to 32 different open reading frames widely distributed throughout the M. pneumoniae genome; 30 of these were dispensable for cytadherence. We confirmed the clonality of selected transformants by Southern blot hybridization and PCR analysis and characterized satellite growth and gliding by microcinematography. For some mutants, satellite growth was absent or developed more slowly than that of the wild type. Others produced lawn-like growth largely devoid of typical microcolonies, while still others had a dull, asymmetrical leading edge or a filamentous appearance of colony spreading. All mutants exhibited substantially reduced gliding velocities and/or frequencies. These findings significantly expand our understanding of the complexity of M. pneumoniae gliding and the identity of possible elements of the gliding machinery, providing a foundation for a detailed analysis of the engineering and regulation of motility in this unusual prokaryote.
The cell wall-less prokaryote Mycoplasma pneumoniae establishes chronic infections of the human respiratory tract that result in bronchitis and atypical or “walking” pneumonia and account for up to 30% of all cases of community-acquired pneumonia (49). With a minimal genome lacking genes for typical transcriptional regulators, two-component systems, and pathways for de novo synthesis of most macromolecular building blocks, including nucleotides and amino acids (8, 21), M. pneumoniae is considered to be among the simplest organisms capable of self-replication; yet this unusual species exhibits remarkable architectural complexity, with a dynamic cytoskeleton and a specialized, membrane-bound polar cell extension or terminal organelle that has an elaborate macromolecular core (2, 12, 20, 33). The terminal organelle functions in diverse cellular processes that include adherence to host epithelium (cytadherence) and cell division (7, 17, 40) and engages as the leading end as M. pneumoniae cells glide over solid surfaces (3). Recent studies have begun to establish the assembly sequence in terminal organelle development and the hierarchy of protein interactions required for core stability and adhesin trafficking (17, 26, 41), but the macromolecular components and engineering of the gliding machinery are unknown. Inspection of the 816-kbp M. pneumoniae genome reveals no homology to proteins that function in bacterial motility of any type in walled species, and even within the genus Mycoplasma there appear to be distinct gliding mechanisms, since known components of the Mycoplasma mobile gliding machinery are not found in M. pneumoniae (23, 42, 47, 48).
Analysis by electron cryotomography suggests that the M. pneumoniae terminal organelle core is conformationally flexible, prompting speculation that this structure powers gliding by cyclic contraction and extension (20). Although the constituents of the terminal organelle core are largely unknown, proteins HMW1, HMW2, and HMW3 are believed to be involved in core architecture, as they localize to the terminal organelle, are required for core development and stability, and partition in the Triton X-100-insoluble cytoskeletal fraction, of which the terminal organelle core is a prominent component (12, 26, 33). However, the impact of the loss of these proteins on cell gliding has not been assessed. Furthermore, about 100 proteins comprise the M. pneumoniae cytoskeletal fraction (36), many of which have no assigned function.
Cytadhesins P1 and P30 are, to date, the only M. pneumoniae proteins for which a role in gliding has been implicated. Gliding velocity and substrate binding are reduced in the presence of P1-specific antibodies (39), and the loss of P30 renders M. pneumoniae noncytadherent and nonmotile (15). A wild-type phenotype is restored in the P30− mutant with transposon delivery of the wild-type MPN453 allele encoding P30 (15). In contrast, the altered P30 allele P30R, differing from P30 by 17 residues in the middle of the protein, confers cytadherence at about 70% of the wild-type level but confers gliding velocity at only 5% of the wild-type level (15). Cytadherence and gliding are also separable in the closely related organism Mycoplasma genitalium, where transposon insertion in MG200 or MG386 significantly reduces cell gliding velocity and frequency but only marginally affects cytadherence (34).
In the current study, we established that the M. pneumoniae terminal organelle proteins HMW1 as well as B and/or C are required for gliding motility. Furthermore, we identified M. pneumoniae genes specifically associated with cell gliding but dispensable for cytadherence. Over 3,500 transformants were screened individually, approximately 100 of which had a satellite growth-altered (SGA) phenotype, indicating a likely gliding defect (15). This manifested as the complete absence or impaired development of satellite growth; the production of a lawn-like growth largely devoid of typical microcolonies; the presence of a dull, asymmetrical leading edge of colony spreading; or a filamentous appearance of spreading. Gene disruptions were defined for each SGA mutant, identifying 47 distinct transposon insertions in 32 genes, which were grouped according to their predicted functions. Hemadsorption (HA) was assessed for each mutant, which identified 30 genes associated with gliding motility but dispensable for cytadherence. Gliding by individual cells was measured by microcinematography for selected mutants, each exhibiting substantially reduced gliding velocities and/or gliding frequencies. Finally, representative transformants were evaluated for likely polar effects on transcription based on transposon orientation relative to flanking genes and the positions of probable promoter sequences.
MATERIALS AND METHODS
Mycoplasma strains and culture conditions.
Wild-type M. pneumoniae strain M129 (31), mutant II-3 with recombinant Tn4001 carrying a wild-type P30 allele (II-3/P30WT) (15), and cytadherence mutants III-4, IV-22 (27), and M6 (29) were described previously (Table 1). Wild-type M129 was transformed with transposon Tn4001.2065 (25) in four independent electroporations according to established procedures (19). Mixed transformant populations were transferred into tissue culture flasks containing 25 ml Hayflick medium (18) plus 18 μg/ml gentamicin and grown to mid-log phase. Gentamicin selection was maintained for subsequent cultures of transformants except as indicated. Polystyrene-adherent cells were recovered in order to enrich for transformants likely to retain cytadherence. These cells were diluted in fresh medium in tissue culture flasks, again enriched for polystyrene adherence, collected in fresh medium, aliquoted, and stored at −80°C.
TABLE 1.
Protein profiles, gliding capacity, and glass binding of wild-type M. pneumoniae and the indicated cytadherence-negative mutants
| Strain | Level of cytadherence-associated proteinsa
|
Mean cell velocity (μm/s) | Mean cell gliding frequency (%) | Mean glass-binding capability (% of wild type) | |||||
|---|---|---|---|---|---|---|---|---|---|
| P1 | B/C | P30 | HMW1 | HMW2 | HMW3 | ||||
| Wild type | +++ | +++ | +++ | +++ | +++ | +++ | 0.32 | 28 | 100 |
| III-4 | +++ | − | +++ | +++ | +++ | +++ | 0 | 0 | 104 |
| IV-22 | − | − | +++ | +++ | +++ | +++ | 0 | 0 | 78c |
| M6 | +++ | +++ | ++b | − | + | + | 0 | 0 | 92 |
Relative levels of the indicated proteins are represented as follows: +++, wild-type levels; ++, slightly reduced levels; +, substantially reduced levels; −, protein is absent.
Protein is truncated.
Significantly lower than wild-type levels (P < 0.05).
Isolation of motility mutants.
Mixed transformant populations were thawed, passed twice through 0.22-μm filters to disperse aggregates, diluted, and plated onto PPLO agar (6). After 7 days at 37°C, plates were overlaid with blood agar, and 2 days later, colonies were visible as hemolytic plaques, making it easier to pick individual transformants. These transformants were selected at random, cultured in 400 μl of Hayflick medium to mid-log phase (6 to 8 days), and stored at −80°C. Individual transformants were later thawed, diluted, and inoculated into 600 μl Hayflick medium containing 3% gelatin in individual wells of 24-well tissue culture plates. Satellite growth around microcolonies was evaluated by phase-contrast microscopy using a Nikon DIAPHOT microscope (×20 objective) at 24-h intervals after 4 to 8 days, relative to positive controls (wild-type M. pneumoniae and strain II-3/P30WT) (15). Transformants exhibiting reduced or altered satellite growth were screened two additional times in separate experiments to verify the altered phenotype before cataloging and storage.
Insertion mapping and filter cloning.
Transposon Tn4001.2065 contains a HindIII site immediately downstream of a promoterless β-galactosidase gene, an Escherichia coli replication origin, and a β-lactamase gene within the IS256L element (25). To map transposon insertion sites, genomic DNA isolated from individual M. pneumoniae transformants was digested with HindIII, diluted, religated, and transformed into E. coli. Plasmid DNA isolated from ampicillin-resistant E. coli clones was sequenced across the insertion site using the transposon-specific, outward-directed primer 5′-CACTCCAGCCAGCTTTCCGGCACCGCTTCT-3′ for comparison with the published genome sequence (8, 21). Each primary transformant stock was then filtered sequentially through 0.45-and 0.22-μm-pore-size filters, serially diluted, and plated onto PPLO agar. Eight progeny colonies were isolated for each stock and reevaluated for satellite growth as described above. Southern hybridization was performed (38) using multiple probes to ensure the presence of Tn4001.2065 in a single copy and that IS256 had not duplicated independently and inserted elsewhere in the genome.
Western immunoblotting.
Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting, as described previously (15), using a monoclonal P30-specific antibody (1) or rabbit antiserum to P1 (50), P24 (10), P65 (35), P41 (10), B (50), C (50), HMW1 (43), HMW2 (53), or HMW3 (44).
Analysis of glass binding and HA.
Binding of mycoplasmas to glass was measured as described previously (15). HA is a convenient model for M. pneumoniae cytadherence and was measured qualitatively as described previously (27) except that sheep blood was used.
Time lapse analysis of satellite growth.
Satellite growth was evaluated for a minimum of three filter-cloned progeny per primary transformant. Cultures were diluted in SP-4 medium (45) plus 3% gelatin, inoculated into four-well borosilicate glass chamber slides (Nunc Nalgene, Naperville, IL), and incubated at 37°C. Satellite growth around colonies was recorded at 12-h intervals (15).
Quantitation of cell gliding.
Average and corrected gliding velocities, gliding frequencies, and percentage of time that cells rested were quantitated, as described previously (15), for individual cells using the Openlab measurements module v3.51-4.0.4 (Improvision, Lexington, MA), with generally 20 to 100 cells per field at the start of image capture and with gliding velocities and frequencies measured for a minimum of 30 cells per transformant from at least two independent experiments (15). A resting period was assigned when no net cell movement of greater than 1 pixel (0.0645 μm) occurred between sequential frames (in most cases, 1 frame per second) (15). Corrected gliding velocities were calculated as the distance traveled by a cell divided by total time of the field interval minus the amount of time spent in resting periods.
RESULTS
Gliding capacity of cytadherence mutants M6, III-4, and IV-22.
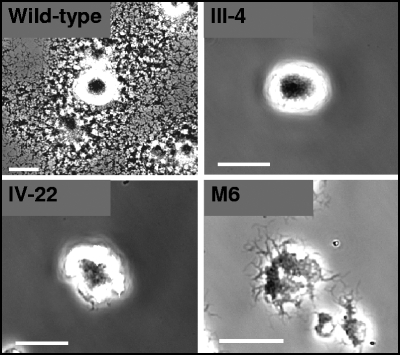
M. pneumoniae cytadherence mutant II-3 is nonmotile, indicating a requirement for the cytadherence-associated protein P30 in cell gliding (15). Proteins B and C, also designated P90 and P40, respectively, are thought to function as a complex with P30 and the adhesin P1 in receptor binding (28, 37, 50). To assess their requirement in gliding, we examined cytadherence mutants III-4 and IV-22 for satellite growth and cell gliding (Table 1). Neither mutant produced satellite growth over 96 h (Fig. 1) or in extended incubations up to 168 h (data not shown), and time lapse digital microcinematography confirmed a nonmotile phenotype. Likewise, no motility was observed for cytadherence mutant M6 (Fig. 1), which produces a truncated P30 and lacks HMW1 (29) and as a result fails to assemble an electron-dense core or localize P1 properly (41, 53). For reasons that are not clear, the leading edge of mutant M6 colonies had a filamentous appearance compared to the smooth edge seen with mutants III-4 and IV-22, indicating that cell motility alone does not determine colony appearance by this technique. We measured glass binding by each mutant under conditions identical to those used to examine cell gliding. Glass binding ranged from 78 to 104% of wild-type levels (Table 1), indicating that, except perhaps for mutant IV-22, the loss of gliding motility was not a function of poor binding to the inert surface.
FIG. 1.
Analysis of M. pneumoniae cytadherence mutant satellite growth. Shown are colony morphologies of wild-type, mutant III-4, mutant IV-22, and mutant M6 cells cultured on glass in SP-4 medium plus 3% gelatin. Images were captured at 96 h postinoculation. Scale bars, 30 μm.
Isolation of SGA transformants.
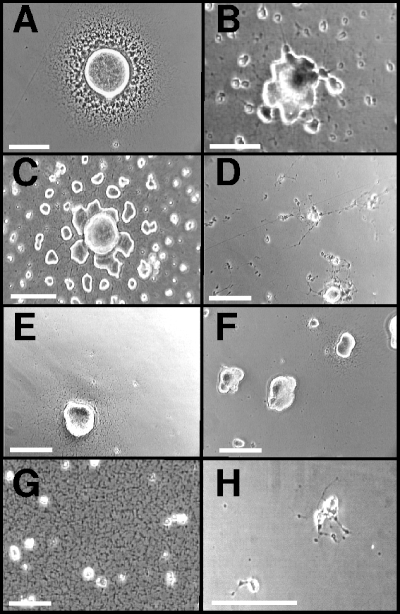
We used transposon mutagenesis to identify additional M. pneumoniae genes associated with gliding motility. Over 3,500 transformants were screened individually for satellite growth; approximately 100 of these transformants exhibited abnormal growth patterns relative to wild-type controls. The nature of the altered satellite growth varied considerably: some transformants had a dull, asymmetrical leading edge of colony spreading (Fig. 2B and C), while others exhibited filamentous spreading (Fig. 2D). Many transformants produced satellite growth radiating symmetrically but at a much lower density than the wild type (Fig. 2E and F), while still others exhibited lawn-like growth devoid of typical microcolonies (Fig. 2G) or barely discernible satellite growth (Fig. 2H). Each SGA insertion mutant was evaluated in three separate experiments to verify the altered phenotype.
FIG. 2.
Representative satellite growth patterns of M. pneumoniae SGA mutants. Satellite growth for selected SGA mutants cultured on polystyrene in SP-4 medium plus 3% gelatin was recorded at 96 h postinoculation. (A) Mutant II-3/WT P30 (positive control) (14); (B) mutant 320-30; (C) mutant 359-130; (D) mutant 311-22; (E) mutant 308-274; (F) mutant 110-112; (G) mutant 254-70; (H) mutant 387-2. Scale bars, 40 μm.
Identification of transposon-disrupted genes.
SGA transformants were sequenced across the transposon insertion site. Several transformants had identical insertion sites and were likely siblings, reinforcing the fidelity of the screening protocol. Sequencing revealed more than one insertion site for about 30% of the SGA transformants, indicating likely mixed populations; these were retained but not examined further here. Likewise, SGA transformants with Tn4001 at intergenic sites were retained but not characterized further here. Among the 47 SGA transformants characterized further (Table 2), 32 putative gliding-associated genes were found to be scattered around the chromosome rather than clustered. These genes were grouped according to the predicted functions of their gene products (8): (i) metabolic enzymes and transport system components, (ii) DNA- or protein-modifying enzymes, and (iii) conserved or genus-specific proteins of unknown function (Table 2). The designation for each mutant reflects the disrupted gene (8) and the residue at which its predicted protein product was disrupted; e.g., transformant 311-22 had a transposon insertion in MPN311 truncating the predicted protein product at residue 22 (Table 2). The first group of 12 mutants included genes for two proteins reported in the M. pneumoniae proteome (22, 36) and annotated as hypothetical but which appear to be permease components of ABC-type transport systems (MPN333 and MPN335). Notably, each protein was disrupted independently at two and three distinct sites, respectively. Also disrupted were the only predicted M. pneumoniae lipase (MPN407), a putative amino acid transporter (MPN308), and ThyA (MPN320), which converts dUMP to dTMP. The only predicted protein phosphatase (MPN247) as well as methyltransferase and/or DNA specificity subunits of separate gene clusters predicted to encode DNA modification systems were among the genes disrupted in the second group of nine mutants. While M. pneumoniae genes of unknown function constitute less than 20% of the genome (8), the third class of 26 mutants accounted for over 50% of the SGA mutants characterized. These included MPN254, encoding an ortholog of a highly conserved competence-induced protein of unknown function (CinA), and MPN311, encoding P41, a cytoskeletal protein of unknown function localizing to the base of the terminal organelle (24). Additional genus-specific genes disrupted included MPN376 and MPN387, encoding previously identified cytoskeletal proteins of unknown function (36). Finally, more than one independent insertion was identified for seven genes, with MPN359 and MPN376 having insertions at eight and three distinct sites, respectively (Table 2), reinforcing the correlation between gene disruption and phenotype and reflecting a level of mutagenesis likely approaching saturation.
TABLE 2.
Insertion site identification for M. pneumoniae SGA mutants
| Mutanta | Siteb | Descriptionc | Homology
|
||
|---|---|---|---|---|---|
| M. genitalium | M. gallisepticum | M. mobile | |||
| Transport and metabolism | |||||
| MPN114-566 | 150449 | Cat, carnitine O-acetyltransferase, hypothetical | Noned | None | None |
| MPN308-274 | 363224 | Amino acid transporter | MG225 | MGA0287 | MMOB2220 |
| MPN320-30 | 380633 | ThyA, thymidylate synthase | MG227 | MGA0699 | MMOB0340 |
| MPN333-32 | 393007 | Predicted ABC transport system, permease component | None | MGA1302 | None |
| MPN333-614 | 394752 | Predicted ABC transport system, permease component | None | MGA1302 | None |
| MPN335-38 | 396248 | Predicted ABC transport system, permease component | None | MGA1302 | None |
| MPN335-377 | 397263 | Predicted ABC transport system, permease component | None | MGA1302 | None |
| MPN335-595 | 397917 | Predicted ABC transport system, permease component | None | MGA1302 | None |
| MPN407-137 | 489736 | Predicted lipase/acylhydrolase | None | None | None |
| MPN493-20 | 600912 | UlaD, 3-keto-L-gulonate-6-phosphate decarboxylase | None | None | None |
| MPN509-145 | 621408 | Mycoplasma-specific export protein | MG288 | None | None |
| MPN510-42 | 624124 | Mycoplasma-specific export protein | MG288 | None | None |
| DNA/protein modification | |||||
| MPN107-68 | 139495 | Predicted DNA methylase | None | None | None |
| MPN107-152 | 139746 | Predicted DNA methylase | None | None | None |
| MPN110-112 | 143608 | Predicted restriction endonuclease | None | None | None |
| MPN153-35 | 202490 | Superfamily I DNA/RNA helicase | MG140 | MGA0793 | None |
| MPN247-100 | 298631 | PrpC, serine/threonine protein phosphatase | MG108 | MGA0461 | MMOB5580 |
| MPN294-96 | 350687 | Similar to ATP-independent intracellular protease | None | MGA0998 | None |
| MPN342-324 | 408833 | Restriction-modification system methyltransferase | None | MGA0537 | None |
| MPN342-362 | 408949 | Restriction-modification system methyltransferase | None | MGA0537 | None |
| MPN607-64 | 727792 | MsrA, peptide methionine sulfoxide reductase | MG408 | MGA0571 | MMOB4950 |
| Unknown function | |||||
| MPN083-62 | 104127 | Uncharacterized mycoplasma-specific lipoprotein | MG067 | MGA1162 | None |
| MPN104-29 | 134672 | Uncharacterized mycoplasma-specific protein | None | None | None |
| MPN254-70 | 305278 | CinA, uncharacterized protein (competence induced) | MG115 | None | None |
| MPN281-61 | 333465 | Uncharacterized mycoplasma-specific lipoprotein | MG185 | MGA0674 | None |
| MPN311-22 | 371006 | P41, uncharacterized mycoplasma-specific protein | MG218.1 | None | None |
| MPN311-161 | 371454 | P41, uncharacterized mycoplasma-specific protein | MG218.1 | None | None |
| MPN358-485 | 429136 | Uncharacterized mycoplasma-specific protein | MG255 | None | None |
| MPN359-12 | 429550 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN359-54 | 429675 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN359-93 | 429792 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN359-130 | 429904 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN359-131 | 429906 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN359-132 | 429909 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN359-160 | 429994 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN359-175 | 430040 | Hypothetical mycoplasma-specific protein | MG256 | None | None |
| MPN376-744 | 450763 | Uncharacterized mycoplasma-specific protein | None | None | None |
| MPN376-626 | 451116 | Uncharacterized mycoplasma-specific protein | None | None | None |
| MPN376-335 | 451988 | Uncharacterized mycoplasma-specific protein | None | None | None |
| MPN387-2 | 465051 | Uncharacterized mycoplasma-specific protein | MG269 | None | None |
| MPN403-114 | 485469 | Hypothetical mycoplasma-specific protein | MG284 | MGA1027e | MMOB2170 |
| MPN404-42 | 485600 | Hypothetical mycoplasma-specific protein | MG285 | MGA1029e | None |
| MPN524-74 | 645829 | Uncharacterized mycoplasma-specific protein | None | None | None |
| MPN582-82 | 704868 | Uncharacterized mycoplasma-specific lipoprotein | MG395 | MGA1162 | None |
| MPN614-33 | 738065 | Hypothetical mycoplasma-specific protein | MG414 | None | None |
| MPN634-134 | 760807 | Uncharacterized mycoplasma-specific protein | None | None | None |
| MPN648-107 | 773430 | Hypothetical mycoplasma-specific protein | MG441 | None | None |
ORF numbering according to reference 8.
Nucleotide after which the transposon inserted (this study).
Based on reference 8 and updated by current BLAST analysis.
Homology assigned when greater than 25% sequence identity exists over at least 50% of the M. pneumoniae protein, except as indicated.
Twenty-five percent identity over greater than 40% of the protein sequence.
Filter cloning and HA.
Primary transformant stocks were filter cloned, with eight progeny isolated and rescreened for satellite growth for each stock. In most cases, filter clones retained an SGA phenotype, but rarely, some progeny reacquired wild-type satellite growth, probably from Tn4001 transposition to a secondary site; only clones retaining an SGA phenotype were analyzed further. Each mutant was uniformly HA positive except 407-137 and 153-35, encoding a predicted lipase and a predicted helicase, respectively; these were not characterized further here (Table 2).
Filter-cloned progeny from 17 selected HA-positive SGA mutants representing 13 gliding-associated genes (Table 3) were evaluated by Southern hybridization using transposon- and IS256-specific probes to ensure the presence of Tn4001 in a single copy with no independent duplication and insertion of IS256 at secondary loci. Each probe hybridized to a specific band of the predicted size based upon the genome sequence for all filter clones examined, with no duplicate integrated copy of the transposon or insertion element evident (data not shown). Clonality was also assessed by PCR using primers flanking each disrupted gene. Rarely, filter clones appearing clonal by Southern analysis yielded a faint PCR product, probably from Tn4001 excision from the original insertion site in a very low percentage of the population. Only transformants clonal by all parameters described above were characterized further.
TABLE 3.
Characterization of cell gliding and glass binding for wild-type M. pneumoniae and selected SGA mutants
| Satellite growth alteration | Insertion mutant(s) | Mean cell gliding frequency (μm/s) ± SEMb | Mean cell velocity (μm/s) ± SEMb | Mean % time resting ± SEMb | Mean glass-binding capability ± SEMc |
|---|---|---|---|---|---|
| Wild type | NAd | 27.8 ± 0.3 | 100.0 ± 9.4 | 29.5 ± 0.5 | 100.0 ± 3.2 |
| Lawn-like growth lacking microcolony formation | MPN247-100 | 2.8 ± 1.9 | 42.3 ± 24.9 | 58.8 ± 14.6 | 101.3 ± 4.0 |
| MPN254-70 | 11.0 ± 3.9 | 53.4 ± 0.5 | 48.2 ± 1.8 | 106.5 ± 12.5 | |
| Filamentous spreading | MPN311-22/161a | 3.7 ± 1.4 | 29.7 ± 4.5 | 54.3 ± 1.2 | 107.9 ± 1.3 |
| Reduced spreading, confluence by 168 h | MPN083-62 | 18.8 ± 3.0 | 56.0 ± 11.6 | 45.3 ± 0.3 | 91.5 ± 7.7 |
| MPN342-324/362a | 11.8 ± 5.8 | 53.6 ± 5.5 | 47.3 ± 4.8 | 95.2 ± 2.9 | |
| MPN359-54/130a | 8.1 ± 5.1 | 43.1 ± 1.3 | 53.0 ± 1.9 | 90.7 ± 4.9 | |
| MPN376-626/744a | 6.8 ± 0.6 | 54.9 ± 12.7 | 50.4 ± 7.3 | 110.1 ± 1.6 | |
| Reduced spreading, no confluence by 168 h | MPN104-29 | 12.4 ± 0.3 | 87.2 ± 5.0 | 31.0 ± 5.3 | 93.7 ± 5.4 |
| MPN281-61 | 8.2 ± 3.6 | 41.8 ± 9.6 | 55.9 ± 4.7 | 78.5 ± 1.5 | |
| MPN387-2 | 0 | 0 | 100 | 100.4 ± 16.8 | |
| MPN403-114 | 7.6 ± 2.6 | 57.7 ± 3.3 | 46.0 ± 4.1 | 103.6 ± 2.2 | |
| MPN404-42 | 11.3 ± 1.9 | 50.8 ± 2.5 | 48.6 ± 3.4 | 78.4 ± 7.8 | |
| MPN524-74 | 12.8 ± 3.2 | 52.8 ± 7.8 | 49.1 ± 4.9 | 68.2 ± 2.0 |
Data were combined for the indicated insertion mutants for these ORFs.
Data are presented as the means ± standard errors of the means from two filter clones for each SGA mutant, with measurements for 30 to 50 individual cells for each mutant except for MPN247 and MPN311 insertion mutants, where mean gliding frequencies were much lower.
In each case, glass binding was measured for the two filter clones from which gliding values were derived and normalized relative to the wild type.
NA, not applicable.
Western immunoblotting analysis of SGA mutants.
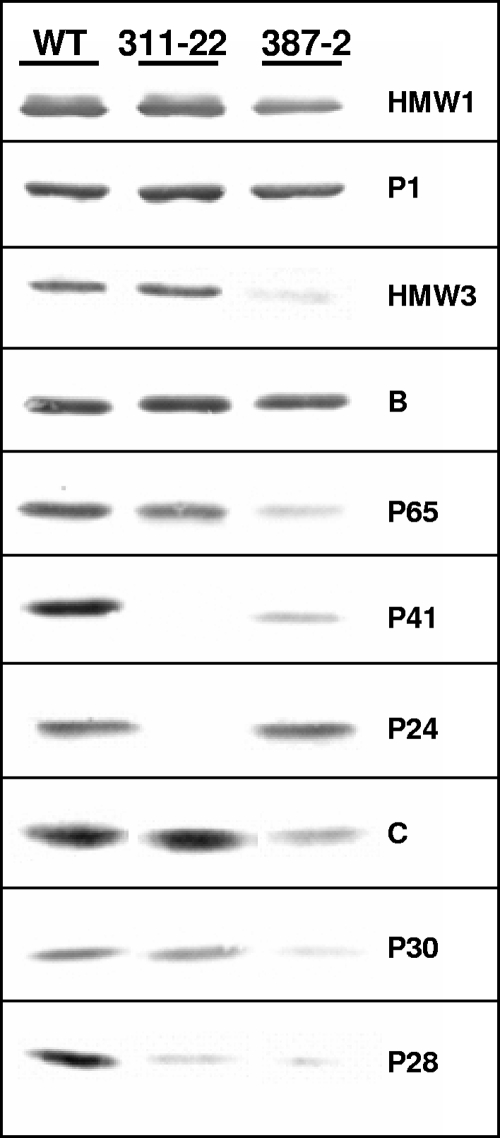
To assess the possible spontaneous or secondary loss of known cytadherence-associated proteins in the 17 SGA transformants analyzed further, several filter-cloned isolates of each transformant were assessed by Western immunoblotting using sera specific for cytadherence-associated proteins B, P1, P30, P41, and HMW1 (data not shown). Only MPN311 and MPN387 insertion mutants exhibited profiles distinct from that of the wild type and were subsequently rescreened with a battery of additional antisera (Fig. 3 and data not shown). As expected, disruption of MPN311 resulted in the loss of protein P41, encoded by that gene. In addition, the products of the genes immediately upstream (P28) and downstream (P24) were reduced. Disruption of MPN387 was accompanied by drastically reduced levels of HMW3, P28, P30, P41, and P65 and moderately reduced levels of HMW1 and C, while B, HMW2, P1, and P24 were largely unaffected (Fig. 3 and data not shown). An excision revertant of the MPN387 mutant had a wild-type profile and satellite growth pattern, suggesting that the loss of the MPN387 gene product was indeed responsible for reduced steady-state levels of these terminal organelle proteins (data not shown).
FIG. 3.
Western immunoblot analysis of selected SGA transformants. WT, wild-type M. pneumoniae profile; 311-22 and 387-2, SGA transformants with insertions in MPN311 and MPN387, respectively. Ten micrograms of protein was loaded per lane, and samples were analyzed on a 4 to 10% polyacrylamide gradient gel. Antisera are indicated on the right.
Time lapse analysis of satellite growth and cell gliding.
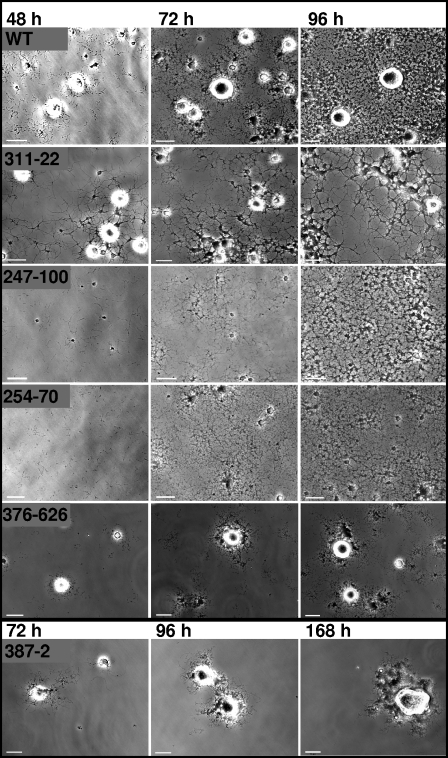
Satellite growth (Fig. 4) and cell gliding (Table 3) were characterized by time lapse microcinematography for the 17 selected insertion mutants. MPN311 insertion mutants produced sparse satellite growth that appeared as thickened, elongated filaments extending from microcolonies. MPN247 and MPN254 mutants both exhibited lawn-like growth lacking microcolonies, raising the possibility that the defect in each mutant affects related cellular processes. For most SGA insertion mutants such as MPN376, satellite growth simply developed at a slower rate but otherwise appeared to be like that of the wild type. However, other mutants including MPN387 failed to establish the satellite growth typical of wild-type M. pneumoniae, even with extended incubation periods of up to 168 h. No individual cell gliding was observed for this mutant over extended frame intervals, although some cell movement was apparent and probably reflected cell growth. Wild-type controls exhibited a gliding frequency of approximately 28%, as expected (15), while gliding frequencies were reduced for all mutants, in most cases ranging from 11 to 13% (or about 40% of wild-type levels) but considerably lower for MPN247, MPN311, MPN359, and MPN403 (Table 3). Likewise, in most cases, average mutant gliding velocities ranged from 36 to 58% of wild-type levels, although no cell gliding was seen with MPN387. However, the slowest velocities did not necessarily correlate with the lowest gliding frequencies. For example, the MPN247 mutant had an extremely low gliding frequency (2.8%, or 10% of wild-type levels) but an intermediate cell velocity (42.3% of the wild-type level), while the MPN104 mutant had a high gliding velocity (87.2% of the wild-type level) but an intermediate gliding frequency (12.4%, or 44% of the wild-type level). This was also the only mutant to exhibit a normal percentage of resting time, suggesting a normal capacity to move once gliding was initiated. Finally, no clear correlation between the specific SGA phenotype and the gliding behavior of individual cells was observed (Table 3).
FIG. 4.
Analysis of satellite growth formation over time for representative SGA mutant phenotypes. Cultures were incubated for 168 h, with images captured at 12-h intervals, and selected frames are shown here for the indicated mutants. Scale bars, 15 μm. WT, wild type.
Analysis of glass binding.
To rule out the possibility that poor binding to the inert surface was responsible for altered gliding, attachment to glass was measured for each of 13 selected SGA mutants (Table 3). Only mutants 281-61, 404-42, and 524-74 exhibited significantly reduced glass binding (P < 0.05), at 78.5, 78.4, and 68.2% of wild-type levels, respectively. All other mutants examined bound to glass at levels ranging from 90 to 110% of wild-type levels.
Assessment of potential polar effects.
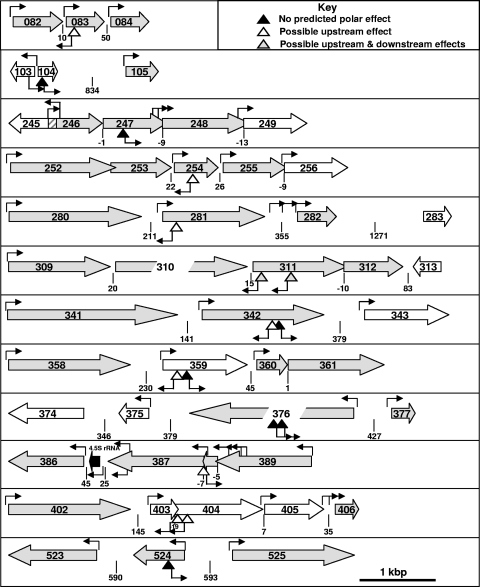
Definitive correlation of gene disruption with altered gliding for each mutant will require the isolation and characterization of excision revertants and genetic complementation, which are not trivial procedures in mycoplasmas (10, 34). However, the likelihood of polar effects of transposon insertion can be assessed on the basis of other parameters. The IS256 at one end of Tn4001.2065 has an outward-directed promoter (Pout) (5, 25) that functions well in M. pneumoniae (13), while the corresponding sequence in the IS256 element at the opposite end of the transposon has been disrupted (25). Pout can direct the transcription of downstream genes in one orientation but might inhibit translation upstream by RNA interference (RNAi) from an antisense transcript in the reverse orientation. M. pneumoniae has a single σ70 RNA polymerase typical of gram-positive species (21); while −35 sequences are divergent in M. pneumoniae, the −10 sequence is conserved, with a consensus of TAxxxT (51, 52). The insertion mutants in Table 3 were evaluated for the likelihood of upstream and downstream polar effects based on the orientation of the transposon with respect to Pout and the presence of the −10 consensus within 300 bp of the 5′ end of flanking genes (Fig. 5). By these criteria, no polar effect on flanking genes was predicted for 7 of the 17 mutants analyzed further, with only the MPN311 mutants clearly predicted to have a polar effect downstream, as supported by the data shown in Fig. 3. Upstream polar effects for the remaining mutants would require translational knockdown by RNAi, although this has not been demonstrated in M. pneumoniae. Genetic complementation studies will be required to rule out potential polar effects associated with translational coupling (50).
FIG. 5.
Schematic diagram of disrupted loci for selected SGA mutants to assess the likelihood of polar consequences of transposon insertions. ORFs (8) are indicated by large open arrows, with gray and white indicating the reported presence or absence, respectively, of the protein product in the M. pneumoniae proteome (22, 46). ORF overlap is indicated by overlapping arrows, except for MPN245/MPN246 overlap, which is cross-hatched. The spacing between ORFs is indicated below each locus. Inverted triangles correspond to transposon insertions, with predicted polar effects as detailed in the key. Small arrows originating from the inverted triangles indicate the orientation of a promoter in IS256 that is functional in M. pneumoniae (5). Other small arrows indicate the location and orientation of predicted promoters based upon the consensus sequence TAxxxT (51, 52). Scale is approximate, as indicated at the bottom of the diagram.
DISCUSSION
Mycoplasmas are considered simple organisms, and yet M. pneumoniae and related species have a complex cytoskeletal structure and a differentiated terminal organelle (2, 12, 20, 33) that function in gliding motility (16), although neither the molecular components nor the mechanics of the gliding machinery have been established. Electron cryotomography imaging reveals a three-dimensional core architecture that suggests conformational flexibility, prompting speculation that alternating contraction and extension of the core propels gliding, utilizing core-tethered surface adhesins for force transmission (20). In the current study, we examined gliding by M. pneumoniae cytadherence mutants III-4 and IV-22 to investigate the requirement for the major cytadhesin P1 and accessory proteins B and C in cell gliding. These proteins are linked at the levels of transcription, translation, protein stability, and subcellular localization and likely function as a multiprotein complex in receptor binding (28, 50). The finding that mutants III-4 and IV-22 are also nonmotile indicates that at a minimum, protein B and/or C (Table 1) is likewise required for gliding motility. HMW1 is required for P1 localization to the terminal organelle (14), and the loss of HMW1 is accompanied by a failure to glide (this study). However, P1 localizes to the terminal organelle in mutant II-3 (37), which lacks P30 and is also nonmotile (1, 15); hence, P1 localization alone is not sufficient for gliding function. Whether P1 and P30 are the core-tethered surface proteins in the model described previously by Henderson and Jensen (20) is unknown and requires a more detailed characterization of the protein linkages between core components and the mycoplasma membrane.
Analysis of M. pneumoniae mutants defective in both gliding and cytadherence does not allow for the identification of gliding-specific components or for the determination of the contribution of gliding to colonization and pathogenesis. Therefore we combined transposon mutagenesis with screening for altered satellite growth and HA to identify M. pneumoniae proteins that are associated with cell gliding but dispensable for cytadherence. Thirty-two different open reading frames (ORFs) were disrupted among 47 transformants, 30 of which were dispensable for HA. Individual cell gliding was quantitated for 13 of these transformants, all of which exhibited reduced gliding velocities and/or frequencies. It remains to be determined whether the impact on gliding for each transformant is primary or secondary.
SGA mutants were grouped according to the predicted functions of the disrupted genes (Table 2). Gliding deficiencies resulting from the disruption of genes associated with metabolism and transport were not unexpected for an organism with limited metabolic capabilities but were not characterized further here. However, an association between defects in gliding motility and the disruption of genes predicted to encode DNA- and protein-modifying enzymes could provide important new insights into M. pneumoniae regulatory mechanisms. For example, promoters that are inactive when methylated might function in hemimethylated DNA during chromosome replication, perhaps providing a means to coordinate the expression of genes encoding components for terminal organelle assembly with cell division. We also disrupted MPN247, encoding the only M. pneumoniae protein phosphatase (PrpC) annotated to date. This disruption is not likely to affect transcription of the downstream cognate kinase, but we cannot rule out the possibility of a polar effect associated with translational coupling, as these ORFs overlap by nine nucleotides. In either case, this mutant is likely to be defective in the ability to modify the phosphorylation state of its protein target(s), perhaps including the terminal organelle phosphoproteins HMW1, HMW2, and P1 (9). The 10-fold reduction in gliding frequency for this mutant was among the largest observed here, suggesting that phosphorylation may control the activation of the gliding motor. This mutant also exhibited a lawn-like growth pattern largely devoid of microcolonies. Perhaps significantly, a loss of PrpC in Bacillus subtilis alters biofilm architecture and increases stationary-phase cell densities by fivefold (32), prompting speculation that protein phosphorylation may play a part in regulating quorum sensing. Additional studies are required to determine if the altered growth pattern and reduced gliding frequency for this mutant reflect a similar function in M. pneumoniae.
Proteins of unknown function represented over 50% of the loci disrupted in SGA mutants, with only MPN254 (CinA) found outside the genus. Expression of cinA is upregulated in Streptococcus pneumoniae as part of the ComX quorum-sensing pathway, where it is thought to function in the uptake of extracellular DNA in response to increasing culture densities (30). A role in DNA uptake may be nutritionally significant for M. pneumoniae, given its inability to synthesize purines and pyrimidines de novo, although it is not clear how this might contribute to the lawn-like growth observed here.
While most gliding-associated genes were identified from single insertion mutants, two or more independent disruptions were observed for several genes; these yielded the same SGA phenotype, reinforcing a cause-and-effect relationship between transposon insertion and phenotype. Eight distinct insertion mutants were isolated for MPN359, none of which were predicted to have polar effects on flanking genes. To date, MPN359 has been found only in M. pneumoniae and the closely related M. genitalium. Its gene product, predicted to have three membrane-spanning domains, was not detected in the proteome (22, 46), perhaps due to its deduced alkaline pI and transmembrane nature. The products of MPN403 and MPN404 were likewise not detected by proteomic analyses, but like MPN359, orthologs of MPN403 and MPN404 are present in M. genitalium, and given the retention of these genes in both species despite considerable genome reduction, we speculate that their products are synthesized.
The disrupted genus-specific genes associated with gliding included MPN083 and MPN281, encoding putative lipoproteins. M. pneumoniae genome annotation indicates a complement of approximately 42 lipoproteins based largely upon homology to putative M. genitalium lipoproteins (8), none with assessed functions. Palmitoylation data suggest that 25 to 30 lipoproteins are synthesized in M. pneumoniae, while 30 of the 42 predicted lipoproteins, including the MPN083 and MPN281 products, were detected in the M. pneumoniae proteome (21, 22). Polar effects downstream are not predicted, but we cannot rule out the possibility of upstream effects due to RNAi. Nevertheless, our findings represent the first genetic evidence of function for these M. pneumoniae lipoproteins. MPN524 and MPN104 contain a central domain of unknown function (DUF) that is unique to M. pneumoniae and is designated DUF16 (21). Approximately 20 additional M. pneumoniae proteins share this motif, which is predicted to adopt a coiled-coil conformation through the DUF motif, but none have been identified in the M. pneumoniae cytoskeletal fraction (36). MPN524 has a 40-amino-acid C-terminal domain that is strongly predicted to form a dimeric coiled-coil structure. This domain is absent in MPN104 and other DUF proteins. Glass binding was reduced with the disruption of MPN524 but not MPN104, raising the possibility that this C-terminal domain has functional significance.
The gliding defects associated with the disruption of MPN376, MPN311, and MPN387 were among the most severe defects observed here. Three independent insertions were identified in MPN376, underscoring the likely correlation between gene disruption and mutant phenotype, and polar effects were not predicted. MPN376 transcripts are among the most abundant in M. pneumoniae (52), and the product is a cytoskeletal element of unknown function (36) but is predicted to have an N-terminal signal peptide and C-terminal membrane-spanning domain. Both gliding velocity and gliding frequency were reduced with the disruption of MPN376, suggesting a requirement for initiation as well as operation of the gliding motor. MPN311 mutants were unique among the SGA variants characterized here in producing filamentous satellite growth. Furthermore, individual cells of this mutant had an unusual elongated morphology, often reaching lengths of 4 to 8 μm, compared to 1 to 2 μm for wild-type cells (data not shown). MPN311 encodes P41, a cytoskeletal protein of unknown function localizing to the base of the terminal organelle (24). MPN311 disruption is not predicted to have downstream effects on MPN312 transcription, but preliminary data indicate that the loss of protein P24, encoded by MPN312, accompanies the disruption of MPN311 and may be due to translational coupling, as MPN311 overlaps the 5′ end of MPN312 by 10 bp. Complementation studies will be required to determine whether the motility and morphology defects associated with MPN311 disruption are due to a loss of P41, P24, or both. Disruption of MPN387 resulted in a complete loss of gliding; some cell movement was observed with extended observation intervals, but it probably reflects cell growth. The transposon insertion site at the 5′ end of MPN387 corresponded to the 3′ end of MPN388; hence, complementation analyses will be required to assess whether truncation of MPN388 contributes to the phenotype of this mutant. The MPN387 product is a cytoskeletal protein (36) that appears to lack membrane-spanning domains and is predicted to assume an elongated coiled-coil conformation. Loss of MPN387 did not appreciably affect the steady-state levels of B, HMW2, P1, or P24, but the levels of other known terminal organelle proteins were moderately or drastically reduced; thus, it was somewhat surprising that this mutant retained an HA-positive phenotype, although erythrocyte binding was not assessed quantitatively. It has been proposed that P1, B, and C are incorporated into the terminal organelle by a pathway separate from that of HMW1, HMW3, P65, and P30 (26). The destabilization of protein C in addition to HMW1, P28, P30, P41, and P65 with the disruption of MPN387 suggests that the MPN387 gene product participates in both putative assembly pathways. Significantly, an MPN387 excision revertant had a wild-type satellite growth pattern and protein profile (data not shown), indicating that the destabilization of cytadherence-associated proteins was indeed a consequence of the transposon insertion and not a secondary defect.
Eighteen of the 32 ORFs associated here with M. pneumoniae gliding are also found in the closely related M. genitalium. Global transposon mutagenesis has established what may approach a minimal genomic complement for laboratory growth in M. genitalium (11); among the 18 gliding-associated ORFs shared with M. genitalium (Table 2), only the MPN247 ortholog was not identified as being dispensable (11). As each of the 482 M. genitalium ORFs are represented in M. pneumoniae, the gliding machineries of both species are likely similar. Nevertheless, transposon mutagenesis and screening for altered satellite growth in M. genitalium yielded only MG200 and MG386 (34), corresponding to MPN119 and MPN567, respectively, in M. pneumoniae; thus, a common subset of gliding-associated genes was not identified for both species. However, in separate studies, we have identified insertion mutants in MPN119 and MPN567, and preliminary analysis indicates an altered gliding phenotype (our unpublished data).
Over 200 mycoplasmas have been described to date, but gliding motility has been reported in only six species, including M. gallisepticum, M. pulmonis, M. amphoriforme, and M. mobile in addition to M. pneumoniae and M. genitalium (4, 17a). Since genes required for gliding in M. mobile (23) are found in M. pulmonis but not in M. pneumoniae, M. genitalium, or M. gallisepticum, we assessed whether the gliding-associated genes identified here for M. pneumoniae are found in M. gallisepticum and M. mobile in addition to M. genitalium (Table 2). Even using relatively low stringency for this determination, only 1 genus-specific gene of unknown function associated with gliding in M. pneumoniae was conserved in M. mobile compared with 12 found in M. genitalium and 5 found in M. gallisepticum, consistent with a gliding mechanism in M. mobile that is distinct from those of the other mycoplasmas compared here.
In conclusion, the current study represents the first widespread genetic analysis of gliding motility in M. pneumoniae. The generation and initial characterization of gliding mutants provide strong evidence associating a number of mycoplasma genes with this novel cellular locomotion in M. pneumoniae and set the stage for future studies of the engineering and regulation of the gliding machinery. Analysis of existing cytadherence mutants has established a clear requirement for protein P30 (15) and now protein HMW1, as well as B and/or C, in M. pneumoniae gliding. Moreover, global transposon mutagenesis identified 30 additional genes in which insertions resulted in altered satellite growth with no effect on HA, as evaluated qualitatively. A quantitative assessment of binding to erythrocytes as well as other cell types by these mutants is still required. Each of the 17 representative SGA mutants analyzed further exhibited substantially reduced gliding frequencies, gliding velocities, or both, identifying, on a genetic basis, a number of mycoplasma proteins with no previously assigned function as having an association with gliding. Furthermore, our findings raise the possibility of regulatory roles in M. pneumoniae for DNA methylation and protein phosphorylation in terminal organelle function in gliding. Finally, the isolation of cytadherence-positive gliding mutants should allow for an elucidation of the contribution of gliding to the colonization of bronchial epithelium and pathogenesis.
Acknowledgments
We thank C. Minion for providing Tn4001.2065; R. Herrmann for antisera; M. Farmer, R. Krause, M. Hamrick, and L. Sellers for technical assistance; and J. Piñol for providing data prior to publication.
This work was supported by Public Health Service grant AI49194 from the National Institute of Allergy and Infectious Diseases to D.C.K.
REFERENCES
- 1.Baseman, J. B., J. Morrison-Plummer, D. Drouillard, B. Puleo-Scheppke, V. V. Tryon, and S. C. Holt. 1987. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr. J. Med. Sci. 23:474-479. [PubMed] [Google Scholar]
- 2.Biberfield, G., and P. Biberfield. 1970. Ultrastructural features of Mycoplasma pneumoniae. J. Bacteriol. 102:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. 32:321-326. [DOI] [PubMed] [Google Scholar]
- 4.Bredt, W. 1979. Motility, p. 141-155. In M.F. Barile and S. Razin (ed.), The mycoplasmas, vol. I. Cell biology. Academic Press, New York, N.Y. [Google Scholar]
- 5.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 6.Chanock, R. M., L. Hayflick, and M. F. Barile. 1962. Growth on artificial medium of an agent associated with atypical pneumonia and its identification as a PPLO. Proc. Natl. Acad. Sci. USA 48:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier, A. M., W. A. Clyde, Jr., and F. W. Denny. 1971. Mycoplasma pneumoniae in hamster tracheal organ culture: immunofluorescent and electron microscopic studies. Proc. Soc. Exp. Biol. Med. 136:569-573. [DOI] [PubMed] [Google Scholar]
- 8.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dirksen, L. B., K. A. Krebes, and D. C. Krause. 1994. Phosphorylation of cytadherence-accessory proteins in Mycoplasma pneumoniae. J. Bacteriol. 176:7499-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisseha, M., H. W. Gohlmann, R. Herrmann, and D. C. Krause. 1999. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J. Bacteriol. 181:4404-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gobel, U., V. Speth, and W. Bredt. 1981. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J. Cell Biol. 91:537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, T. W., K. A. Krebes, and D. C. Krause. 1996. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol. Microbiol. 19:1085-1093. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, T. W., M. J. Willby, and D. C. Krause. 1998. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J. Bacteriol. 180:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasselbring, B. M., J. L. Jordan, and D. C. Krause. 2005. Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J. Bacteriol. 187:6281-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasselbring, B. M., and D. C. Krause. Unpublished data.
- 17.Hasselbring, B. M., J. L. Jordan, R. W. Krause, and D. C. Krause. Unpublished data.
- 17a.Hatchel, J. M., R. S. Balish, M. L. Duley, and M. F. Balish. 2006. Ultrastructure and gliding motility of Mycoplasma amphoriforme, a possible human respiratory pathogen. Microbiology 152:2181-2189. [DOI] [PubMed] [Google Scholar]
- 18.Hayflick, L. 1965. Tissue cultures and mycoplasmas. Tex. Rep. Biol. Med. 23(Suppl. 1):285. [PubMed] [Google Scholar]
- 19.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170-175. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, G. P., and G. J. Jensen. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60:376-385. [DOI] [PubMed] [Google Scholar]
- 21.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaffe, J. D., H. C. Berg, and G. M. Church. 2004. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4:59-77. [DOI] [PubMed] [Google Scholar]
- 23.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenri, T., S. Seto, A. Horino, Y. Sasaki, T. Sasaki, and M. Miyata. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knudtson, K. L., and F. C. Minion. 1993. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene 137:217-222. [DOI] [PubMed] [Google Scholar]
- 26.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 27.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Layh-Schmitt, G., and R. Herrmann. 1994. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect. Immun. 62:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Layh-Schmitt, G., H. Hilbert, and E. Pirkl. 1995. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J. Bacteriol. 177:843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipman, R. P., and W. A. Clyde, Jr. 1969. The interrelationship of virulence, cytadsorption, and peroxide formation in Mycoplasma pneumoniae. Proc. Soc. Exp. Biol. Med. 131:1163-1167. [DOI] [PubMed] [Google Scholar]
- 32.Madek, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46:571-586. [DOI] [PubMed] [Google Scholar]
- 33.Meng, K. E., and R. M. Pfister. 1980. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J. Bacteriol. 144:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pich, O. Q., R. Burgos, M. Ferrer-Navarro, E. Querol, and J. Piñol. 2006. Mycoplasma genitalium mg200 and mg386 genes are involved in gliding motility but not in cytadherence. Mol. Microbiol. 60:1509-1519. [DOI] [PubMed] [Google Scholar]
- 35.Proft, T., and R. Herrmann. 1994. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol. Microbiol. 13:337-348. [DOI] [PubMed] [Google Scholar]
- 36.Regula, J. T., G. Boguth, A. Gorg, J. Hegermann, F. Mayer, R. Frank, and R. Herrmann. 2001. Defining the mycoplasma ‘cytoskeleton’: the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147:1045-1057. [DOI] [PubMed] [Google Scholar]
- 37.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of a 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens, M. K., and D. C. Krause. 1991. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike Triton shell. J. Bacteriol. 173:1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens, M. K., and D. C. Krause. 1992. Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J. Bacteriol. 174:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tully, J. G., R. F. Whitcomb, H. F. Clark, and D. L. Williamson. 1977. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195:892-894. [DOI] [PubMed] [Google Scholar]
- 46.Ueberle, B., R. Frank, and R. Herrmann. 2002. The proteome of the bacterium Mycoplasma pneumoniae: comparing predicted open reading frames to identified gene products. Proteomics 2:754-764. [DOI] [PubMed] [Google Scholar]
- 47.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uenoyama, A., and M. Miyata. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 187:5578-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waldo, R. H., III, and D. C. Krause. 2006. Synthesis, stability, and function of cytadhesin P1 and accessory protein B/C complex of Mycoplasma pneumoniae. J. Bacteriol. 188:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldo, R. H., III, P. L. Popham, C. E. Romero-Arroyo, E. A. Mothershed, K. K. Lee, and D. C. Krause. 1999. Transcriptional analysis of the hmw gene cluster of Mycoplasma pneumoniae. J. Bacteriol. 181:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiner, J., III, R. Herrmann, and G. F. Browning. 2000. Transcription in Mycoplasma pneumoniae. Nucleic Acids Res. 28:4488-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willby, M. J., M. F. Balish, S. M. Ross, K. K. Lee, J. L. Jordan, and D. C. Krause. 2004. HMW1 is required for stability and localization of HMW2 to the attachment organelle of Mycoplasma pneumoniae. J. Bacteriol. 186:8221-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]