Abstract
The general stress response of the bacterium Bacillus subtilis is regulated by a partner-switching mechanism in which serine and threonine phosphorylation controls protein interactions in the stress-signaling pathway. The environmental branch of this pathway contains a family of five paralogous proteins that function as negative regulators. Here we present genetic evidence that a sixth paralog, YtvA, acts as a positive regulator in the same environmental signaling branch. We also present biochemical evidence that YtvA and at least three of the negative regulators can be isolated from cell extracts in a large environmental signaling complex. YtvA differs from these associated negative regulators by its flavin mononucleotide (FMN)-containing light-oxygen-voltage domain. Others have shown that this domain has the photochemistry expected for a blue-light sensor, with the covalent linkage of the FMN chromophore to cysteine 62 composing a critical part of the photocycle. Consistent with the view that light intensity modifies the output of the environmental signaling pathway, we found that cysteine 62 is required for YtvA to exert its positive regulatory role in the absence of other stress. Transcriptional analysis of the ytvA structural gene indicated that it provides the entry point for at least one additional environmental input, mediated by the Spx global regulator of disulfide stress. These results support a model in which the large signaling complex serves to integrate multiple environmental signals in order to modulate the general stress response.
In Bacillus subtilis and related gram-positive bacteria, the general stress response is controlled by the alternative transcription factor σB (reviewed in references 24 and 49). When activated by diverse growth-limiting stresses, expression of the σB regulon confers resistance against future, potentially lethal stresses. These diverse stress signals are conveyed to σB via a network that functions by the “partner-switching” mechanism, in which serine and threonine phosphorylation governs interactions among network components (2, 17, 20, 58). Genome analysis indicates that orthologs of these partner-switching components are widely distributed among the eubacteria (34, 43, 47, 49), and recent experimental evidence confirms that the basic features of the mechanism are conserved in evolutionarily distant branches of the gram-positive and gram-negative lineages (5, 27, 35, 37). Study of this mechanism in B. subtilis should therefore uncover themes applicable to a broad array of signaling pathways. Here we address the question of how dissimilar environmental signals are sensed and integrated to activate a single transcription factor, σB.
A model of the B. subtilis signaling network is shown in Fig. 1A. In this model, the partner switch comprises the RsbV antagonist (an anti-anti-σ factor), the RsbW switch protein (an anti-σ factor), and the σB target protein. In addition to its anti-σ activity, RsbW also possesses a serine kinase activity that can phosphorylate and disable its RsbV antagonist (2, 7, 17). In unstressed cells, RsbV is found primarily in the phosphorylated form and cannot interact with RsbW anti-σ, which then binds σB in an inactive complex. The release of σB is effected differently by the three classes of stress that activate the response. Two of these classes—energy stress and environmental stress—are transmitted by independent pathways that terminate with differentially regulated serine phosphatases specific for RsbV-P (56-58). When activated by its particular stress, either phosphatase can dephosphorylate RsbV-P, allowing it to bind RsbW and force the release of σB. The third class of stress—cold adaptation—is transmitted by an RsbV-independent pathway that is not well understood and may involve differences in RsbW and σB synthesis or stability under prolonged cold stress (10).
FIG. 1.
Model of the σB signal transduction network. (A) Two independent signaling pathways converge on RsbV anti-anti-σ and RsbW anti-σ, the direct regulators of σB activity. Phosphorylated RsbV (RsbV-P) is the antagonist form in unstressed cells. When activated by the upstream elements shown in panel B, the environmental signaling phosphatase RsbU dephosphorylates RsbV-P, allowing it to bind RsbW and induce the release of σB. (B) In the environmental signaling branch, RsbS and RsbT are paralogs of RsbV and RsbW, respectively. RsbS is the antagonist form in unstressed cells, and RsbRA, RsbRB, RsbRC, and RsbRD are redundant coantagonists that make up a large environmental signaling complex with RsbS. This complex binds the RsbT phosphatase regulator and holds it inactive. Following environmental stress, RsbT phosphorylates RsbRA and RsbS, releasing RsbT to activate the RsbU phosphatase. The RsbX feedback respectively. RsbS is the antagonist form in unstressed cells, and RsbRA, RsbRB, RsbRC, and RsbRD are redundant coantagonists that make up a large environmental signaling complex with RsbS. This complex binds the RsbT phosphatase regulator and holds it inactive. Following environmental stress, RsbT phosphorylates RsbRA and RsbS, releasing RsbT to activate the RsbU phosphatase. The RsbX feedback phosphatase returns the system to its prestress condition. Phosphorylation of RsbRB, RsbRC, and RsbRD is not shown but is thought to resemble that of RsbRA. Here we present evidence that the blue-light receptor protein YtvA is also a constituent of the large environmental signaling complex. (C) Members of the RsbR family share a C-terminal STAS domain (shaded) with the smaller RsbS antagonist but have different N-terminal domains: either a nonheme globin domain thought to be important for signaling (RsbR coantagonist proteins) or a LOV domain with an FMN chromophore that forms a light-dependent photoadduct with LOV-C62 (YtvA blue-light receptor). Residues at conserved positions within the STAS domain are charged (RsbS D26, YtvA E168 and E202) or are phosphorylated by RsbT (RsbS S59, RsbRA T171 and T205).
Our focus is the environmental pathway, which activates the RsbU phosphatase in response to acid, ethanol, heat, or salt stress (30, 57, 58). This pathway, shown in Fig. 1B, is regulated by a second partner switch consisting of the RsbS antagonist, the RsbT switch protein, and the RsbU target. RsbS and RsbT are paralogs of RsbV and RsbW, and they likewise regulate their target by direct protein-protein interaction (16, 30, 31, 58). However, there are two significant differences between the switches shown in panels A and B. First, the output of the S-T switch is positive rather than negative, with RsbT activating its RsbU target when a stress signal is perceived, and second, the RsbS antagonist alone is insufficient to bind the RsbT switch protein in an inactive complex. Also required is at least one of four apparently redundant coantagonist proteins belonging to the RsbR family (1, 11, 33), each of which has a C-terminal sulfate transporter anti-sigma (STAS) domain (3) equivalent to the entire length of the smaller RsbS antagonist (Fig. 1C). These STAS domains contain the conserved serine residue critical for RsbS function (30, 58) or the two conserved threonine residues implicated in RsbRA and RsbRB function (20, 33). The N-terminal region of RsbRA contains a nonheme globin domain proposed to function in signal transduction, and sequence comparisons suggest that the other members of the RsbR family possess similar N-terminal, globin-like domains (44).
Notably, RsbRA, RsbRB, and RsbS copurify from cell extracts in a large complex with a mass of >700 kDa (11, 33), and RsbRC and RsbRD have also been identified as constituents of this same complex (A. L. Weigel, T. J. Kim, S. Neissen, J. R. Yates, and C. W. Price, unpublished data). The supramolecular properties of this complex have been suggested to facilitate the sensing or transmission of environmental stress signals. According to the model shown in Fig. 1B, in the absence of environmental stress the RsbR coantagonists and the RsbS antagonist jointly bind the RsbT switch protein, preventing its association with RsbU (11, 33, 44). Upon environmental stress, the RsbR family members and RsbS are increasingly phosphorylated by RsbT (1, 32). These phosphorylation events are thought to correlate with release of RsbT, which binds and activates the RsbU environmental phosphatase.
YtvA is also a member of the RsbR family (1), but it lacks the conserved threonine residues within its C-terminal STAS domain (Fig. 1C). Instead, YtvA has charged glutamate residues at these positions, presumably mimicking the phosphorylated state of the other family members. Moreover, in place of the N-terminal globin-like domain, YtvA has a light-oxygen-voltage (LOV) domain similar to those found in plant phototropins, which are signaling proteins that contain N-terminal, blue-light-sensing LOV domains coupled to C-terminal kinase output domains (28). A subset of the Per-Arnt-Sim (PAS) superfamily (55), these plant LOV domains bind a flavin mononucleotide (FMN) chromophore, and upon blue-light illumination this chromophore becomes covalently linked with a conserved cysteine residue within the LOV domain (12, 14, 51, 52). The resulting structural or dynamic change is then transmitted to the adjacent serine/threonine kinase domain, likely with the involvement of a conserved α helix C terminal to the LOV domain (15, 22, 23, 52). Notably, Losi and colleagues have found that the LOV domain of YtvA undergoes a photocycle that closely resembles that manifested by the LOV photoreceptors of plant phototropins (39). Like the phototropins, purified YtvA also binds an FMN chromophore which forms a photoadduct with a conserved cysteine upon blue-light illumination, and this change appears to alter the interaction between the adjacent LOV and STAS domains (6, 40, 41).
An earlier genetic analysis by Akbar et al. (1) only implicated YtvA as a positive regulator in the environmental signaling branch of the σB signal transduction network. Here we present genetic and biochemical experiments which squarely place YtvA in this pathway. Our data further suggest that YtvA provides the ability to sense two discrete environmental parameters—one via control of ytvA expression and the other via control of YtvA domain interaction.
MATERIALS AND METHODS
Bacterial strains and genetic methods.
The B. subtilis strains used in this study are listed in Table 1. Standard recombinant DNA methods, including transformation of B. subtilis PB2 and its derivatives, were as previously described (20).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Reference or construction |
|---|---|---|
| ORB4342 | amyE::pSN56 pheA1 trpC2 | 45 |
| ORB4556 | amyE::pSN56 pheA1 trpC2 trxB-lacZ | P. Zubera |
| PB2 | trpC2 | 168 Marburg strain |
| PB198 | amyE::ctc-lacZ trpC2 | 9 |
| PB423 | rsbTΔ1 amyE::ctc-lacZ trpC2 | 30 |
| PB495 | rsbUΔ2 amyE::ctc-lacZ trpC2 | 56 |
| PB545 | rsbRBΔ1::kan trpC2 | 1 |
| PB593 | (6His-10myc) rsbRA10 trpC2 | 33 |
| PB743 | amyE::ctc-lacZ trpC2/pDG148 | pDG148 → PB198b |
| PB801 | amyE::ctc-lacZ trpC2/pTG5659 | pTG5659 → PB198 |
| PB806 | rsbTΔ1 amyE::ctc-lacZ trpC2/pTG5659 | pTG5659 → PB423 |
| PB907 | rsbUΔ2 amyE::ctc-lacZ trpC2/pTG5659 | pTG5659 → PB495 |
| PB963 | amyE::pALW6 trpC2 | pALW6 → PB2 |
| PB965 | amyE::pALW7 trpC2 | pALW7 → PB2 |
| PB967 | amyE::pALW8 trpC2 | pALW8 → PB2 |
| PB980 | amyE::pSN56 pheA1 trpC2 thrC::pALW9 | pALW9 → ORB4342 |
| PB1003 | amyE::ctc-lacZ trpC2/pMB5876 | pMB5876 → PB198 |
| PB1004 | amyE::ctc-lacZ trpC2/pMB5877 | pMB5877 → PB198 |
| PB1005 | amyE::ctc-lacZ trpC2/pMB5878 | pMB5878 → PB198 |
| PB1006 | amyE::ctc-lacZ trpC2/pMB5879 | pMB5879 → PB198 |
Personal communication.
Arrow indicates transformation of donor plasmid into recipient strain.
The plasmids used to overexpress wild or mutant ytvA in the pDG148 vector are shown in Table 2. The wild ytvA fragment (extending from 29 bp upstream of the translational initiation triplet ATG to 11 bp downstream from the termination triplet TAA) was PCR amplified from chromosomal DNA of strain PB2 with oligonucleotides YtvASalI (5′-GGGTCGACTAGCAAGCTGACGGCCTAAG-3′, containing a SalI site) and YtvASphI (5′-GGGCATGCGGATCTTTTTACATAATCGG-3′, containing an SphI site). This PCR product was cloned into the SalI and SphI sites of the pDG148 polylinker, placing ytvA expression under the control of the vector's isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter to create pTG5659. Two deletion and two missense mutations within ytvA were made by a four-primer method (26) and likewise cloned into the pDG148 vector; ytvAΔ1 removed the LOV domain (residues 25 to 126) to create pMB5876, ytvAΔ2 removed the LOV domain and a predicted downstream α helix (residues 25 to 157) to create pMB5877, and ytvA C62A and C62S altered a cysteine residue essential for light-induced conformational changes in other LOV domains, creating pMB5878 and pMB5879, respectively. The DNA sequence was confirmed in these and all following constructs.
TABLE 2.
Integrative and replicative plasmids used for strain construction
| Plasmid | Alteration or relevant feature | Reference |
|---|---|---|
| pALW6 | ytvA −76 to +276 fragment in pDH32a | This study |
| pALW7 | ytvA −20 to +276 fragment in pDH32a | This study |
| pALW8 | ytvA −339 to +276 fragment in pDH32a | This study |
| pALW9 | ytvA −339 to +276 fragment in pDG793a | This study |
| pDG148 | Pspac promoter, multicopy replicative plasmid | 54 |
| pDG793 | lacZ fusion vector, thrC integration | 21 |
| pDH32 | lacZ fusion vector, amyE integration | 48 |
| pMB5876 | ytvAΔ1 in pDG148 (LOV deleted) | This study |
| pMB5877 | ytvAΔ2 in pDG148 (LOV and α helix deleted) | This study |
| pMB5878 | ytvA C62A (TGT → GCT) in pDG148 | This study |
| pMB5879 | ytvA C62S (TGT → AGT) in pDG148 | This study |
| pSN56 | Pspank-hy promoter expressing spxDD, amyE integration | 45 |
| pTG5659 | ytvA coding region in pDG148 | This study |
Numbers indicate fragment endpoints relative to ytvA +1 determined by RACE-PCR.
In order to provide an indirect measurement of σB activity, many of the strains in Table 1 harbor a single-copy transcriptional fusion between the well-characterized σB-dependent ctc promoter and a lacZ reporter gene, constructed in the pDH32 vector that integrates at the amyE locus. This same vector was used to locate sequences important for ytvA expression. For these ytvA constructions, three fragments were generated by PCR amplification of PB2 chromosomal DNA, with a unique upstream primer for each fragment and a common +276 downstream primer (5′-GCAGGATCCGTGTGTTTCCCCT-3′; extending to +276 relative to the +1 site of the ytvA message). These upstream and downstream primers contained BamHI sites to allow subsequent subcloning of the fragments into the BamHI site of pDH32. The upstream primers were (i) −339 (5′-GCAGGATCCCCATCATCACCTTCC-3′), generating pALW8; (ii) −76 (5′-TTTTAGTGGATCCGAGGACAAGCC-3′), generating pALW6; and (iii) −20 (5′-AGAGGATCCGTTGTGCTACGGTTAC-3′), generating pALW7. Linearized pALW6, pALW7, and pALW8 were integrated into the amyE locus of wild-type strain PB2 to make PB963, PB965, and PB967, respectively. The longest ytvA fragment (−339 to +276) was also inserted into the BamHI site of the pDG793 fusion vector, generating pALW9. Linearized pALW9 was integrated into the thrC locus of the SpxDD overproducing strain ORB4342 to make PB980.
β-Galactosidase accumulation assays.
Shake cultures were grown at 37°C to mid-exponential phase in buffered Luria broth medium (BLB) lacking salt (8) and then diluted 1:25 into fresh BLB. One of two additions was made to this second culture during the early exponential phase of growth, depending on the experiment, i.e., 1 mM IPTG (to induce expression of wild and mutant proteins under control of the Pspac or Pspank-hy promoter) or 0.3 M NaCl (for environmental stress experiments). Samples were collected at the indicated times and treated essentially as described by Miller (42), as previously described (32). Protein levels were determined with the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA). Activity was defined as ΔA420 × 1,000 per minute per milligram of protein.
Purification of hexahistidine-tagged RsbRA from cell extracts.
The two-step affinity and sizing purification is described elsewhere (33). Strain PB593 (rsbRA10) was cultured at 37°C in 2 liters of BLB lacking salt. At mid-exponential phase, cells were harvested by centrifugation, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), and then broken by sonication. Tagged RsbRA was initially purified by immobilized-metal affinity chromatography, with 5 ml of a 50% Ni-nitrilotriacetic acid agarose gel slurry according to the QIAexpressionist protocol (QIAGEN Inc., Valencia, CA). Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting, and those with the highest RsbRA levels were combined. The 2.5-ml pooled sample was applied to a Sephacryl S-300 Hi-prep HR column (1.6 by 60 cm; Amersham Pharmacia Biotech, Piscataway, NJ). The column was run at a flow rate of 1 ml/min; fractions were collected every minute for assay by Western blot analysis.
Western blotting experiments.
Mouse anti-RsbRA and -RsbS antibodies were provided by William Haldenwang (18). Rabbit anti-RsbRB and anti-YtvA antibodies were made by the UC Davis Animal Resources Service, with purified recombinant proteins. Specificities of all four antibodies were confirmed by Western blot analysis of wild and mutant cell extracts (not shown). Proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories), and treated as previously described (33). Briefly, after exposure to a primary antibody, the membranes were washed and incubated with an immunoglobulin G (IgG) peroxide-conjugated secondary antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA), either anti-mouse (for RsbRA and RsbS) or anti-rabbit (for RsbRB and YtvA). Bound antibody was detected with the ECL Plus Western blotting kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. For experiments comparing steady-state levels of wild and altered forms of YtvA, cells were grown to early exponential phase and broken by sonication. Equal amounts of protein from these extracts were subjected to SDS-PAGE and analyzed by Western blotting.
Immunoprecipitation experiments.
PB2 (wild-type) and PB545 (rsbRBΔ1::km) cells were grown and harvested as described above for the purification of tagged RsbRA protein. Immunoprecipitation is described elsewhere (33) and used anti-RsbRB antibody and a protein G PLUS agarose suspension (Santa Cruz Biotechnology). The agarose beads were collected and washed, and the bound proteins were eluted in SDS-PAGE sample buffer (12 mM Tris-HCl [pH 6.8], 5% glycerol, 0.4% SDS, 2.88 mM 2-mercaptoethanol, 0.02% bromophenol blue) by heating to 100°C for 5 min. Eluted proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and then probed with specific antibodies, as described for the Western blotting experiments.
RACE-PCR experiments.
Rapid amplification of cDNA ends (RACE)-PCR (19) was used to identify the 5′ end of the ytvA message, essentially as described previously (50). PB2 (wild type) was grown as described above for the β-galactosidase accumulation assays. Total RNA was extracted with the QIAGEN RNeasy kit (QIAGEN) according to the manufacturer's instructions, with the exception of increasing the lysozyme concentration to 15 mg/ml and including a homogenization step in which the sample was passed through a 20-gauge needle. Two ytvA-specific primers were used; oligo-1 (5′-GATGTTGTCCACTTCTGC-3′) was located about 200 bp downstream from the predicted translational initiation triplet, and oligo-2 (5′-TGTGTGTTTCCCCTGTAAGAAGCG-3′) was nested upstream of oligo-1. Moloney murine leukemia virus reverse transcriptase (New England Biolabs, Beverly, MA) and 20 pmol of oligo-1 were used to reverse transcribe 1 μg of total RNA according to the manufacturer's instructions. Reaction mixtures were purified with the QIAquick PCR purification kit (QIAGEN). Poly(dA) tails were added to first-strand synthesis products with terminal transferase (New England Biolabs) according to the manufacturer's instructions. The subsequent PCR used Taq polymerase (Promega, Madison, WI), oligo-2, and a primer complementary to the poly(A) tail (5′-CACCCGTATGGATCTCGACT16V-3′, where V = A, C, or G). The PCR product was gel purified and directly sequenced.
RESULTS
YtvA functions in the environmental signaling branch.
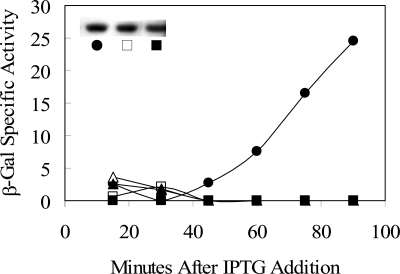
Previous genetic analysis suggested that YtvA is a positive regulator of the σB general stress transcription factor, with a ytvA loss-of-function mutation causing a twofold decrease in σB activity following environmental stress and a lesser reduction following energy stress (1). As shown in Fig. 2, we extended these previous results by using a genetic-epistasis test that located YtvA in the environmental branch of the σB signaling network. First, overexpression of ytvA (by means of an inducible promoter on a multicopy plasmid) led to increased σB activity in logarithmically growing cells, even in the absence of the stress signals commonly needed to elicit this increase. This result supports the hypothesis of Akbar et al. (1) that YtvA acts as a positive regulator of σB. Second, this increased activity was abolished in strains missing one or another of the two most downstream regulators in the environmental signaling branch: the RsbT kinase or the RsbU phosphatase (Fig. 2). We infer from this latter result that YtvA acts upstream from RsbT and RsbU. This inference is strongly supported by the biochemical experiments described in the next section.
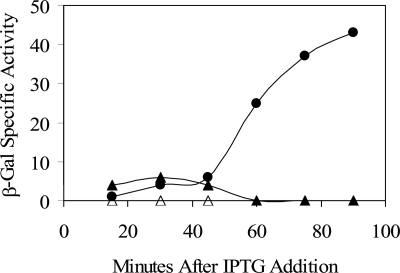
FIG. 2.
YtvA overexpression induces σB activity. Effects of YtvA overexpression were measured with a σB-dependent ctc-lacZ transcriptional fusion present in single copy on the B. subtilis chromosome. IPTG was added to logarithmically growing cultures to induce ytvA expression from the Pspac promoter of multicopy plasmid pTG5659. Samples were taken at the indicated times and assayed for β-galactosidase (β-Gal) activity as described in Materials and Methods. Filled circles, PB801 (wild type with pTG5659); open triangles, PB806 (rsbTΔ1 with pTG5659; filled triangles, PB907 (rsbΔU2 with pTG5659). All strains were grown in BLB supplemented with 10 μg/ml kanamycin to prevent loss of the multicopy plasmid; cells remained in logarithmic growth through the time shown. Values are corrected for the basal activity of the PB743 control (wild type with the pDG148 vector alone).
YtvA can be purified from cell extracts in a complex together with its paralogous environmental signaling regulators.
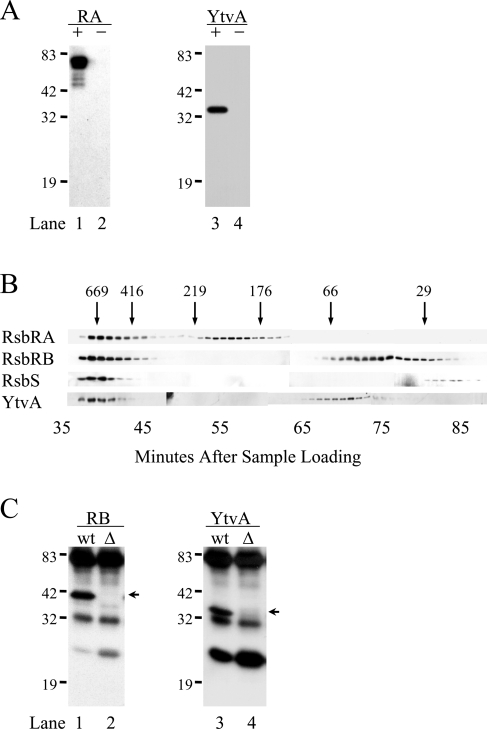
The genetic epistasis experiment indicated that YtvA acts in the upstream part of the environmental signaling branch, which comprises the YtvA paralogs RsbRA, RsbRB, RsbRC, RsbRD, and RsbS (1, 30). Earlier, Kim et al. (33) used two independent methods to isolate from cell extracts a >700-kDa environmental signaling complex containing RsbRA, RsbRB, and RsbS. RsbRC and RsbRD have also been identified as constituents of this same complex (unpublished data). On the basis of the functional association inferred from the genetic analysis, we hypothesized that YtvA was also physically associated with its paralogs in the signaling complex. With the same methods and materials described by Kim et al. (33), we now show that YtvA indeed copurified with the environmental signaling complex, together with RsbRA, RsbRB, and RsbS.
We first reexamined the complex previously purified from extracts of an unstressed strain encoding a hexahistidine-tagged version of the RsbRA coantagonist protein (33). This PB593 strain carried the modified rsbRA10 allele at the rsbRA chromosomal locus and was indistinguishable from the wild type with respect to environmental signaling (data not shown). Here we used SDS-PAGE and Western blotting to probe the same nickel affinity column fractions described in reference 33. As shown in Fig. 3A, a protein with the mobility and antigenicity of YtvA copurified with the tagged RsbRA protein (the + lanes); this YtvA signal was not detected in a parallel purification from the wild-type strain in which RsbRA was untagged (the − lanes). The RsbRA-containing fractions of the nickel affinity column had earlier been applied to a Sephacryl S-300 sizing column (33), and here we used SDS-PAGE and a Western blotting assay to probe these same sizing column fractions. As shown in Fig. 3B, a significant amount of RsbRA, RsbRB, RsbS, and YtvA was found in the early sizing fractions, eluting with the void volume before the thyroglobulin marker (669 kDa). This cofractionation through two successive purification steps—nickel affinity chromatography and a sizing column—supports the idea that RsbRA, RsbRB, RsbS, and YtvA form part of a stable complex in vivo.
FIG. 3.
Association of YtvA with RsbRA and RsbRB in cell extracts. (A) Two cell extracts were prepared: one from strain PB593, encoding a His-tagged version of RsbRA (+), and the other from wild-type control strain PB2 with no tag (−). After nickel affinity chromatography, proteins were analyzed by SDS-PAGE and Western blotting with specific antibodies. Lanes 1 and 2, anti-RsbRA (RA); lanes 3 and 4, anti-YtvA (YtvA). Positions of molecular mass standards are shown on the left (sizes are in kilodaltons). (B) RsbRA-containing fractions from the nickel affinity step shown in panel A were applied to a Sephacryl S300 sizing column; the resulting fractions were analyzed by SDS-PAGE and Western blotting with anti-RsbRA, -RsbRB, -RsbS, or -YtvA antibody. Arrows indicate the elution volumes of the molecular mass standards (sizes are in kilodaltons). (C) Proteins were coimmunoprecipitated from cell extracts with anti-RsbRB antibody and a protein G-agarose conjugate. The precipitated proteins were separated by SDS-PAGE and analyzed by Western blotting with antibody specific for the protein of interest. For each antibody, proteins precipitated from two different cell extracts are shown: wt indicates the PB2 wild type, and Δ indicates the PB545 control bearing a null rsbRB allele. Lanes 1 and 2, anti-RsbRB antibody (RB); lanes 3 and 4, anti-YtvA antibody. Each pair of lanes contains one specific signal for the protein detected, shown by the arrows on the right, and also nonspecific signals for protein G and the IgG light and heavy chains. Positions of molecular mass standards are indicated on the left (sizes are in kilodaltons). These cell extracts, column fractions, and immune precipitates are the same as those shown in Fig. 4 and 5 of reference 33, and elements of those figures are reprinted here (with the permission of the publisher) for comparison with the additional anti-YtvA antibody data in panels A, B, and C.
The results shown in Fig. 3A and B were obtained with a modified form of RsbRA and nickel affinity chromatography as the first purification step. We therefore wanted to confirm these results in an unmodified strain, with both a different purification method and a different point of entry into the complex. As shown in Fig. 3C, immunoprecipitation analysis showed that a protein with the mobility and antigenic signature of YtvA specifically interacted with RsbRB in wild-type cells but not in rsbRB mutant cells. We had previously used immunoprecipitation to demonstrate that RsbRA and RsbS also interacted with RsbRB (33). Thus, two independent approaches, affinity chromatography and coimmunoprecipitation, point toward the physical association of RsbRA, RsbRB, RsbS, and YtvA in cell extracts.
Transcriptional analysis of ytvA indicates control by the disulfide stress regulator Spx.
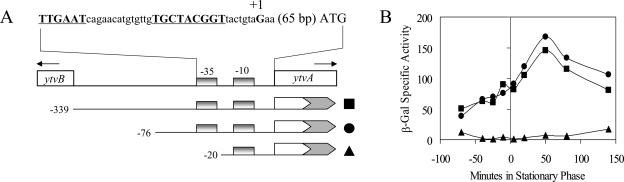
Because ytvA overexpression from our artificial construction led to increased σB activity (Fig. 2), we asked what transcriptional controls might be exerted on ytvA expression at its normal chromosomal locus. With RNA from unstressed cells, RACE-PCR experiments located a 5′ end for ytvA message 68 nucleotides upstream from the ATG initiation triplet (Fig. 4A). This 5′ end was preceded by sequences resembling an extended σA promoter (25), with a good match at −35 (TTGAat) and a lesser match at extended −10 (TGnTAcggT). A series of single-copy transcriptional fusions covering the ytvB-ytvA intercistronic interval indicated that the region containing these sequences is required for promoter activity and that this activity increases as logarithmically growing cells enter stationary phase (Fig. 4A and B).
FIG. 4.
Transcriptional organization of the ytvB-ytvA interval. (A) RACE-PCR located the 5′ end of the ytvA message 68 nucleotides upstream from the ATG initiation triplet, at the G residue labeled +1. This residue is preceded by sequences resembling an extended σA promoter (underlined). DNA fragments from the region were used to construct single-copy transcriptional fusions to lacZ (shaded), all with the same 3′ end (at +276 within ytvA) but with different 5′ ends, as shown: filled squares, PB967 (−339); filled circles, PB963 (−76); filled triangles, PB965 (−20). (B) β-Galactosidase (β-Gal) accumulation assays for the three fusions shown in panel A. Logarithmically growing cells were allowed to enter stationary phase and sampled at the indicated times.
Earlier transcriptional profiling studies by Nakano et al. (45) suggested that ytvA was among the set of genes whose expression is upregulated by the Spx global regulator of disulfide stress. Spx is the first example of a new class of transcriptional regulators, and it acts in either a positive or a negative fashion by directly contacting the α subunit of RNA polymerase and not, apparently, the DNA of the target promoter (reviewed in reference 59). Spx concentrations are normally modulated by controlled proteolysis (46), and for their profiling studies Nakano et al. (45) used a strain that overexpressed a proteolytically stable form of the protein, SpxDD. To confirm and extend the results of Nakano et al., we employed this same strain to ask whether overexpression of SpxDD would lead to increased activity from our ytvA reporter fusion.
Overexpression of SpxDD caused immediate induction of the trxB-lacZ control fusion (Fig. 5A), eventually attaining sixfold accumulation over the control. This induction is comparable to the ninefold increase noted by Nakano et al. (45) in a message-based assay. In contrast, overexpression of SpxDD led to induction of the ytvA-lacZ fusion (Fig. 5B) only upon entry into stationary phase, eventually reaching 2.5-fold accumulation over the control. Clearly, the effect of SpxDD on ytvA expression differs from the trxB control in that it requires the presence of an additional signal, generated here in stationary phase. We conclude that the Spx regulator of disulfide stress does indeed control ytvA expression, directly or indirectly, but with an additional level of control that distinguishes it from the classic disulfide stress-induced gene trxB (45).
FIG. 5.
Spx activates trxB and ytvA fusion expression. Strains bearing single-copy fusions of trxB-lacZ (A) or ytvA-lacZ (B) were engineered to express the SpxDD disulfide stress regulator from a Pspank-hy promoter integrated at amyE (45). SpxDD expression was induced in logarithmically growing cells by adding IPTG at time zero. Cells entered stationary phase at 50 min, indicated by the vertical dotted line. Panel A, ORB4556 (trxB-lacZ) with (filled circles) or without (open circles) added IPTG. Panel B, PB980 (ytvA-lacZ) with (filled squares) or without (open squares) added IPTG. Both fusions were assayed in the ORB4342 background to ensure that the strains were otherwise isogenic (Table 2). β-Gal, β-galactosidase.
Is the blue-light photochemistry of the LOV domain required for the positive role of YtvA?
The experiments shown in Fig. 2 indicate that increased expression of ytvA leads to activation of the general stress response via the environmental signaling pathway. We next wished to test whether this property of YtvA depends on its LOV domain and associated FMN chromophore, which together undergo a conformational shift when illuminated by blue light (6, 39, 40). Because the experiments shown in Fig. 2 were done under laboratory fluorescent lighting, which is known to emit blue light (53), the observed overexpression phenotype could reflect the effect of YtvA already activated via its LOV domain. In support of this view, Ávila-Pérez et al. (4) have found that YtvA overexpression does indeed increase σB activity under blue-light illumination but not in the dark.
We therefore used a genetic analysis to determine the influence of the LOV domain on the demonstrated positive function of YtvA. For these experiments, we made two point mutations within the LOV domain that altered conserved cysteine 62 to either serine (C62S) or alanine (C62A). In both YtvA (39) and related plant phototropins (51), this cysteine residue forms an adduct with the FMN chromophore in response to blue light. Among the plant phototropins, alteration of this conserved cysteine to either serine or alanine abrogates both adduct formation and the blue-light photocycle but does not affect either LOV domain folding (23) or its ability to noncovalently bind FMN (51); the alanine substitution is also known to prevent activation of the adjacent serine/threonine kinase domain in vitro and in vivo (13). In addition to the C62S and C62A substitutions, we made two in-frame deletion mutations within ytvA—Δ1, which removed the LOV domain (residues 25 to 126), and Δ2, which removed both the LOV domain and much of a predicted α helix (residues 127 to 157) analogous to that implicated in signaling between the LOV and kinase domains of plant phototropins (22, 23).
With the simple overexpression assay shown in Fig. 2, we found that none of the mutant YtvA proteins was able to activate σB (Fig. 6). However, the basis for this defective phenotype was not the same for the two types of mutation—substitution or deletion. For the mutant proteins bearing either the C62S or the C62A alteration, Western blotting experiments found that their steady-state levels were the same as in the wild type (Fig. 6, inset). We therefore conclude that the conserved cysteine 62 residue is required for YtvA to activate σB in the absence of another stress. However, for the mutant proteins lacking the entire LOV domain, or the LOV domain and part of the adjacent α helix, we were unable to detect any mutant YtvA at all (not shown). From this, we conclude that the LOV domain is important for YtvA synthesis or stability in vivo.
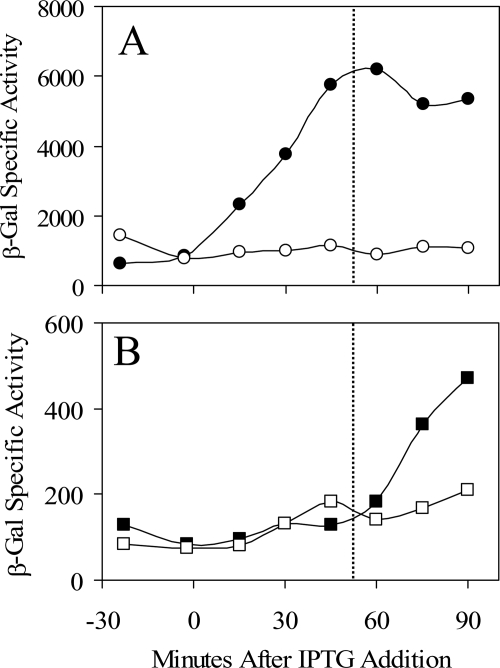
FIG. 6.
The C62 residue is required for the positive effect of YtvA overexpression in the absence of stress. The effects of wild and mutant YtvA proteins were measured with a σB-dependent ctc-lacZ transcriptional fusion. IPTG was added to logarithmically growing cultures to induce ytvA expression from the Pspac promoters of the different multicopy plasmids (time zero), and samples were assayed for β-galactosidase (β-Gal) activity. Filled circles, PB801 (with pTG5659: wild ytvA); open triangles, PB1003 (with pMB5876: ytvAΔ1); filled triangles, PB1004 (with pMB5877: ytvAΔ2); open squares, PB1005 (with pMB5878: ytvAC62A); filled squares, PB1006 (with pMB5879: ytvAC62S). Strains were grown in BLB supplemented with 10 μg/ml kanamycin to prevent plasmid loss; values were corrected for the basal activity of the PB743 control (with the pDG148 vector alone). The inset shows YtvA levels in PB801 (filled circle), PB1005 (open square), and PB1006 (filled square) assayed by Western blotting.
Is environmental stress activation of σB affected by YtvA overexpression?
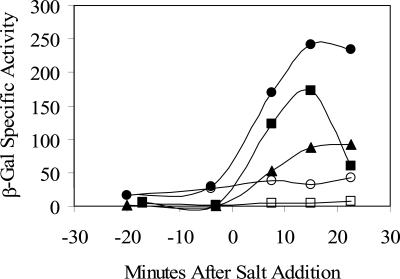
Loss of YtvA function is known to cause a twofold decrease in σB activation in response to salt or ethanol stress (1). We next asked whether activation of σB via an environmental stress—specifically, salt stress—is influenced by overexpression of the wild or C62S form of YtvA. In the experiment shown in Fig. 7, overexpression (by IPTG addition) was accomplished earlier in logarithmic growth compared to the experiment shown in Fig. 6. Nonetheless, equivalent results were obtained in the absence of salt stress, in that the strain overexpressing wild-type YtvA showed greater σB activity compared to the strain overexpressing the C62S mutant (compare open circles and squares). In contrast to this clear difference in the absence of salt, the strains overexpressing the wild and C62S mutant forms of YtvA each manifested a comparable enhancement of σB activation in the presence of salt (closed circles and squares), relative to that of the wild-type control, in which YtvA was not overexpressed (closed triangles). We conclude that activation of σB in response to salt stress is enhanced by YtvA overexpression and that conserved cysteine 62 of the LOV domain is not absolutely required for this enhanced activation.
FIG. 7.
The C62 residue is not required for the positive effect of YtvA overexpression in the presence of salt stress. The effects of wild and mutant YtvA proteins were measured with a σB-dependent ctc-lacZ transcriptional fusion. IPTG was added to logarithmically growing cultures to induce ytvA expression from multicopy plasmids (at time −100 min), and NaCl was added to induce the environmental stress response (at time zero). Samples were assayed for β-galactosidase (β-Gal) activity. PB743 (pDG148 vector alone) with (filled triangles) added NaCl, PB801 (pTG5659: wild ytvA) with (filled circles) or without (open circles) NaCl, and PB1006 (pMB5879: ytvAC62S) with (filled squares) or without (open squares) NaCl are shown. Growth was in BLB supplemented with 2.5 μg/ml neomycin to prevent plasmid loss. Values were corrected for the basal activity of the PB743 control (pDG148 vector alone) with no added salt.
DISCUSSION
Genome analysis has found an increasing number of bacterial proteins with domains that closely resemble the LOV blue-light photoreceptor domains of plant phototropins (15, 38). Moreover, for three of these bacterial proteins there is now experimental evidence that their LOV domains manifest a photochemistry much like that of their counterparts in plant phototropins: the LOV kinase of Caulobacter crescentus (38), the SB2-LOV protein of Pseudomonas putida (36), and the YtvA protein of B. subtilis (39), which is the subject here. Many of the predicted bacterial LOV domains are associated with potential output domains, including histidine kinase, GGDEF, EAL, and helix-turn-helix domains, leading to the suggestion that the LOV domain provides a means for blue-light intensity to regulate diverse adaptive responses (15, 38). However, thus far no clear role has been established for any of these proteins, and consequently there has been no experimental evidence that blue-light sensing via a LOV domain impacts any bacterial signaling pathway.
YtvA of B. subtilis provides an attractive system to probe this blue-light functionality. An earlier genetic analysis by Akbar et al. (1) suggested that YtvA is a positive regulator in the environmental signaling branch of the σB signal transduction network. We have presented genetic (Fig. 2) and biochemical data (Fig. 3) which functionally and physically place YtvA directly in this pathway. To the idea of a large, stable environmental signaling complex that contains all four members of the RsbR family of coantagonist proteins together with the RsbS antagonist (11, 33), we can now add the YtvA positive regulator (Fig. 1B). Previous genetic and biochemical analyses have shown that the RsbRA, RsbRB, and RsbS regulators are immediately involved in the transmission of environmental stress signals (1, 11, 30, 33, 58), and the supramolecular properties of the large signaling complex are suggested to facilitate the sensing, as well as the modulation, of these environmental signals (11, 33).
It is in the context of this large complex that YtvA provides the potential entry point for a signal of light intensity, and we have shown that the C62 residue of the LOV domain, which is critical for FMN-C4 adduct formation in related blue-light photoreceptors (13, 14, 51), is also required for the positive role of YtvA in the absence of another stress (Fig. 6). Consistent with the findings reported here, Ávila-Pérez and colleagues (4) used blue- and red-light illumination of growing cells to clearly demonstrate that YtvA function is required for blue-light activation of σB in vivo. Thus, the combined genetic, biochemical, and physiological evidence indicates that YtvA functions as a blue-light-responsive positive regulator in the environmental signaling pathway that activates B. subtilis σB.
How could this blue-light sensing relate to the role of σB in controlling the general stress response? Blue light is a convenient wavelength range to indicate the presence of sunlight, whose spectrum also contains a significant UV component. Moreover, blue light itself is potentially harmful because its absorption by porphyrins can produce damaging singlet oxygen species (29, 38). Because a number of genes in the σB regulon encode products which protect cellular DNA against mutation, and which protect cellular protein, lipid, and DNA against oxidative damage (24, 49), the ability of YtvA to modulate σB activity could well contribute to survival in the soil environment.
And how might YtvA achieve its regulatory effect? On one level, a simple increase in ytvA expression could increase the stoichiometry of the YtvA positive regulator relative to its companion RsbRA, -RB, -RC, and -RD coantagonist proteins that function as negative regulators in the large environmental signaling complex. In the experiments reported here, we mainly used significant overexpression of ytvA to exaggerate this condition and thereby study the role of YtvA in σB activation. Likewise, the cell has the means to more subtly alter ytvA expression and achieve an analogous result. As shown in Fig. 5, the Spx regulator of disulfide stress (together with an as-yet-uncharacterized second signal) can activate ytvA expression between two- and threefold. However, on another level, a key control of YtvA function appears to be posttranslational, in response to blue light.
We infer this posttranslational control from the following: (i) blue-light illumination of YtvA induces a photocycle that alters the interaction between the N-terminal LOV domain and the C-terminal STAS domain (6, 39, 40); (ii) this cycle requires the formation of a photoadduct between the FMN chromophore and the conserved C62 residue of the LOV domain (6, 39); and (iii) alteration of this C62 residue to either serine or alanine abolished one positive regulatory activity of YtvA in vivo (Fig. 6). We therefore propose that a blue-light-induced structural or dynamic change is important for YtvA function and suggest that this change alters the interactions between YtvA and its potential binding partners within the large signaling complex.
In summary, YtvA provides a nexus that, at the very least, integrates signals of blue-light intensity, disulfide stress, and an uncharacterized third signal to modulate σB activity via the large environmental signaling complex. Intriguingly, overexpression of YtvA also increases the magnitude of the general stress response elicited by other environmental stress signals, such as salt stress (Fig. 7). Because this augmentation does not appear to require the integrity of the C62 residue of YtvA, we infer that it does not require a blue-light signal and can therefore occur in the dark. The mechanism by which YtvA increases signaling through the environmental stress pathway in the light and in the dark remains to be established. Nonetheless, the retention of clear YtvA orthologs in Listeria monocytogenes and Oceanobacillus iheyensis (38), close relatives of B. subtilis whose genomes appear to encode similar environmental signaling complexes, suggests that for all of these bacteria blue-light sensing is an important component of the general stress response in the natural environment.
Acknowledgments
We thank Peter Zuber for providing strains ORB4342 and ORB4556, William Haldenwang for the kind gift of anti-RsbRA and anti-RsbS antibodies, Remco Kort and colleagues for sharing results prior to publication, and the two anonymous reviewers for helpful comments and suggestions.
This research was supported by Public Health Service grant GM42077 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 2000. The STAS domain—a link between anion transporters and antisigma-factor antagonists. Curr. Biol. 10:R53-R55. [DOI] [PubMed] [Google Scholar]
- 4.Ávila-Pérez, M., K. Hellingwerf, and R. Kort. 2006. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 188:6411-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-σ factor antagonists control σF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 6.Bednarz, T., A. Losi, W. Gartner, P. Hegemann, and J. Heberle. 2004. Functional variations among LOV domains as revealed by FT-IR difference spectroscopy. Photochem. Photobiol. Sci. 3:575-579. [DOI] [PubMed] [Google Scholar]
- 7.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor σB by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brigulla, M., T. Hoffmann, A. Krisp, A. Völker, E. Bremer, and U. Völker. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C. C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657-1669. [DOI] [PubMed] [Google Scholar]
- 12.Christie, J. M., M. Salomon, K. Nozue, M. Wada, and W. R. Briggs. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96:8779-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie, J. M., T. E. Swartz, R. A. Bogomolni, and W. R. Briggs. 2002. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32:205-219. [DOI] [PubMed] [Google Scholar]
- 14.Crosson, S., and K. Moffat. 2002. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14:1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosson, S., S. Rajagopal, and K. Moffat. 2003. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42:2-10. [DOI] [PubMed] [Google Scholar]
- 16.Delumeau, O., S. Dutta, M. Brigulla, G. Kuhnke, S. W. Hardwick, U. Völker, M. D. Yudkin, and R. J. Lewis. 2004. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J. Biol. Chem. 279:40927-40937. [DOI] [PubMed] [Google Scholar]
- 17.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufour, A., U. Voelker, A. Voelker, and W. G. Haldenwang. 1996. Relative levels and fractionation properties of Bacillus subtilis σB and its regulators during balanced growth and stress. J. Bacteriol. 178:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frohman, M. 1994. On beyond classic RACE (rapid amplification of cDNA ends). PCR Methods Appl. 4:540-558. [DOI] [PubMed] [Google Scholar]
- 20.Gaidenko, T. A., X. Yang, Y. M. Lee, and C. W. Price. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J. Mol. Biol. 288:29-39. [DOI] [PubMed] [Google Scholar]
- 21.Guérout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 22.Harper, S. M., J. M. Christie, and K. H. Gardner. 2004. Disruption of the LOV-Jα helix interaction activates phototropin kinase activity. Biochemistry 43:16184-16192. [DOI] [PubMed] [Google Scholar]
- 23.Harper, S. M., L. C. Neil, and K. H. Gardner. 2003. Structural basis of a phototropin light switch. Science 301:1541-1544. [DOI] [PubMed] [Google Scholar]
- 24.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 25.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 27.Hua, L., P. S. Hefty, Y. J. Lee, Y. M. Lee, R. S. Stephens, and C. W. Price. 2006. Core of the partner switching signalling mechanism is conserved in the obligate intracellular pathogen Chlamydia trachomatis. Mol. Microbiol. 59:623-636. [DOI] [PubMed] [Google Scholar]
- 28.Huala, E., P. W. Oeller, E. Liscum, I. Han, E. Larsen, and W. R. Briggs. 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 298:2120-2123. [DOI] [PubMed] [Google Scholar]
- 29.Jori, G., and J. D. Spikes. 1984. Photobiochemistry of porphyrins, p. 183-318. In K. C. Smith (ed.), Topics in photobiology. Plenum Press, New York, N.Y.
- 30.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang, C. M., K. Vijay, and C. W. Price. 1998. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol. Microbiol. 30:189-196. [DOI] [PubMed] [Google Scholar]
- 32.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 186:6124-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135-150. [DOI] [PubMed] [Google Scholar]
- 34.Koonin, E. V., L. Aravind, and M. Y. Galperin. 2000. A comparative-genomic view of the microbial stress response, p. 417-444. In G. Storz and R. Hennge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 35.Kozak, N. A., S. Mattoo, A. K. Foreman-Wykert, J. P. Whitelegge, and J. F. Miller. 2005. Interactions between partner switcher orthologs BtrW and BtrV regulate type III secretion in Bordetella. J. Bacteriol. 187:5665-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krauss, U., A. Losi, W. Gartner, K. E. Jaeger, and T. Eggert. 2005. Initial characterization of a blue-light sensing, phototropin-related protein from Pseudomonas putida: a paradigm for an extended LOV construct. Phys. Chem. Chem. Phys. 7:2804-2811. [DOI] [PubMed] [Google Scholar]
- 37.Lee, E. J., Y. H. Cho, H. S. Kim, B. E. Ahn, and J. H. Roe. 2004. Regulation of σB by an anti- and an anti-anti-σ factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 186:8490-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Losi, A. 2004. The bacterial counterparts of plant phototropins. Photochem. Photobiol. Sci. 3:566-574. [DOI] [PubMed] [Google Scholar]
- 39.Losi, A., E. Polverini, B. Quest, and W. Gartner. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Losi, A., B. Quest, and W. Gartner. 2003. Listening to the blue: the time-resolved thermodynamics of the bacterial blue-light receptor YtvA and its isolated LOV domain. Photochem. Photobiol. Sci. 2:759-766. [DOI] [PubMed] [Google Scholar]
- 41.Losi, A., E. Ternelli, and W. Gartner. 2004. Tryptophan fluorescence in the Bacillus subtilis phototropin-related protein YtvA as a marker of interdomain interaction. Photochem. Photobiol. 80:150-153. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. M. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Mittenhuber, G. 2002. A phylogenomic study of the general stress response sigma factor σB of Bacillus subtilis and its regulatory proteins. J. Mol. Microbiol. Biotechnol. 4:427-452. [PubMed] [Google Scholar]
- 44.Murray, J. W., O. Delumeau, and R. J. Lewis. 2005. Structure of a nonheme globin in environmental stress signaling. Proc. Natl. Acad. Sci. USA 102:17320-17325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakano, S., E. Küster-Schöck, A. D. Grossman, and P. Zuber. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:13603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano, S., M. M. Nakano, Y. Zhang, M. Leelakriangsak, and P. Zuber. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. USA 100:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pané-Farré, J., R. J. Lewis, and J. Stülke. 2005. The RsbRST stress module in bacteria: a signalling system that may interact with different output modules. J. Mol. Microbiol. Biotechnol. 9:65-76. [DOI] [PubMed] [Google Scholar]
- 48.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. ASM Press, Washington, D.C.
- 49.Price, C. W. 2002. General stress response, p. 161-178. In A. L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 50.Price, C. W., P. Fawcett, H. Cérémonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 51.Salomon, M., J. M. Christie, E. Knieb, U. Lempert, and W. R. Briggs. 2000. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39:9401-9410. [DOI] [PubMed] [Google Scholar]
- 52.Salomon, M., W. Eisenreich, H. Dürr, E. Schleicher, E. Knieb, V. Massey, W. Rüdiger, F. Müller, A. Bacher, and G. Richter. 2001. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc. Natl. Acad. Sci. USA 98:12357-12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seibert, M., P. J. Wetherbee, and D. D. Job. 1975. The effects of light intensity and spectral quality on growth and shoot initiation in tobacco callus. Plant Physiol. 56:130-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 55.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 57.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 59.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]