Abstract
IS16 is a thiol-sensitive, Q-deficient mutant strain of Escherichia coli. Here, we show that IS16 harbors a mutation in the ubiG gene encoding a methyltransferase required for two O-methylation steps of Q biosynthesis. Complementation of IS16 with either ubiG or ubiXK-12 reverses this phenotype, suggesting that UbiX may interact with UbiG.
Ubiquinone (coenzyme Q or Q) is a prenylated, redox-active lipid that functions as an electron carrier in the respiratory electron transport chain in mitochondria of eukaryotes and the plasma membranes of most prokaryotes (5, 8). In addition to the role of Q in the electron transport chain, the redox poise of the quinone/hydroquinone pool (Q/QH2) acts as a signal for the global two-component ArcB/ArcA (anoxic redox control) system in Escherichia coli (9). Q also functions in the process of disulfide bond formation in E. coli periplasm (2).
Previous work has shown that IS16, a Q-deficient mutant strain of E. coli, is thiol hypersensitive and unable to grow on succinate. Expression of the ubiX gene from E. coli K-12 was found to rescue the IS16 Q-deficient phenotypes (30). E. coli has two distinct genes, ubiD and ubiX, thought to be involved in the decarboxylation of 3-octaprenyl-4-hydroxybenzoate (19, 23). The IS16 mutant strain was found to contain an ubiX gene sequence identical to that of its parental strain THU (an E. coli 15 strain), encoding a single-amino-acid substitution (S98R) relative to the ubiX sequence from E. coli K-12. It was proposed that strain IS16 harbored a second mutation in ubiD, a gene considered to be isofunctional with ubiX (30). The physical location of ubiD on the E. coli chromosome was established (31). The ubiD gene sequence was determined for both THU and IS16 strains; however, we found no mutation in the ubiD gene of either strain, indicating that the mutation must reside elsewhere.
Here, we show that IS16 harbors a mutation in the ubiG gene encoding a methyltransferase required for two O-methylation steps of Q biosynthesis. We show that Q biosynthesis in IS16 is restored by expression of either the E. coli ubiG or the ubiXK-12 gene, providing genetic evidence for an interaction of UbiG and UbiX in Q biosynthesis in E. coli (Table 1 shows a list of strains).
TABLE 1.
Genotypes and sources of E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| HW272 | Parental strain of GD1 | 28 |
| GD1 | ubiG::Kan | 13 |
| THU | Derivative of strain 15, thy-43 his-67 pyr-37 | 7 |
| IS16 | Mutagenized derivative of THU (ubiG T395A) | 29; this study |
| Plasmids | ||
| pUbiGTHU | Harbors ubiGTHU (Glu8) | This study |
| pAHG | Harbors ubiGK12 (Val8) | 13 |
| pHZ1 | Harbors ubiXTHU (Arg98) | 29 |
| pPZ2 | Harbors ubiXK12 (Ser98) | 29 |
Identification of an ubiG mutation in the IS16 mutant strain.
To identify the metabolic block of Q synthesis in strain IS16, a plasmid library was constructed from genomic DNA of E. coli THU. Chromosomal DNA of strain THU was partially digested by Sau3AI, and 3- to 12-kb fragments were inserted into the BamHI sites of pUC18 plasmids (Amersham Pharmacia Biotech Inc., Piscataway, NJ). IS16 cells were transformed with this library, and transformants were screened for the ability to grow on media containing succinate. The complementing chromosomal insert contained a 6,023-bp hybrid segment of DNA resulting from the ligation of two chromosomal Sau3AI fragments into the vector; one fragment contained a portion of gyrA, complete ubiG, and a portion of xfaL, while the second fragment contained araB, complete araA, and a small stretch of araD. To determine whether IS16 harbors a mutation in the ubiG gene, the sequence was amplified from IS16 and THU genomic DNA. Sequence analysis revealed that the ubiG gene in IS16 contained a unique nucleotide substitution, T395A, which resulted in an amino acid change, L132Q. The L132Q mutation in IS16 lies adjacent to methyltransferase motif II (Fig. 1). The four motifs shown in Fig. 1 are present in a large family of AdoMet-dependent methyltransferases (18).
FIG. 1.
Alignment of E. coli UbiG, S. cerevisiae Coq3, human Coq3, and rat COMT amino acid sequences across methyltransferase motifs I, post-I, II, and III. The box designates the L132Q ubiG mutation in the IS16 mutant strain. This amino acid substitution is adjacent to methyltransferase motif II. Sequence analysis revealed that ubiG genes from THU and IS16 share five nucleotide variances in comparison to the ubiG gene from K-12. These variances include T23A, resulting in V8E, and four silent changes, G90T, C109T, A294G, and C321T. Secondary structural elements (β1, β2, β4, and β5) and important active site residues involved in the binding of ligands are indicated for the rat soluble COMT. a, AdoMet; m, magnesium; s, substrate (27).
The structure of UbiG is not currently available; hence, it is difficult to ascertain the functional role of this amino acid substitution. However, based on the known crystal structure of the rat catechol O-methyltransferase (COMT), the post-motif II region is known to comprise the active site of the enzyme, with specific residues that contact AdoMet and the catechol substrate (29). The alignment between E. coli UbiG, Saccharomyces cerevisiae Coq3p, human hCoq3p, and rat COMT amino acid sequences over methyltransferase motifs I, post-I, II, and III is shown in Fig. 1. The UbiG L132Q mutation occurs at the position corresponding to K144 in COMT, which is shown to be involved in substrate binding. It is reasonable to assume that the L132Q mutation may impact the substrate binding specificity of the UbiG polypeptide. Use of PHYRE, a protein fold recognition server (http://www.sbg.bio.ic.ac.uk/phyre/), identified mycolic acid cyclopropane synthases from Mycobacterium tuberculosis as protein structures closely related to the UbiG polypeptide sequence. Cyclopropane synthases catalyze the transfer of the methyl group from AdoMet to a double bond of the acyl substrate. Residues 137 to 144 of mycolic acid cyclopropane synthases are involved in cofactor binding (15), are located in the post-motif II region, and also implicate the UbiG L132Q substitution in UbiG as potentially affecting cofactor or substrate binding.
IS16 can be complemented by either ubiG or ubiX.
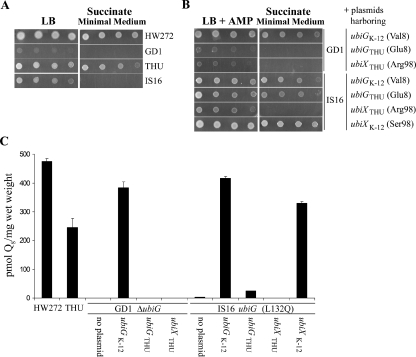
The ubiG gene from E. coli THU was amplified by PCR and inserted into pNoTA/T7. The resulting plasmid, pUbiGTHU, restored growth on succinate and partially restored production of Q8 in the IS16 mutant (Fig. 2). Rescue of Q-deficient phenotypes can be achieved with levels of Q that are significantly lower than the amount normally present in wild-type cells (3, 6, 17). Lower steady-state levels for the UbiGTHU polypeptide than for UbiGK-12 were observed when each construct was expressed in the GD1 mutant strain (data not shown). Both IS16 and ubiG null mutant strain GD1 were rescued by ubiGK-12. Based on this, it seems likely that the differential rescue may be due to efficiency of expression: ubiGK-12 in pAHG is expressed from the yeast CYC1 promoter, while ubiGTHU is expressed from its native promoter sequence. A plasmid harboring ubiX from E. coli THU, pHZ1, failed to rescue either IS16 or GD1, while pPZ2, a plasmid harboring ubiX from E. coli K-12, rescued IS16, as observed previously (30). pPZ2 failed to rescue GD1 (data not shown). For the determination of E. coli Q8 content, cultures were grown in Davis minimal media at 37°C overnight and collected by centrifugation. E. coli cells (0.1 to 0.2 g wet weight) were extracted and quinones separated by reverse-phase high-pressure liquid chromatography (HPLC) and quantified with an electrochemical detector as described previously (16). Q10 (Sigma-Aldrich, St. Louis, MO) was added as an internal standard (final concentration of 20 pmol/μl). The areas of the peaks corresponding to Q8 and Q10 samples and standards were determined with Gilson Unipoint version 5.1 software.
FIG. 2.
Succinate growth and Q8 levels in E. coli strains. (A) Serial 10-fold dilutions (starting optical density at 600 nm, 0.2) of the indicated strains were plated on LB and succinate minimal medium plates supplemented with thymine, histidine, and uracil and incubated at 37°C for 1 day. (B) Serial 10-fold dilutions (starting optical density at 600 nm, 0.2) of the indicated strains were plated on LB-AMP and succinate minimal medium plates supplemented with thymine, histidine, and uracil and incubated at 37°C for 1 day. E. coli strains harbored plasmids expressing the designated genes as follows: pAHG, UbiGK-12; pUbiGTHU, UbiGTHU; pHZ1, UbiXTHU; and pPZ2, UbiXK-12. (C) Quinones were extracted and separated by reverse-phase HPLC, and Q8 content was quantified. A standard for Q8 was prepared from E. coli lipid extracts and verified by mass spectrophotometric methods. External standard curves were created for Q8 (600, 300, 75, and 30 pmol) and Q10 (300, 150, and 37.5 pmol). The amounts of Q8 were corrected by recovery of the Q10 internal standard.
O-Methyltransferase activity in the IS16 E. coli mutant is restored by expression of either E. coli ubiG or ubiX.
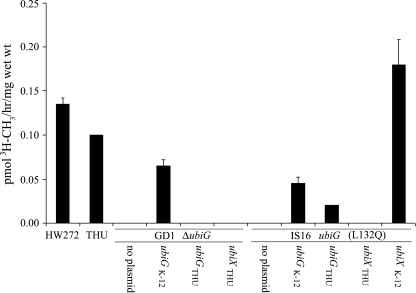
The purified E. coli UbiG polypeptide has been shown to function as a soluble enzyme and catalyze the O methylation of three different farnesylated analogs of intermediates in Q biosynthesis (24). To examine the effect of the L132Q mutation on UbiG O-methyltransferase activity in the IS16 mutant, we employed a cell permeabilization assay because this method has been shown to preserve O-methyltransferase activity in analyses of another E. coli ubiG mutant (14). In vitro assays of O-methyltransferase activity employed the farnesylated analogs of the E. coli intermediate (5-farnesyl-2-hydroxyphenol), demethyl-Q3 (2-farnesyl-5-hydroxy-6-methoxy-3-methyl-1,4 benzoquinone), or the yeast intermediate (3,4-dihydroxy-5-farnesylbenzoic acid) and S-adenosyl-[methyl-3H]l-methionine as previously described (22, 24). O-methyltransferase activity assays were linearly dependent on time of incubation, substrate concentration, and amount of permeabilized cells. O-methyltransferase activity with 3,4-dihydroxy-5-farnesylbenzoic acid (Fig. 3, open bars) or 5-farnesyl-2-hydroxyphenol (Fig. 3, closed bars) was readily detected in the HW272 and THU E. coli parental strains, while activity was either not detected or present at significantly lower levels in the GD1 and IS16 ubiG mutant strains. Complementation of IS16 with either ubiX or ubiG from K-12 or ubiG from THU rescued O-methyltransferase activity with the demethyl-Q3 substrate (Fig. 4). However, O-methyltransferase activity in the ubiG disruption mutant GD1 was restored only by ubiG from K-12.
FIG. 3.
O methylation of early Q intermediates is defective in IS16 and is restored by expression of E. coli UbiXK-12. Permeabilized E. coli cells were prepared from HW272 (wild-type, parental strain of GD1), GD1 (ubiG disruption mutant), THU (parental strain of IS16), IS16 (UbiG-L132Q), and IS16:pPZ2 (1S16 harboring UbiXK-12 on a plasmid) and incubated with farnesylated analogs of 1 mM 3,4-dihydroxy-5-farnesylbenzoic acid (open bars), 250 μM 5-farnesyl-2-hydroxyphenol (closed bars) for 10 min. S-Adenosyl-[methyl-3H]l-methionine (6.9 μM, 81.5 Ci/mmol; PerkinElmer Life Sciences) was added to the reaction mixture. Following incubation at 37°C for 45 min, lipids were extracted and separated by reverse-phase HPLC (BetaBasic C18 column, 5 μM, 4.6 by 250 mm; Thermo Electron Corporation). Radioactivity present in fractions 6 and 7 (open bars) or fractions 10 to 12 (closed bars) is expressed in pmol of CH3 groups/hr/mg wet weight and eluted with the 3H-labeled product standard. Error bars represent standard deviations obtained from two O-methyltransferase assays of the same sample, and data shown represent two independent experiments.
FIG. 4.
The final O-methylation reaction is defective in the IS16 mutant and is restored by expression of either UbiG or UbiXK-12. In vitro O-methyltransferase assays were carried out as described for Fig. 3, except that 50 μM 2-farnesyl-5-hydroxy-6-methoxy-3-methyl-1,4 benzoquinone was used as a substrate, and 1 mM NADH was included in all incubations to allow formation of the hydroquinone. At the end of the reaction incubation, 25 μl of freshly prepared 1% ammonium cerium (IV) nitrate was added to the reaction mixture prior to lipid extraction (to oxidize hydroquinone products). The elution positions of the methylated products (fractions 11 and 12) were similar to that of the 3H-labeled product standard and are expressed in pmol of CH3 groups/hr/mg wet weight. Error bars represent standard deviations obtained from two O-methyltransferase assays of the same sample, and data shown represent two independent experiments. E. coli strains harbored plasmids expressing the designated genes as follows: pAHG, UbiGK-12; pUbiGTHU, UbiGTHU; pHZ1, UbiXTHU; and pPZ2, UbiXK-12.
Steady-state levels of UbiG polypeptide in the IS16 mutant strain are similar to those in the wild type.
The steady-state levels of UbiG were examined to determine whether the deficiency in O-methyltransferase activity in the IS16 strain was attributed to UbiG polypeptide levels. UbiG steady-state levels for E. coli HW272, GD1, THU, IS16, and IS16:pPZ2 (plasmid harboring ubiXK-12) cells were assessed by Western blot analysis as previously described (12). Levels of UbiG polypeptide in IS16 were similar to those in the parental strain THU (Fig. 5).
FIG. 5.
Steady-state levels of UbiG and cytochrome o oxidase in E. coli strains. Purified E. coli protein His6-UbiG (24) was used to generate antisera in rabbits (Cocalico Biologicals, Inc.). Steady-state levels of UbiG were analyzed for E. coli HW272 (wild-type, parental strain of GD1), GD1 (ubiG disruption mutant), THU (parental strain of IS16), IS16 (UbiG-L132Q), and IS16:pPZ2 (IS16 complemented with UbiXK-12) cells. Cytochrome o oxidase was used as a loading control.
Complementation studies revealed that multiple copies of ubiXK-12 restore growth on succinate, Q8 levels (30), and O-methyltransferase activity in IS16 (Fig. 2, 3, and 4). This is a surprising finding because the O-methyltransferase activity of the purified UbiG polypeptide does not require other Ubi polypeptides (24). It is possible that overexpression of UbiX may stimulate another O-methyltransferase with overlapping substrate specificity to UbiG. We consider this unlikely; the rescue is unique to the L132Q UbiG point mutant. Interactions between Ubi polypeptides were not identified by use of tagged constructs to identify multisubunit complexes (7). However, an interaction was detected between UbiX and FldA. FldA plays an essential role in the synthesis of isoprenoid precursors in E. coli (26) and suggests that synthesis of the isoprenoid tail occurs in complex with ring modifications. Based on the genetic evidence presented here, UbiX, a protein thought to be involved in a decarboxylation step in Q biosynthesis, may be required to stabilize the catalytic activity of UbiG L132Q by direct interaction or by channeling substrates (Fig. 6). These results provide support for a polypeptide complex involved in E. coli Q biosynthesis, first described by Knoell (20, 21). Knoell demonstrated that a complex of membrane-associated polypeptides in E. coli converts 2-octaprenylphenol to Q8 in vitro.
FIG. 6.
Genetic evidence for the interaction of UbiX and UbiG. IS16 harbors a mutation in UbiG (L132Q) that catalyzes the O-methylation step in E. coli coenzyme Q biosynthesis. Lack of growth on succinate, Q deficiency, thiol hypersensitivity (29), and inactive O-methyltransferase activity in IS16 are restored with wild-type ubiXK-12, suggesting that UbiX may be interacting with UbiG (L132Q). X, UbiX; G, UbiG.
Similarly, a growing body of evidence suggests that a complex of Coq polypeptides is involved in Q biosynthesis in S. cerevisiae. Gel filtration chromatography shows that Coq3p, Coq4p, Coq6p, and Coq7p coelute as a high-molecular-weight complex (22, 28). Coq3p, Coq4p, and Coq7 comigrate as high-molecular-mass complexes as assessed by two-dimensional blue native analysis (22, 28). O-Methyltransferase activity is decreased in coq null mutants relative to that in atp2 and cor1 respiratory deficient mutants (13). Deletions in any of the COQ genes affect the steady-state levels of Coq3p, Coq4p, and Coq6p (11). Unlike yeast coq3-coq9 mutants that accumulate an early predominant intermediate, 3-hexaprenyl-4-hydroxybenzoic acid (HHB) (1, 4, 12, 25), E. coli ubi mutants tend to accumulate immediate precursor substrates at the blocked steps (10). However, no ubiG mutant has been reported to accumulate 3,4-dihydroxy-5-octaprenyl-benzoic acid (14, 27), possibly due to the instability of the catechol moiety. We are currently investigating the lipid quinone intermediate formed in the IS16 strain to further characterize the polypeptide Q-biosynthetic complex in the THU genetic background strain of E. coli.
Acknowledgments
We thank J. N. Shepherd for the farnesylated Q intermediate analogs and W. W. Poon for the generation of the UbiG antibody. We thank N. Lee for the purified E. coli protein His6-UbiG and R. H. Kaback for the generous gift of the E. coli cytochrome oxidase antibody. We also thank members of the Clarke laboratory for helpful suggestions in this study.
This work was supported in part by National Institutes of Health grant GM45952.
REFERENCES
- 1.Baba, S. W., G. I. Belogrudov, J. C. Lee, P. T. Lee, J. Strahan, J. N. Shepherd, and C. F. Clarke. 2004. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 279:10052-10059. [DOI] [PubMed] [Google Scholar]
- 2.Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. [DOI] [PubMed] [Google Scholar]
- 3.Bar-Tana, J., B. J. Howlett, and R. Hertz. 1980. Ubiquinone synthetic pathway in flagellation of Salmonella typhimurium. J. Bacteriol. 143:637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belogrudov, G. I., P. T. Lee, T. Jonassen, A. Y. Hsu, P. Gin, and C. F. Clarke. 2001. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q. synthesis. Arch. Biochem. Biophys. 392:48-58. [DOI] [PubMed] [Google Scholar]
- 5.Brandt, U., and B. Trumpower. 1994. The protonmotive Q cycle in mitochondria and bacteria. Crit. Rev. Biochem. Mol. Biol. 29:165-197. [DOI] [PubMed] [Google Scholar]
- 6.Branicky, R., P. A. Nguyen, and S. Hekimi. 2006. Uncoupling the pleiotropic phenotypes of clk-1 with tRNA missense suppressors in Caenorhabditis elegans. Mol. Cell. Biol. 26:3976-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 8.Collins, M. D., and D. Jones. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45:316-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, F., and I. G. Young. 1978. Isolation and characterization of intermediates in ubiquinone biosynthesis. Methods Enzymol. 53:600-609. [DOI] [PubMed] [Google Scholar]
- 11.Gin, P., and C. F. Clarke. 2005. Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J. Biol. Chem. 280:2676-2681. [DOI] [PubMed] [Google Scholar]
- 12.Gin, P., A. Y. Hsu, S. C. Rothman, T. Jonassen, P. T. Lee, A. Tzagoloff, and C. F. Clarke. 2003. The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J. Biol. Chem. 278:25308-25316. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, A. Y., T. Q. Do, P. T. Lee, and C. F. Clarke. 2000. Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim. Biophys. Acta 1484:287-297. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, A. Y., W. W. Poon, J. A. Shepherd, D. C. Myles, and C. F. Clarke. 1996. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry 35:9797-9806. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C. C., C. V. Smith, M. S. Glickman, W. R. Jacobs, Jr., and J. C. Sacchettini. 2002. Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis. J. Biol. Chem. 277:11559-11569. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, A., P. Gin, B. N. Marbois, E. J. Hsieh, M. Wu, M. H. Barros, C. F. Clarke, and A. Tzagoloff. 2005. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J. Biol. Chem. 280:31397-31404. [DOI] [PubMed] [Google Scholar]
- 17.Jonassen, T., and C. F. Clarke. 2000. Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J. Biol. Chem. 275:12381-12387. [DOI] [PubMed] [Google Scholar]
- 18.Katz, J. E., M. Dlakic, and S. Clarke. 2003. Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2:525-540. [DOI] [PubMed] [Google Scholar]
- 19.Kawamukai, M. 2002. Biosynthesis, bioproduction and novel roles of ubiquinone. J. Biosci. Bioeng. 94:511-517. [DOI] [PubMed] [Google Scholar]
- 20.Knoell, H. E. 1979. Isolation of a soluble enzyme complex comprising the ubiquinone-8 synthesis apparatus from the cytoplasmic membrane of Escherichia coli. Biochem. Biophys. Res. Commun. 91:919-925. [DOI] [PubMed] [Google Scholar]
- 21.Knoell, H. E. 1979. Ubiquinone synthesis in vitro starting from 2-octaprenyl phenol. FEBS Lett. 97:155-158. [DOI] [PubMed] [Google Scholar]
- 22.Marbois, B., P. Gin, K. F. Faull, W. W. Poon, P. T. Lee, J. Strahan, J. N. Shepherd, and C. F. Clarke. 2005. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 280:20231-20238. [DOI] [PubMed] [Google Scholar]
- 23.Meganathan, R. 2001. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203:131-139. [DOI] [PubMed] [Google Scholar]
- 24.Poon, W. W., R. J. Barkovich, A. Y. Hsu, A. Frankel, P. T. Lee, J. N. Shepherd, D. C. Myles, and C. F. Clarke. 1999. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 274:21665-21672. [DOI] [PubMed] [Google Scholar]
- 25.Poon, W. W., B. N. Marbois, K. F. Faull, and C. F. Clarke. 1995. 3-Hexaprenyl-4-hydroxybenzoic acid forms a predominant intermediate pool in ubiquinone biosynthesis in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 320:305-314. [DOI] [PubMed] [Google Scholar]
- 26.Puan, K. J., H. Wang, T. Dairi, T. Kuzuyama, and C. T. Morita. 2005. fldA is an essential gene required in the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis. FEBS Lett. 579:3802-3806. [DOI] [PubMed] [Google Scholar]
- 27.Stroobant, P., I. G. Young, and F. Gibson. 1972. Mutants of Escherichia coli K-12 blocked in the final reaction of ubiquinone biosynthesis: characterization and genetic analysis. J. Bacteriol. 109:134-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran, U. C., B. Marbois, P. Gin, M. Gulmezian, T. Jonassen, and C. F. Clarke. 2006. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast COQ7 polypeptide in coenzyme Q biosynthesis. J. Biol. Chem. 281:16401-16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidgren, J., L. A. Svensson, and A. Liljas. 1994. Crystal structure of catechol O-methyltransferase. Nature 368:354-358. [DOI] [PubMed] [Google Scholar]
- 30.Zeng, H., I. Snavely, P. Zamorano, and G. T. Javor. 1998. Low ubiquinone content in Escherichia coli causes thiol hypersensitivity. J. Bacteriol. 180:3681-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, H., and G. T. Javor. 2000. Identification of the ubiD gene on the Escherichia coli chromosome. J. Bacteriol. 182:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]