Abstract
Expression of yeast mitochondrial aconitase (Aco1) in a Bacillus subtilis aconitase null mutant restored aconitase activity and glutamate prototrophy but only partially restored sporulation. Late sporulation gene expression in the Aco1-expressing strain was delayed.
Bacillus subtilis aconitase, encoded by the citB gene (6), is both an enzyme of the Krebs citric acid cycle and an RNA binding protein (1, 6). The RNA binding function is similar to that of the eukaryotic cytoplasmic aconitase IRP-1 (iron regulatory protein 1) (2, 4, 8, 9), but the physiological role of B. subtilis aconitase RNA binding is not known (1). A citB null mutant requires glutamate (Glt) for growth and is severely defective in sporulation, having a sporulation efficiency 106-fold lower than that of the wild type (5). Part of the reason for this defect is intracellular and extracellular accumulation of citrate, which chelates divalent cations, reduces the pH, and prevents activation of the master transcriptional regulator of sporulation Spo0A, resulting in a blockage of sporulation at stage 0 (5). However, alleviation of these metabolic defects results in only partial restoration of sporulation (5, 11, 13, 20), suggesting a nonenzymatic role for aconitase in sporulation, possibly through utilization of its RNA binding function.
To assess the effect of the loss of aconitase RNA binding on sporulation, we sought to create a B. subtilis mutant strain that has aconitase enzyme activity but no RNA binding activity. To do so, we expressed a heterologous aconitase, the Saccharomyces cerevisiae mitochondrial aconitase (Aco1), which is devoid of RNA binding activity (12, 16, 17), in a citB null mutant strain and tested the ability of the yeast enzyme to substitute for B. subtilis aconitase during growth and sporulation.
The coding sequence of Aco1 was cloned downstream of a B. subtilis promoter (Pspac*) and ribosome binding site, in plasmid pKM67 (13), and integrated into the chromosome of a citB null mutant, MAB160 [trpC2 pheA1 Ω(citB::spc)] (5), at the nonessential amyE locus. Transformants were selected for glutamate prototrophy (Glt+) or for chloramphenicol resistance (Camr), a vector marker. Camr transformants appeared during overnight incubation at 30°C, but most were Glt−. Glt+ transformants appeared only after 48 to 72 h at 30°C; the prolonged incubation period suggested that a mutation in the recipient cells might have been required to permit the appearance of the Glt+ transformants. Glt+ transformants grew in minimal medium at 30°C but not at 37°C. Note that laboratory strains of S. cerevisiae grow much better at 30°C than at 37°C (D. Dawson, personal communication).
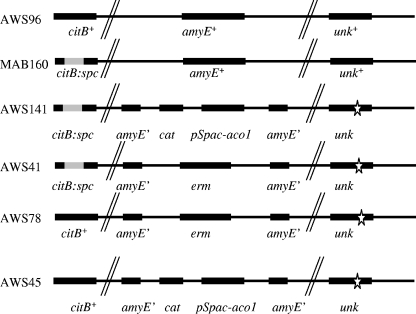
To investigate whether a mutation had occurred in the Glt+ transformants, chromosomal DNA from a Glt+ Camr transformant was isolated and introduced again into MAB160. Transformants were selected either for the Glt+ or for the Camr phenotype and were then tested for the unselected marker. All Glt+ transformants were Camr, but only 5% of Camr transformants were Glt+. In addition, the frequency of primary Glt+ transformants was 20-fold lower than for Camr transformants. The simplest explanation for this result is that two individual pieces of DNA must integrate into the citB null mutant in order to obtain a strain expressing enough Aco1 activity to yield the Glt+ phenotype. One such strain was called AWS141 [trpC2 pheA1 Ω(citB::spc) amyE::(Pspac*-aco1 cat) unk]. (unk indicates the uncharacterized mutation that enables Aco1 activity in B. subtilis). Preliminary results suggest that this unlinked mutation stabilizes Aco1 protein (data not shown). To obtain a strain lacking aco1 but otherwise isogenic, AWS141 was transformed with pJPM82 (3), selecting for erythromycin resistance. The resulting strain, AWS41 [trpC2 pheA1 Ω(citB::spc) amyE::erm unk], was deleted for aco1, was Glt−, and retained the mutation that Aco1 needs to be active in B. subtilis (Fig. 1). The retention of the unk mutation was confirmed by determining the cotransformation ratios of Glt+ and Camr integrants of MAB160 and AWS41 transformed with the aco1-containing derivative of pKM67 (data not shown).
FIG. 1.
Genotypic description of strains. The citB locus was either wild type or contained a spectinomycin resistance gene insertion, which results in a B. subtilis aconitase null mutant. The amyE locus was either wild type or contained various integrated DNAs, either the yeast mitochondrial aconitase gene (aco1) and an antibiotic resistance gene or an antibiotic resistance gene alone. unk represents an unlinked chromosomal mutation required for Aco1 function in B. subtilis.
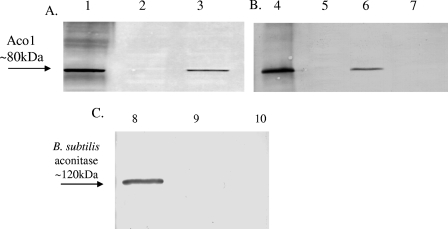
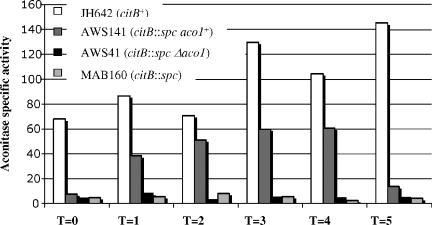
AWS141 grew at 30°C in minimal medium without the addition of Glt, albeit at a lower rate than the wild type (the wild type had a doubling time of 145 min, while AWS141 had a doubling time of 180 min), while AWS41 was unable to grow without Glt (data not shown). Immunoblotting with antibody specific to Aco1 verified synthesis of the yeast protein in strain AWS141 but not in strain JH642, AWS41, or MAB160 (Fig. 2), while immunoblotting with antibody specific to B. subtilis aconitase confirmed that only JH642 synthesized this protein. The specific activity of Aco1, determined as previously described (6), during exponential phase in minimal medium was similar to that of B. subtilis aconitase; the wild type had a specific activity of 58.6 U/mg, while AWS141 had a specific activity of 49.8 U/mg. In nutrient broth medium, AWS141 grew at a rate similar to that of the wild type (data not shown). Aco1 specific activity, although present and substantial, did not quite reach the same level as for wild-type B. subtilis aconitase activity, while AWS41 had no detectable enzyme activity (Fig. 3).
FIG. 2.
Immunoblots with antibodies to yeast mitochondrial aconitase, Aco1, or to B. subtilis aconitase. Growth was at 30°C in DSM (A and C) (7) or minimal medium (B). Cells were isolated upon entry into stationary phase, and cell extracts were analyzed by immunoblotting with Aco1 antibody (A and B) or with antibody to B. subtilis aconitase (C). Lanes 1 and 4 show S. cerevisiae cell extract. The remaining lanes contain cell extracts from B. subtilis: lane 2, MAB160 (citB::spc); lane 3, AWS141 (citB::spc Pspac*-aco1 unk); lane 5, wild type (citB+); lane 6, AWS141(citB::spc Pspac*-aco1 unk); lane 7, AWS41 (citB::spc Δ aco1 unk); lane 8, JH642 (citB+); lane 9, AWS141 (citB::spc Pspac*-aco1 unk); lane 10, MAB160 (citB::spc). B. subtilis aconitase is a protein of 99 kDa but has the mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis of a protein of approximately 120 kDa (6).
FIG. 3.
Kinetics of appearance of aconitase activity during stationary phase. Strains JH642, AWS141, MAB160, and AWS41 were grown in DSM, and cells were isolated at the time points indicated, in hours (T = 0, time of entry into stationary phase). Cell extracts were prepared and analyzed for aconitase activity (6), reported as units per milligram of protein.
Spore formation was partially restored in strain AWS141 compared to MAB160 or AWS41, as shown by heat resistance assays of cells harvested approximately 20 h after entry into stationary phase (19) (Table 1). Still, fewer than 10% of AWS141 cells had sporulated at that time. After an additional 20 h of incubation, 45% of the remaining AWS141 cells were spores, compared to 98% of wild-type cells (Table 1). These results suggest that AWS141 is competent to sporulate but either initiates or completes sporulation with a delay compared to cells expressing B. subtilis aconitase.
TABLE 1.
Sporulation of B. subtilis strains expressing yeast aconitasea
| Strain | Relevant genotype | Total titer | Spore titer | Sporulation efficiencyb | Relative sporulation efficiencyc |
|---|---|---|---|---|---|
| T = 20 h | |||||
| JH642 | citB+ | 3.9 × 108 | 2.7 × 108 | 0.69 | 1.0 |
| MAB160 | citB::spc | 4.9 × 107 | 10 | 2.7 × 10−7 | 3.7 × 10−8 |
| AWS45 | citB+Δaco1 unk | 3.9 × 108 | 2.6 × 108 | 0.67 | 0.98 |
| AWS141 | citB::spc Pspac*-aco1 unk | 1.74 × 108 | 1.44 × 107 | 0.08 | 0.05 |
| AWS41 | citB::spc Δaco1 unk | 4.0 × 107 | 30 | 7.5 × 10−7 | 1.1 × 10−7 |
| T = 40 h | |||||
| JH642 | citB+ | 5.34 × 108 | 5.24 × 108 | 0.98 | 1.0 |
| MAB160 | citB::spc | 3.44 × 107 | 2.2 × 103 | 6.5 × 10−5 | 4.14 × 10−6 |
| AWS45 | citB+Δaco1 unk | 5.1 × 108 | 4.56 × 108 | 0.90 | 0.87 |
| AWS141 | citB::spc Pspac*-aco1 unk | 1.4 × 108 | 6.34 × 107 | 0.45 | 0.12 |
| AWS41 | citB::spc Δaco1 unk | 2.3 × 107 | 8.9 × 103 | 3.9 × 10−4 | 1.7 × 10−5 |
The indicated B. subtilis strains were grown in DSM at 30°C (7). Samples removed at T = 20 (20 h after entry into stationary phase) or at T = 40 were titered before (total titer) and after (spore titer) incubation for 10 min at 80°C.
Sporulation efficiency is defined as the ratio of the spore titer to the total titer.
Relative sporulation efficiency is defined as the spore titer of a given strain relative to that of the parental strain, JH642.
To make sure the delay was specific to Aco1 expression and not to the uncharacterized mutation that stabilizes Aco1, we introduced the citB+ allele into strains AWS41 and AWS141. We used wild-type chromosomal DNA to transform AWS41 to Glt+ at 37°C and verified the simultaneous loss of spectinomycin resistance. The resulting strain, AWS45, deleted for aco1, was Glt+ due to the enzyme activity of B. subtilis aconitase and contained the enabling mutation (Fig. 1). In addition, we used wild-type DNA to transform strain AWS141 to Glt+ with selection at 42°C, a temperature at which Aco1 is inactive. The resulting strain, AWS78 (Fig. 1), produced both Aco1 and B. subtilis aconitase, as seen by immunoblotting (data not shown). Importantly, both AWS45 and AWS78 formed heat-resistant spores at a rate similar to that of wild-type cells (data not shown). These results indicate that neither Aco1 nor the Aco1-enabling mutation has an inhibitory effect on sporulation.
We performed microarray analysis to examine the temporal expression patterns of sporulation-specific genes in order to determine the stage of sporulation at which AWS141 is blocked or delayed. Experimental details can be found in the accompanying paper (19). Since AWS141 was created in an MAB160 background, a new wild-type strain, AWS96, was constructed by transforming MAB160 with JH642 (trpC2 pheA1) chromosomal DNA and citB+ transformants were selected by their Glt+ phenotype; simultaneous loss of spectinomycin resistance was verified. Transcript levels in AWS96 and AWS141 strains were compared at different time points during late stationary phase with RNA isolated from vegetative wild-type cells used as a reference.
Expression of Aco1 allowed citB null mutant cells to bypass their stage 0 defect. That is, genes such as spoIIG, spoIIB, and spoIIA, under the control of the earliest sporulation transcription factor, Spo0A, were induced to the same extent upon entry into stationary phase in AWS141 and AWS96 cells (Table 2). In addition, other transcripts characteristic of stages II and III of sporulation, for example, spoIIIAC, spoIVA, spoVR, spoIIR, and spoIIQ, were also induced in both AWS141 and AWS96 (Table 2). However, AWS141 was reduced in expression of some σG-dependent (i.e., late forespore-specific) genes and severely defective in expression of almost all σK-dependent (i.e., late mother-cell-specific) genes (Table 3). Thus, Aco1 enzyme activity allows AWS141 to bypass the original stage 0 defect seen in a citB null mutant but does not permit timely expression of many late sporulation genes.
TABLE 2.
Microarray analysis of stage II and stage III sporulation genesa
| Gene | Expression ratio
|
Transcription mode | |
|---|---|---|---|
| AWS96 | AWS141 | ||
| spoIIAA | 7.5 | 9.7 | Spo0A dependent |
| spoIIAB | 7.7 | 14.4 | Spo0A dependent |
| spoIIB | 5.1 | 4.3 | Spo0A dependent |
| spoIIGA | 2.6 | 9.0 | Spo0A dependent |
| sigG | 4.7 | 5.6 | σF dependent |
| spoIIQ | 3.3 | 5.4 | σF dependent |
| spoIIR | 2.5 | 2.1 | σF dependent |
| spoIIIAC | 2.0 | 3.1 | σE dependent |
| spoIIIAB | 1.9 | 3.7 | σE dependent |
| spoIIIAE | 3.0 | 3.1 | σE dependent |
| spoIIIAH | 1.7 | 2.2 | σE dependent |
| spoIIID | 15.2 | 4.6 | σE dependent |
| spoIVA | 10.0 | 6.9 | σE dependent |
| spoIVFA | 3.4 | 4.2 | σE dependent |
| spoVID | 3.8 | 2.5 | σE dependent |
| spoVR | 3.1 | 4.2 | σE dependent |
RNA was isolated at h 4 of stationary phase from wild-type [AWS96 (citB+)] or Aco1-expressing [AWS141 (citB::spc Pspac*-aco1)] cells. RNA was labeled with Cy3 and mixed with RNA isolated during the exponential growth phase from wild-type (AWS96) cells that had been labeled with Cy5. The mixed RNA was annealed to arrays of 4,106 B. subtilis open reading frames. The values reported are the ratios of annealing of T4 RNA to exponential-phase RNA. For details of the microarray analysis, see reference 19.
TABLE 3.
Microarray analysis of wild-type (AWS96) and Aco1 (AWS141) cells at h 8 of stationary phasea
| Gene | Expression ratio
|
Transcription mode | |
|---|---|---|---|
| AWS96 | AWS141 | ||
| gerBB | 4.1 | 2.6 | σG dependent |
| spoIVB | 1.8 | 2.1 | σG dependent |
| spoVAB | 2.1 | 2.4 | σG dependent |
| sspD | 11.4 | 6.1 | σG dependent |
| sspH | 16.9 | 1.4 | σG dependent |
| sspI | 4.8 | 1.8 | σG dependent |
| sspJ | 12.1 | 1.3 | σG dependent |
| sspK | 4.5 | 1.7 | σG dependent |
| cgeA | 27.5 | 2.5 | σK dependent |
| cgeB | 52.0 | 1.4 | σK dependent |
| cgeC | 8.7 | 1.6 | σK dependent |
| cgeD | 5.7 | 1.4 | σK dependent |
| cotA | 8.8 | 2.3 | σK dependent |
| cotB | 99.5 | 2.6 | σK dependent |
| cotC | 7.3 | 1.9 | σK dependent |
| cotD | 48.0 | 1.3 | σK dependent |
| cotG | 74.2 | 1.8 | σK dependent |
| cotS | 3.2 | 0.8 | σK dependent |
| cotSA | 27.7 | 3.2 | σK dependent |
| cotT | 36.3 | 2.1 | σK dependent |
| cotW | 130.7 | 5.4 | σK dependent |
| cotX | 95.0 | 1.7 | σK dependent |
| cotY | 134.7 | 2.2 | σK dependent |
| gerE | 6.8 | 1.2 | σK dependent |
Aside from the time of isolation, the method is as described in Table 2, footnote a.
Our results demonstrate that yeast mitochondrial aconitase is expressed and functional in B. subtilis. In general, expression of functional eukaryotic proteins in B. subtilis has rarely been reported (10, 15, 21). Functional Aco1 required growth at 30°C, perhaps to limit inclusion body formation, and an uncharacterized B. subtilis mutation (preliminary work suggests that the mutation affects the activity of a protease [18]). A variety of additional potential difficulties, e.g., 4Fe-4S cluster assembly, proper localization (Aco1 is primarily localized to the mitochondria in S. cerevisiae [14]), and folding, did not prevent Aco1 from providing enough aconitase activity to allow B. subtilis to grow without glutamate. Aco1 activity, however, was unable to substitute for B. subtilis aconitase in support of the later stages of sporulation. These results suggest that an activity of aconitase other than its classical enzymatic activity is required for efficient spore formation and specifically for timely and efficient expression of late sporulation gene expression. We cannot exclude the possibility that Aco1 did not provide enough catalytic activity to fully restore sporulation to a citB null mutant, but this seems highly unlikely. The stage 0 block in a citB null mutant is overcome by supplanting the need for catalytic activity (5), suggesting that aconitase enzyme activity is necessary only for sporulation initiation. Moreover, in the accompanying paper (19), we report that mutagenesis of B. subtilis aconitase in a manner designed to reduce RNA binding leads to a similar delay in late sporulation gene expression. The mutant aconitase had sixfold-higher catalytic activity than did wild-type aconitase, yet the sporulation phenotypes of the Aco1-expressing strain and the B. subtilis mutant aconitase strain were very similar (19). The delay in the mutant strain was correlated with reduced σK-dependent accumulation of gerE mRNA, GerE protein, and mRNAs for GerE-dependent genes. Moreover, B. subtilis aconitase was shown to bind gerE mRNA with much higher affinity than did the mutant protein (19). While it is tempting to suggest that the sporulation defects of the Aco1-expressing strain and the B. subtilis aconitase mutant strain are both due simply to the absence of RNA binding activity, we cannot rule out the possibility that expression of Aco1 as the cell's only aconitase leads to a more complicated defect than does expression of a mutant form of B. subtilis aconitase.
Acknowledgments
We thank A. Dancis for the gift of pYACON and antibodies to yeast aconitase. We thank E. Kuester-Schoeck and R. Britton for assistance with microarrays. We also thank C. Squires, D. Lazinski, J. Coburn, D. Hava, and C. Cornillez-Ty for helpful discussions.
This work was supported by a research grant (GM036718) from the U.S. Public Health Service.
REFERENCES
- 1.Alen, C., and A. L. Sonenshein. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. USA 96:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basilion, J. P., T. A. Rouault, C. M. Massinople, R. D. Klausner, and W. H. Burgess. 1994. The iron-responsive element-binding protein: localization of the RNA-binding site to the aconitase active-site cleft. Proc. Natl. Acad. Sci. USA 91:574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky, B. R., and A. L. Sonenshein. 1997. Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J. Bacteriol. 179:1035-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constable, A., S. Quick, N. K. Gray, and M. W. Hentze. 1992. Modulation of the RNA-binding activity of a regulatory protein by iron in vitro: switching between enzymatic and genetic function? Proc. Natl. Acad. Sci. USA 89:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig, J. E., M. J. Ford, D. C. Blaydon, and A. L. Sonenshein. 1997. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J. Bacteriol. 179:7351-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingman, D. W., and A. L. Sonenshein. 1987. Purification of aconitase from Bacillus subtilis and correlation of its N-terminal amino acid sequence with the sequence of the citB gene. J. Bacteriol. 169:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouet, A., and A. L. Sonenshein. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray, N. K., S. Quick, B. Goossen, A. Constable, H. Hirling, L. C. Kuhn, and M. W. Hentze. 1993. Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur. J. Biochem. 218:657-667. [DOI] [PubMed] [Google Scholar]
- 9.Haile, D. J., T. A. Rouault, J. B. Harford, M. C. Kennedy, G. A. Blondin, H. Beinert, and R. D. Klausner. 1992. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc. Natl. Acad. Sci. USA 89:11735-11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartzell, P. L. 1997. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc. Natl. Acad. Sci. USA 94:9881-9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin, S. 1995. Role of the Krebs cycle in sporulation of Bacillus subtilis. Ph.D. thesis. Tufts University, Boston, Mass.
- 12.Kaptain, S., W. E. Downey, C. Tang, C. Philpott, D. Haile, D. G. Orloff, J. B. Harford, T. A. Rouault, and R. D. Klausner. 1991. A regulated RNA binding protein also possesses aconitase activity. Proc. Natl. Acad. Sci. USA 88:10109-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuno, K., T. Blais, A. W. Serio, T. Conway, T. M. Henkin, and A. L. Sonenshein. 1999. Metabolic imbalance and sporulation in an isocitrate dehydrogenase mutant of Bacillus subtilis. J. Bacteriol. 181:3382-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson, J. B., Jr., and P. A. Srere. 1985. Organization of Krebs tricarboxylic acid cycle enzymes. Biochem. Med. 33:149-157. [DOI] [PubMed] [Google Scholar]
- 15.Roosild, T. P., J. Greenwald, M. Vega, S. Castronovo, R. Riek, and S. Choe. 2005. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science 307:1317-1321. [DOI] [PubMed] [Google Scholar]
- 16.Rouault, T. A., C. D. Stout, S. Kaptain, J. B. Harford, and R. D. Klausner. 1991. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: functional implications. Cell 64:881-883. [DOI] [PubMed] [Google Scholar]
- 17.Rouault, T. A., C. K. Tang, S. Kaptain, W. H. Burgess, D. J. Haile, F. Samaniego, O. W. McBride, J. B. Harford, and R. D. Klausner. 1990. Cloning of the cDNA encoding an RNA regulatory protein—the human iron-responsive element-binding protein. Proc. Natl. Acad. Sci. USA 87:7958-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serio, A. W. 2005. The role of aconitase, a bi-functional protein, in iron regulation and sporulation of Bacillus subtilis. Ph.D. thesis. Tufts University, Boston, Mass.
- 19.Serio, A. W., K. B. Pechter, and A. L. Sonenshein. 2006. Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J. Bacteriol. 188:6396-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swift, K. E. 1999. Characterization of a Bacillus subtilis citrate synthase mutant. M.S. thesis. Tufts University, Boston, Mass.
- 21.Westers, L., H. Westers, and W. J. Quax. 2004. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim. Biophys. Acta 1694:299-310. [DOI] [PubMed] [Google Scholar]