Abstract
A Francisella tularensis live vaccine strain mutant (sodBFt) with reduced Fe-superoxide dismutase gene expression was generated and found to exhibit decreased sodB activity and increased sensitivity to redox cycling compounds compared to wild-type bacteria. The sodBFt mutant also was significantly attenuated for virulence in mice. Thus, this study has identified sodB as an important F. tularensis virulence factor.
Reactive oxygen intermediates (ROI) are generated as a result of the reduction of oxygen during respiration and exposure to radiation or redox cycling compounds and phagocytosis (8). ROI, including superoxide (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (OH·), can cause oxidative damage to DNA, RNA, proteins, and lipids (10, 25). Virtually all organisms possess mechanisms to counter these potentially toxic ROI generated during aerobic growth. The primary cellular antioxidant defense in prokaryotes and eukaryotes is the metalloenzyme family of superoxide dismutases (SODs), which dismutate O2− to molecular oxygen (O2) and hydrogen peroxide (H2O2) at rates nearly sufficient to limit diffusion (11, 12, 25). H2O2 then is detoxified by catalase and/or members of the peroxidase family of enzymes. SODs are classified into three types depending on their prosthetic metal cofactors—manganese (MnSOD) (16, 26), iron (FeSOD) (18, 27, 29), and copper-zinc (CuZnSOD) (2, 6)—encoded by the sodA, sodB, and sodC genes, respectively. In addition, nickel- and iron-zinc-containing isozymes have been identified in bacteria (19, 20). FeSOD has only been reported for prokaryotes, whereas MnSOD and CuZnSOD are found in both prokaryotes and eukaryotes. Most bacteria possess MnSOD or FeSOD in their cytoplasm, while CuZnSOD has been identified in the periplasm of pathogenic or endosymbiotic bacteria (2, 31). In addition to their detoxifying function during aerobic growth, bacterial SODs have been shown to be important virulence factors. SODs protect Salmonella enterica serovar Choleraesuis (28), Staphylococcus aureus (15), and Mycobacterium tuberculosis from the bactericidal effects of the host macrophages and polymorphonuclear leukocytes. The present report addresses the role of a recently reported FeSOD in Francisella tularensis in the aerobic survival of the organism and the pathogenesis of tularemia.
F. tularensis, the etiological agent of tularemia, is a facultative intracellular pathogen that primarily infects macrophages. F. tularensis requires oxygen for growth and has been shown to possess ROI-scavenging enzymes such as SODs, peroxidases, and catalases (21, 30). F. tularensis, like many gram-negative bacteria, contains two SODs: an FeSOD and a CuZnSOD. The genome sequence of the F. tularensis live vaccine strain (LVS) has been shown to possess a single functional copy of the sodB gene (FTL_1791), which encodes a 21.9-kDa FeSOD protein (21). The FeSOD of F. tularensis LVS exhibits 100% homology with that of the virulent type A F. tularensis SchuS4 strain and 59% identity with the FeSOD of Legionella pneumophila. In L. pneumophila, deletion of the sodB gene has been shown to result in the loss of cell viability (27). In agreement with these findings, our initial attempts to generate a sodB null mutant of F. tularensis were unsuccessful. A merodiploid strain containing both the wild-type copy of the gene and the suicide vector integrated in the chromosomal DNA were obtained following the first recombination event, but the merodiploid strain reverted to the wild-type phenotype following a second recombination event. This suggested that the sodB gene may be essential for the viability of the F. tularensis LVS (V. M. Pavlov, personal communication). In the current study, an alternative approach was used to attenuate FeSOD expression and was based on the observation that codon substitutions at the initiating ATG (ATG→GTG) result in a significantly reduced translational efficiency (23). A mutant, sodBFt, was generated by this approach, and decreased activity of FeSOD was found to be associated with increased sensitivity to paraquat and hydrogen peroxide, the redox cycling compounds, as well as with reduced virulence in both C57BL/6 and BALB/c mice.
Construction and characterization of the F. tularensis sodB mutant strain.
A previously described allelic replacement strategy was used to construct a sodB mutant of F. tularensis LVS (7). A region approximately 650 bp upstream of the sodB gene was amplified by PCR using the primer pair Xba-sodB (5′CACTCTAGATTATCCATTTGGTATTGAGG-3′) and Pst-sodB (5′-ATGCTGCAGTACTCTCCTTAAATTCGTG-3′), which included XbaI and PstI sites (underlined), respectively. The full-length sodB gene (∼600 bp) was amplified using the primer pair PstF-sodB (5′ GACCTGCAGTGAAATTTGAATTACCAAAAC 3′), including the PstI site and the substituted GTG initiation codon (underlined), and Sal-sodB (5′ TATGTCGACAACCTAATCAGCGAATTGC 3′), including the SalI site (underlined) (Fig. 1A). The amplified 670- and 600-bp fragments were cloned separately into a PCR cloning vector (pTOPO; Invitrogen Corporation, Carlsbad, CA) and excised by digestion with restriction enzymes XbaI/PstI and SalI/PstI, respectively. The digested fragments were ligated simultaneously to an XbaI/SalI-digested pPV shuttle vector (7) to yield pPV-sodB. The presence of a GTG initiation codon in place of ATG and a PstI site upstream of the start codon in the mutated sodB gene in pPV-sodB was confirmed by sequencing. The PstI site upstream of the start codon was used as a marker for screening of mutant colonies. pPV-sodB was transformed in Escherichia coli S17-1 and transferred into F. tularensis LVS via conjugation (7). The mutants were selected on modified chocolate agar plates (1.5% peptone, 0.1% sodium chloride, 0.4% dipotassium hydrogen phosphate, 0.1% potassium dihydrogen phosphate, 1% d-glucose, 1.5% agar) supplemented with Isovitalex, l-cysteine hydrochloride, 1% hemoglobin, 2 μg/ml chloramphenicol, and 100 μg/ml polymyxin B (the latter component was included for counterselection of the donor E. coli). Colonies obtained on the chloramphenicol plates following the first recombination event were analyzed by PCR using the Xba-sodB and Sal-sodB primers, followed by the digestion of PCR products with PstI enzyme. The positive mutant colonies were selected for a second recombination event by plating on medium containing 5% sucrose (7). Chloramphenicol sensitivity and sucrose resistance following the second recombination event were confirmed by PCR and PstI digestion of the amplified products (Fig. 1B). The positive mutant clone is referred to as sodBFt.
FIG. 1.
Generation and screening of sodB mutants of F. tularensis LVS. (A) Schematic representation of the strategy used for generation and identification of sodBFt mutants. Substitution of GTG for ATG and a PstI site introduced upstream of the start codon for screening of sodBFt mutants are indicated. Locations of various primers used for the generation of mutants are indicated by arrows. (B) Confirmation of sodBFt mutants. A colony PCR was performed using the flanking primers Xba-sodB and Sal-sodB (95οC for 1 min, 53οC for 1 min, and 72οC for 1 min for 30 cycles, followed by a final extension at 72οC for 5 min); the amplified products were digested with PstI and run on a 1.2% agarose gel. Lanes 1 and 8, molecular mass markers (1-kb DNA ladder; Invitrogen Corporation, Carlsbad, CA). Lanes 2 and 4, PCR products amplified from two sodBFt mutant colonies after the first and second recombination events, respectively, and their digestion with PstI (lanes 3 and 5). Lanes 6 and 7, PCR product amplified from the wild-type F. tularensis LVS and its digestion with PstI. Digestion of PCR products amplified from the sodBFt mutant colonies with PstI confirmed mutation of the sodB gene.
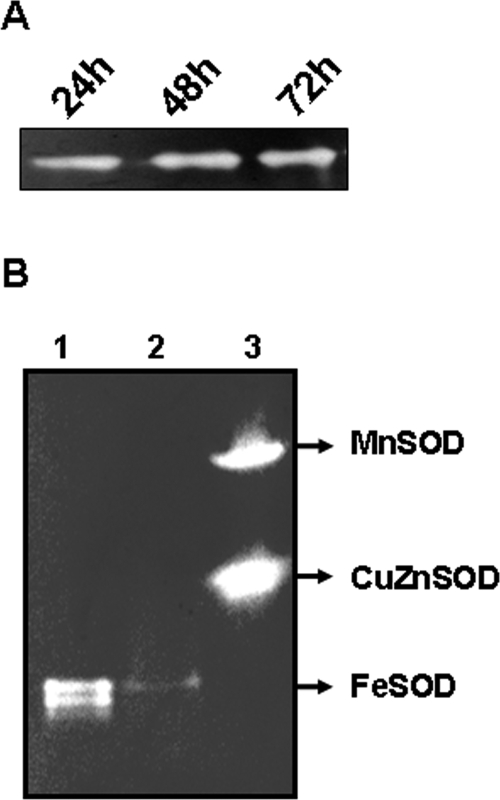
The cellular localization of FeSOD and CuZnSOD in F. tularensis LVS was predicted using PsortB software (http://www.psort.org/psortb/). The subcellular location of FeSOD could not be predicted on the basis of software analysis; however, CuZnSOD was predicted to localize to the periplasm. The expression of FeSOD by F. tularensis LVS at various stages of growth was determined by a previously described indirect SOD activity assay (3). The FeSOD was found to be constitutively expressed at various stages of growth (Fig. 2A); however, expression of CuZnSOD was not observed under any set of in vitro growth conditions. Similar constitutive expression of FeSOD was observed when the bacteria were grown either on chocolate agar plates or in Mueller-Hinton broth (MHB) (Difco Laboratories, Detroit, MI). To determine further whether the mutated sodB gene could be expressed in a distantly related non-Francisella host with dissimilar endogenous enzymes, pPV-sodB was transformed in E. coli DH5α cells and the expression of F. tularensis FeSOD was examined. The mutated sodB was functionally expressed in E. coli, proving that the mutated sodB gene was active (data not shown). Expression of F. tularensis FeSOD was greater than that of endogenous FeSOD in the parent E. coli. This may be due to the differences between the two species in codon usage or the association of FeSOD subunits. It was also observed that increased FeSOD expression was associated with reduced MnSOD expression in the pPV-sodB-transformed E. coli cells (data not shown).
FIG. 2.
FeSOD activity assay. Crude cell extracts of F. tularensis LVS and sodBFt grown on modified chocolate agar plates were prepared by resuspending 10 mg of the bacterial culture in 1.0 ml of lysis solution (50 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, and 0.2 mg lysozyme per ml). The lysates were clarified, and the protein concentration of each lysate was determined with a bicinchoninic protein assay kit (Pierce, Rockford, IL). Protein (15 μg) was loaded on a 10% native polyacrylamide gel electrophoresis gel and run at 150 V for 90 min at 4°C. The gel was stained for SOD activity by the method of Beauchamp and Fridovich (3). (A) Zymogram demonstrating the expression of active FeSOD of F. tularensis LVS following 24, 48, and 72 h of growth on modified chocolate agar plates. (B) FeSOD activity of sodBFt mutants. Lane 1, F. tularensis LVS; lane 2, sodBFt mutant; lane 3, a fibrosarcoma cell line transfected with MnSOD was used as a positive control. Note the markedly reduced expression of FeSOD in the sodBFt mutant.
SOD activity in the sodBFt mutant.
SOD activity was measured in the sodBFt mutant and compared with that of wild-type F. tularensis LVS. Markedly reduced expression of FeSOD was observed for the sodBFt mutant compared to that seen with the wild-type strain (Fig. 2B). However, this reduced FeSOD activity was not compensated for by induction of CuZnSOD activity in the sodBFt mutant, suggesting that these two isozymes have different biological roles. In fact, CuZnSOD was not detected using a sensitive indirect assay in either the LVS or the sodBFt mutant strain. Different functions related to distinct localization of these isozymes have been suggested previously (2, 31).
Effect of sodB mutation on growth and oxidative stress response.
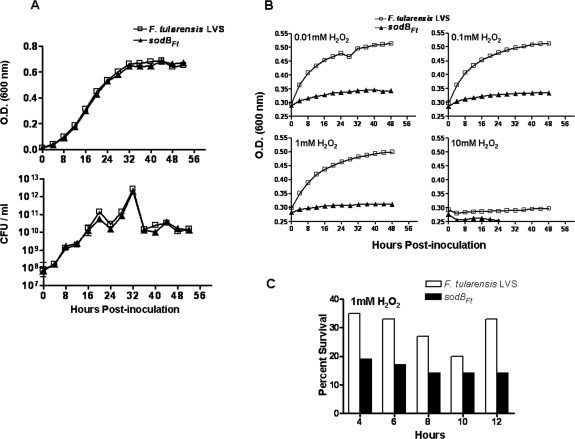
Comparisons of various growth characteristics were made between the sodBFt strain and the corresponding wild-type F. tularensis LVS. On modified chocolate agar plates, the sodBFt mutant exhibited small colonies and required 48 h before individual colonies were clearly visible. In contrast, individual colonies of the wild-type strain were observed within 30 to 36 h. However, the sodBFt mutant showed a growth rate identical to that observed for the wild-type strain when incubated in MHB under aerobic conditions with constant shaking (Fig. 3A) or under stationary growth conditions (data not shown). An unaltered growth rate in MHB suggests that despite markedly reduced FeSOD activity, the sodBFt mutants are capable of handling steady-state production of O2− under normal growth conditions. An altered growth rate due to the complete loss of FeSOD has been reported previously for E. coli and Bordetella pertussis (4, 18). The sodBFt mutant was further evaluated for its growth characteristics under oxidative stress conditions by being grown in the presence of H2O2 in MHB. The sodBFt mutant showed extreme sensitivity when grown in the presence of various concentrations of H2O2 in MHB under aerobic conditions (Fig. 3B).
FIG. 3.
(A) Growth curve of F. tularensis LVS and sodBFt. MHB (25 ml) in a 125-ml flask was inoculated with 1 ml of an overnight culture of F. tularensis LVS or sodBFt and agitated at 175 rpm and 37°C in a shaking incubator. Aliquots were removed at the times indicated to measure optical density at 600 nm (OD600), and 10-fold dilutions were plated on modified chocolate agar for the determination of CFU levels. (B) Effect of sodB mutation on sensitivity to H2O2. F. tularensis LVS and sodBFt mutants were grown in MHB to an OD of 0.2. The indicated concentrations of H2O2 were then added, and the cultures were allowed to grow for 48 h in a shaking incubator. OD readings were taken every 4 h. (C) Aliquots were removed from the cultures grown in the presence of 1 mM of H2O2 at the times indicated and plated on modified chocolate agar plates to determine the CFU counts. Percent survival was calculated using the mean of the results obtained with duplicate samples at the timed intervals and compared to data at 0 h. All results are representative of two independent experiments.
Increased production of O2− can accelerate DNA damage by increasing the intracellular pool of free iron (Fe2+) (17). The Fe2+ levels are elevated when E. coli is exposed to redox cycling compounds, and mutants that lack cytoplasmic SODs contain almost 10 times the amount of Fe2+ found in wild-type cells (24). H2O2 in the presence of Fe2+ can undergo Fenton chemistry to form highly reactive and damaging OH· radicals, leading to increased DNA damage. In contrast, most unbound iron in wild-type cells is in its Fe3+ form and is unavailable for Fenton reactions. F. tularensis LVS, with a full complement of FeSOD, would effectively detoxify superoxide, which would not be available for the reduction of Fe3+ to Fe2+, thus limiting OH· production (14). Thus, the increased sensitivity of sodBFt mutants to H2O2 could be due to increased concentrations of intracellular iron, resulting in DNA damage.
Additionally, inactivation of certain key enzymes by elevated levels of O2− and H2O2· could also impair growth of the sodBFt mutant. To test this notion, F. tularensis LVS and the sodBFt mutant were grown in MHB in the presence of 1 mM of H2O2 and plated on chocolate agar and the number of CFU was determined at 2-h intervals. We observed an exponential decrease in the viability of both the F. tularensis LVS and the sodBFt mutant during the initial stages of growth (consistent with Fe2+ toxicity), following which the number of viable cells of the sodBFt mutant remained constant until 10 h of growth (consistent with enzyme inactivation). The decrease in viability of the sodBFt strain was markedly greater than that seen with the F. tularensis LVS (Fig. 3C). This result suggests that both mechanisms may contribute to the sensitivity of sodBFt mutants to H2O2. In addition, the hypersensitivity of the sodBFt mutant to H2O2, despite normal catalase activity, suggests that part of the damage may be mediated by OH· radicals likely formed in the presence of excess H2O2 and O2− (9). The sodBFt strain also exhibited increased sensitivity to paraquat, as visualized in a disc diffusion assay by an enlarged zone of growth inhibition (47.25 ± 3.50 mm) compared to wild-type strain results (35.30 ± 2.80 mm) (Fig. 4A and B). Kanamycin was used as an experimental control. Since both F. tularensis LVS and the sodBFt mutant are sensitive to kanamycin, an identical zone of growth inhibition was observed for both strains (Fig. 4A, insets). This indicated that the sensitivity of the sodBFt mutant to paraquat was not due to differences in the growth rate between the two strains or the concentration and rate of diffusion of paraquat on chocolate agar plates. The sodBFt mutant also was tested for its sensitivity to paraquat in liquid culture. When the sodBFt mutant was grown in the presence of 1 mM of paraquat in MHB, although the number of viable bacteria remained constant, a complete inhibition of growth was observed (Fig. 4C). Production of O2− through the NADPH-mediated reduction of paraquat is a major factor in paraquat toxicity (13). Paraquat induces an intracellular influx of O2−, and a deficiency in FeSOD is likely responsible for the observed increase in paraquat toxicity and decrease in the growth rate of the sodBFt mutant. Although Psortb software analysis could not predict the localization of FeSOD, the sensitivity of the sodBFt strain to paraquat suggests that it may have a cytoplasmic location. Thus, F. tularensis sodB-encoded FeSOD confers resistance to the redox cycling agents paraquat and H2O2. The exact molecular and metabolic targets that are responsible for the lethality and slow growth rate observed with the sodBFt mutant are unknown and thus warrant further investigation.
FIG. 4.
Effect of sodB mutation on sensitivity to paraquat. (A) The left and right panels represent F. tularensis LVS and sodBFt mutant strains, respectively. Briefly, 1 × 109 F. tularensis LVS or sodBFt bacteria were plated on modified chocolate agar plates. Sterile filter paper discs were placed on each plate, and 5 μl of 1.25, 2.5, 5.0, and 10 mg/ml concentrations of freshly prepared paraquat was added to individual discs. The plates were incubated for 48 to 72 h at 37°C, and the zones of growth inhibition were observed. Insets show the zone of growth inhibition observed with kanamycin, which was used as an experimental control. (B) Quantitation of the zone of growth inhibition. The zone of growth inhibition around the discs impregnated with a 10 mg/ml concentration of paraquat was quantitated using AlphaEase FC software (Alpha Innotech Inc., San Leandro, CA). Data represent the means ± standard errors of the means for the zone of inhibition determined for each strain (n = 5 plates per strain) and the cumulative results of two independent experiments. Comparisons between the strains were made using a nonparametric Mann-Whitney test (P < 0.0001). (C) F. tularensis LVS and the sodBFt mutant strain were grown in MHB to an OD600 of 0.2. Paraquat (1 mM) was then added to the cultures, and cultures were allowed to grow at 37°C with constant shaking. Aliquots were removed at the times indicated, diluted 10-fold, and plated on modified chocolate agar plates to determine the number of CFU. Data are representative of two independent experiments.
sodB contributes to the virulence of F. tularensis.
The effect of the sodB mutation on virulence was determined by infecting C57BL/6 and BALB/c mice intranasally with 1 × 104 CFU of the wild-type F. tularensis LVS strain or the sodBFt mutant. Mice were observed over a period of 21 days for clinical symptoms and mortality. All but 1 of 12 C57BL/6 mice infected with the wild-type strain died by day 15, while only two deaths were observed after sodBFt infection (Fig. 5A). One hundred percent of the BALB/c mice infected with the wild-type strain were dead by day 11 postinfection, in contrast to only 40% mortality observed with the sodBFt strain (Fig. 5A). To further investigate the nature of this impaired virulence, C57BL/6 mice were infected with a sublethal dose of wild-type and sodBFt mutant bacteria. The mice were sacrificed at different time points, and bacterial burden was quantitated in the lungs, liver, and spleen. Mice infected with the sodBFt mutant not only showed a reduced bacterial burden but also demonstrated more rapid elimination of the bacteria from the lungs, liver, and spleen compared to those infected with the wild-type strain (Fig. 5B). The fact that the sodB mutant was rapidly eliminated from mice is consistent with its rapid clearance by macrophages. The sodBFt mutant also was characterized with respect to its ability to replicate in MH-S cells, a murine alveolar macrophage cell line. In contrast to the wild-type strain results, following invasion the sodBFt mutant exhibited a 1-log reduction in recovery from MH-S cells after 4 h of coincubation. These results suggest that the sodBFt mutant is attenuated for virulence in mice.
FIG. 5.
Effect of sodB mutation on virulence in mice. (A) Survival of C57BL/6 (n = 12) and BALB/c (n = 10) mice infected intranasally with 1 × 104 CFU of F. tularensis LVS or sodBFt bacteria. Results are expressed as Kaplan-Meier curves and P values determined using a log rank test. (B) Kinetics and growth of F. tularensis LVS and sodBFt in C57BL/6 mice. C57BL/6 mice were inoculated intranasally with 5 × 103 CFU of F. tularensis LVS or sodBFt. Three mice were killed at each indicated time point, and homogenates of the lungs, liver, and spleen were plated for the determination of bacterial burden. Results represent the means ± standard errors of CFU counts (n = 3 per time point). **, P < 0.01; ***, P < 0.001 (using the nonparametric Mann-Whitney test). ND, not detected. All results are representative of two independent experiments.
In accordance with the recent definition of a virulence factor (5), FeSOD meets the criteria for inclusion as such since it promotes F. tularensis survival, which in turn results in greater host damage and higher mortality than is observed with mice infected with wild-type F. tularensis LVS. FeSOD may have a dual role in protecting F. tularensis from oxidative stress. First, FeSOD binds iron with high affinity and limits the availability of iron that is needed for production of the highly lethal OH·. Second, dismutation of O2− prevents cellular damage of DNA, proteins, and lipids associated with O2− toxicity (10, 25). FeSOD dismutation activity can also protect bacteria from peroxynitrite (ONOO−) toxicity by minimizing the interaction of O2− with nitric oxide (NO) (27). Indeed, ONOO− has been shown to have a major role in the gamma-interferon-induced killing of F. tularensis LVS by murine macrophages (1, 22). Superoxide-dependent ONOO− toxicity may be an important factor in defective invasion and colonization and in rapid clearance of the sodBFt mutant by C57BL/6 and BALB/c mice.
In summary, this study has demonstrated that the sodB gene of F. tularensis encodes a functional FeSOD that is largely responsible for aerotolerance and protection from internal oxidative stress. Alteration of the sodB gene results in reduced expression of FeSOD and an associated sensitivity to oxidative stress, suggesting that sodB may be essential for bacterial survival under conditions of oxidative stress. The increased survival of mice infected with the sodBFt mutant supports the hypothesis that FeSOD plays a role in virulence; however, linking virulence to one gene is simplistic. Further, the 100% similarity in the sequences and locations of the sodB gene in the genomes of LVS and SchuS4 raises the possibility that FeSOD has a similar role in the virulence of the SchuS4 strain. A more rigorous assessment of the role of SODs in virulence would require the generation of both sodB and sodC mutants of the SchuS4 strain. Experiments to generate these mutants and to assess the efficacy of the sodBFt mutant as a putative vaccine candidate are under way.
Acknowledgments
We thank Heather Page for excellent technical support.
This work was supported by National Institutes of Health grant PO1 AI056320 and a fellowship grant from The National Academies National Research Council.
REFERENCES
- 1.Anthony, L. S. D., P. J. Morrissey, and F. E. Nano. 1992. Growth inhibition of Francisella tularensis live vaccine strain by IFN-gamma-activated macrophages is mediated by reactive nitrogen intermediates derived from l-arginine metabolism. J. Immunol. 148:1829-1834. [PubMed] [Google Scholar]
- 2.Battistoni, A., F. Pacello, S. Folcarelli, M. Ajello, G. Donnarumma, R. Greco, M. Grazia Ammendolia, D. Touati, G. Rotilio, and P. Valenti. 2000. Increased expression of periplasmic Cu,Zn superoxide dismutase enhances survival of Escherichia coli invasive strains within nonphagocytic cells. Infect. Immun. 68:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 4.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall, A., and L.-A. Pirofski. 1999. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 67:3703-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gee, J. M., M. W. Valderas, M. E. Kovach, V. K. Grippe, G. T. Robertson, W.-L. Ng, J. M. Richardson, M. E. Winkler, and R. M. Roop II. 2005. The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect. Immun. 73:2873-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golovliov, I., A. Sjostedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222:273-280. [DOI] [PubMed] [Google Scholar]
- 8.Halliwell, B., and J. M. C. Gutteridge. 1990. Role of free radicals and catalytic metal ions in human disease: an overview, p. 1-85. In A. J. Parker and A. N. Glazer (ed.), Oxygen radicals in biological system. Academic Press, New York, N.Y. [DOI] [PubMed]
- 9.Halliwell, B., and J. M. C. Gutteridge. 1984. Free radicals, lipid peroxidation and cell damage. Lancet 10:1095. [DOI] [PubMed] [Google Scholar]
- 10.Halliwell, B., and J. M. C. Gutteridge. 1997. Lipid peroxidation in brain homogenates: the role of iron and hydroxyl radicals. J. Neurochem. 69:1330. [DOI] [PubMed] [Google Scholar]
- 11.Hassan, H. M. 1984. Determination of microbial damage caused by oxygen free radicals, and the protective role of superoxide dismutases. Methods Enzymol. 105:404-412. [DOI] [PubMed] [Google Scholar]
- 12.Hassan, H. M. 1989. Microbial superoxide dismutases. Adv. Genet. 26:65-97. [DOI] [PubMed] [Google Scholar]
- 13.Hassan, H. M., and I. Fridovich. 1979. Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J. Biol. Chem. 254:10846-10852. [PubMed] [Google Scholar]
- 14.Imlay, J. A., and S. Linn. 1987. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J. Bacteriol. 169:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karavolos, M. H., M. J. Horsburgh, E. Ingham, and S. J. Foster. 2003. Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology 149:2749-2758. [DOI] [PubMed] [Google Scholar]
- 16.Keele, B. B. J., J. M. McCord, and I. Fridovich. 1970. Superoxide dismutase from Escherichia coli B. A new manganese containing enzyme. J. Biol. Chem. 245:6176-6181. [PubMed] [Google Scholar]
- 17.Keyer, K., and J. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 93:13635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khelef, N., D. DeShazer, R. L. Friedman, and N. Guiso. 1996. In vivo and in vitro analysis of Bordetella pertussis catalase and Fe-superoxide dismutase mutants. FEMS Microbiol. Lett. 142:231-235. [DOI] [PubMed] [Google Scholar]
- 19.Kim, E.-J., H.-J. Chung, B. Suh, Y. C. Hah, and J.-H. Roe. 1998. Expression and regulation of the sodF gene encoding iron- and zinc-containing superoxide dismutase in Streptomyces coelicolor Müller. J. Bacteriol. 180:2014-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, E. J., H. J. Chung, B. Suh, Y. C. Hah, and J. H. Roe. 1998. Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Muller. Mol. Microbiol. 27:187-195. [DOI] [PubMed] [Google Scholar]
- 21.Larsson, P., P. C. F. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Hälltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. E. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 22.Lindgren, H., L. Stenman, A. Tarnvik, and A. Sjostedt. 2005. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 7:467-475. [DOI] [PubMed] [Google Scholar]
- 23.Martin, R. G., and J. L. Rosner. 2004. Transcriptional and translational regulation of the marRAB multiple antibiotic resistance operon in Escherichia coli. Mol. Microbiol. 53:183-191. [DOI] [PubMed] [Google Scholar]
- 24.McCormick, M. L., G. R. Buettner, and B. E. Britigan. 1998. Endogenous superoxide dismutase levels regulate iron-dependent hydroxyl radical formation in Escherichia coli exposed to hydrogen peroxide. J. Bacteriol. 180:622-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poyart, C., E. Pellegrini, O. Gaillot, C. Boumaila, M. Baptista, and P. Trieu-Cuot. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadosky, A. B., J. W. Wilson, H. M. Steinman, and A. H. Shuman. 1994. The iron superoxide dismutase of Legionella pneumophila is essential for viability. J. Bacteriol. 176:3790-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sansone, A., P. R. Watson, T. S. Wallis, P. R. Langford, and J. S. Kroll. 2002. The role of two periplasmic copper- and zinc-cofactored superoxide dismutases in the virulence of Salmonella choleraesuis. Microbiology 148:719-726. [DOI] [PubMed] [Google Scholar]
- 29.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimaniuk, N. I., N. V. Pavlovich, and B. N. Mishan'kin. 1992. Superoxide dismutase activity in representatives of the genus Francisella. Zhurnal Mikrobiologii Epidemiologii Immunobiologii 5-6:7-9. (In Russian.) [PubMed]
- 31.Wu, C. H. H., J. J. Tsai-Wu, Y. T. Huang, C. Y. Lin, G. G. Lioua, and F. J. Lee. 1998. Identification and subcellular localization of a novel Cu,Zn superoxide dismutase of Mycobacterium tuberculosis. FEBS Lett. 439:192-196. [DOI] [PubMed] [Google Scholar]