Abstract
Anthrax toxin and capsule, determinants for successful infection by Bacillus anthracis, are encoded on the virulence plasmids pXO1 and pXO2, respectively. Each of these plasmids also encodes proteins that are highly homologous to the signal sensor domain of a chromosomally encoded major sporulation sensor histidine kinase (BA2291) in this organism. B. anthracis Sterne overexpressing the plasmid pXO2-61-encoded signal sensor domain exhibited a significant decrease in sporulation that was suppressed by the deletion of the BA2291 gene. Expression of the sensor domains from the pXO1-118 and pXO2-61 genes in Bacillus subtilis strains carrying the B. anthracis sporulation sensor kinase BA2291 gene resulted in BA2291-dependent inhibition of sporulation. These results indicate that sporulation sensor kinase BA2291 is converted from an activator to an inhibitor of sporulation in its native host by the virulence plasmid-encoded signal sensor domains. We speculate that activation of these signal sensor domains contributes to the initiation of B. anthracis sporulation in the bloodstream of its infected host, a salient characteristic in the virulence of this organism, and provides an additional role for the virulence plasmids in anthrax pathogenesis.
The etiological agent of anthrax, Bacillus anthracis, is a uniquely pervasive and persistent environmental pathogen due to its ability to form dormant spores that are resistant to adverse environmental conditions such as extremes of temperature, UV radiation, and antimicrobial chemical agents (9, 22). The spore is essential to the organism not only for its persistence in the environment but also for the ability of this organism to infect its hosts. Infection is initiated when spores are introduced into the host body and phagocytosed by macrophages, or perhaps other phagocytic cells (5, 10, 11). It is believed that this is followed by germination of the spores into vegetative cells, with subsequent toxin gene expression and capsule production, resulting in the onset of anthrax disease (11).
Interestingly, while the spore is required to initiate the infection, once vegetative growth is established, sporulation does not occur in the bloodstream of the infected host (17). This might be explained by the observation that macrophages can take up spores and destroy them as soon as they start to germinate, while encapsulated vegetative cells are able to evade the immune system (14, 16). Thus, while the transition to and maintenance of vegetative growth, which accompanies toxin and capsule production and progression of the disease, are advantageous to the pathogenic lifestyle of B. anthracis, sporulation within the host may not be.
The observation that sporulation and progression of the anthrax disease are potentially mutually exclusive events requires that regulatory networks must exist to ensure that while one is occurring, the other does not. The major deciding factor in orchestrating which of these events occurs is the level of phosphorylated Spo0A (Spo0A∼P) response regulator-transcription factor. Spo0A is the phosphorylation target of the Bacillus species' phosphorelay signal transduction system that controls sporulation initiation (7). In addition to its role in upregulating the expression of genes required to initiate sporulation, in B. anthracis, phosphorylated Spo0A indirectly regulates expression of the anthrax toxin genes pagA (protective antigen), cya (edema factor), and lef (lethal factor) via its negative regulation of the transition state regulator AbrB (3, 23). Thus, while some low level of Spo0A∼P is required for repression of AbrB and maximal anthrax toxin production, too much Spo0A∼P would result in the onset of sporulation, which has been speculated to be antithetical to successful pathogenesis (6). The regulatory mechanism(s) that results in the appropriate levels of Spo0A∼P formation in B. anthracis during an infection has yet to be elucidated.
Given the pivotal role played by Spo0A∼P in the decision between sporulation and virulence in B. anthracis, surprisingly little was known until recently of the signals or the sporulation sensor kinase(s) that feeds into the sporulation phosphorelay in this organism. Functional analysis of nine putative sporulation sensor histidine kinase-encoding genes recently identified in B. anthracis indicated several with likely roles in sporulation. Of particular interest is the chromosomally encoded sensor histidine kinase BA2291 (Ames strain designation). Deletion of the gene for BA2291 results in a delay in sporulation in B. anthracis, and this protein is able to complement sporulation kinase-deficient mutants (ΔkinA ΔkinB mutants) of Bacillus subtilis when introduced in a single copy, supporting its role as a bona fide sporulation histidine kinase (6).
In this communication we report the identification and characterization of two virulence plasmid-encoded proteins with strong similarity to the sensor domain of BA2291 and with a role in regulating the activity of this sporulation kinase.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All B. subtilis strains used in this study are derivatives of JH642 (trpC2 phe-1). B. subtilis strains JH11422 (ΔkinA::cat) and JH16567 (ΔkinA::cat ΔkinB::tet) were transformed with plasmid pCm::Spc (25) in order to replace the chloramphenicol resistance gene with the spectinomycin resistance gene, giving rise to strains JH19190 and JH19191, respectively. These strains were transformed with plasmid pJAK2291 (6) so that the BA2291 gene and its promoter were integrated into the chromosome by double crossover recombination at the amyE gene selecting for chloramphenicol resistance. The resulting strains were named JH19192 and JH19193, respectively. All B. anthracis strains are derivatives of the Sterne strain 34F2 (pX01+ pX02−). The construction of B. anthracis ΔBA4223 and ΔBA2291 strains was described by Brunsing et al. (6). The transformation of B. anthracis strains with pHT315 and its derivatives was done as previously described (15). The transformation of B. subtilis strains was done as described by Anagnostopoulos and Spizizen (1).
Bacterial strains were grown in Schaeffer's sporulation medium (SM) (24) or Luria-Bertani (LB) medium with the appropriate antibiotics. For B. subtilis, spectinomycin was used at 50 μg/ml, and chloramphenicol was used at 5 μg/ml. For both B. anthracis and B. subtilis strains harboring plasmid pHT315 and its derivatives, erythromycin and lincomycin were used at 5 and 25 μg/ml, respectively.
Spore assays.
Images of live sporulating cells were captured after growth in 5 ml SM broth supplemented with erythromycin and lincomycin for 17 h at 37°C with shaking. Sporulation phenotypes were examined on SM agar plates by streaking isolated colonies of the desired strains onto SM agar plates containing erythromycin and lincomycin. The plates were incubated at 37°C for 48 h.
Liquid sporulation assays were carried out in SM supplemented with erythromycin and lincomycin. Cultures (5 ml) were grown for 48 h at 37°C. Cells were plated as duplicate serial dilutions before and after treatment with chloroform (10%, vol/vol, final concentration). The percentage of sporulation was calculated as the ratio of the spore count after CHCl3 treatment to the total viable count.
Plasmid construction.
Construction of pXO1-118 and pXO2-61 expression vectors in pHT315 (copy number, approximately 15) (2) was carried out by PCR amplification of the genes using genomic DNA of B. anthracis 34F2 or plasmid pXO2, respectively, as the template. The respective amplification reactions were carried out with the following pairs of oligonucleotide primers (the restriction site used for cloning is underlined): 5′-CGATGGATATCGGTGTTAGCATGTC-3′ and 5′-ATTGAGAATTCTATAACTCCCAAAAATTTC-3′; and 5′-ATCACCTGCAGTTTATTATTCTGAAATATTTTAATAG-3′ and 5′-CAATAAAGCTTAACAATCATGCTTTTTGTTC-3′. The fragment containing the pXO1-118 gene was digested with EcoRI and EcoRV and cloned in pHT315 digested with EcoRI and SmaI, obtaining plasmid pHT315-118. The fragment carrying the pXO2-61 gene was digested with PstI and HindIII and cloned in similarly digested plasmid pHT315, obtaining plasmid pHT315-61. The fidelity of the PCR was verified by DNA sequence analysis.
Construction of BA2291 overexpression vector.
The coding sequence for BA2291 was amplified by PCR from the chromosome of B. anthracis 34F2 using the following primers: 5′-TATTCGTCATATGGAAATGGAGGGAATG-3′ and 5′-GACCCTTCGAAGCTTAGAAGCAGTTATACTTAC-3′. The PCR product was digested with NdeI and HindIII and ligated into the same sites of vector pET28 (Novagen), resulting in a fusion to six histidine codons at the 5′ end of the gene (plasmid pET28-BA2291). The insertion sequence was verified by sequencing analysis.
Expression and purification of BA2291.
pET28-BA2291 was overexpressed in Escherichia coli BL21(DE3) in 1 liter of LB broth containing kanamycin at 30 μg/ml. The culture was grown at 37°C with shaking to an optical density at 600 nm of approximately 0.6. Expression was induced by the addition of a 0.4 mM final concentration of isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were incubated for an additional 3 hours at 37°C. Approximately 5.9 g (wet weight) of cells was harvested by centrifugation and resuspended in binding buffer (50 mM Tris-HCl [pH 8.0], 0.3 M NaCl, 10 mM β-mercaptoethanol). Cells were broken by two passages through a French pressure cell at 16,000 lb/in2, and the cell extract was cleared of the cellular debris and membrane fraction by ultracentrifugation. The resulting cleared lysate was incubated with 3 ml of preequilibrated Ni-nitrilotriacetic acid nickel resin (QIAGEN) for 16 h at 4°C on an orbital rocker. Unbound protein was removed by washing the resin with 150 column volumes of binding buffer followed by 50 column volumes of binding buffer containing 30 mM imidazole. Pure protein was eluted in binding buffer containing 250 mM imidazole and collected in 1-ml fractions. Fractions containing the most pure preparations of BA2291 (98% purity) as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were pooled and concentrated by ultrafiltration with a membrane with a molecular weight cutoff of 30,000. The amino-terminal six-His tag was removed by digestion with thrombin (10 mg of N-terminal six-His-BA2291 and 24 U of thrombin) during dialysis in 1 liter of 50 mM Tris-Cl (pH 8.0), 10% glycerol, and 1 mM dithiothreitol using Spectra/Por dialysis tubing with a molecular weight cutoff of 12,000 to 14,000. Digestion was carried out for 16 h at 4°C. The digested protein was stored at a final concentration of 0.6 mg/ml (14.6 μM) at −80°C.
Autophosphorylation and phosphotransfer assays.
Phosphorylation reactions and purification of B. subtilis KinA, Spo0F, and Spo0F∼P were performed as previously described (19, 26). Autophosphorylation assays of KinA and BA2291 used 1 μM and 5 μM concentrations of proteins, respectively. Assays for KinA to Spo0F phosphotransferase activity used the enzymes at 0.2 and 2 μM final concentrations, respectively. When BA2291 was included in these assays, it was used at a final concentration of 5 μM. These assays were carried out in a 30-μl reaction volume at room temperature. Aliquots of 12 μl were removed and added to 2.4 μl of SDS-PAGE sample buffer at 0 min and 60 min of incubation. Samples were analyzed on 15% SDS-PAGE gels. The gels were dried, exposed to a PhosphorImager screen, and analyzed by using ImageQuant software (Molecular Dynamics).
RESULTS
Bioinformatic identification of virulence plasmid-encoded sensor domains.
Whole-genome sequence analysis of B. anthracis resulted in the identification of two virulence plasmid-encoded proteins with significant sequence similarity to the sensor domain only of the BA2291 sporulation histidine sensor kinase. Proteins encoded by pXO1-118 (GenBank accession number AAT28889.2) (18) and pXO2-61 (GenBank accession number AAT29005.2) share 62% identical and conserved residues in predicted amino acid sequence with the sensor domain of BA2291 (residues 1 to 161) (Fig. 1). The pXO1-118 gene is located in very close proximity to (358 nucleotides) and divergently transcribed from the gene encoding the trans-acting virulence gene regulator AtxA on the pathogenicity island of virulence plasmid pXO1. The pXO2-61 protein is encoded by a gene adjacent to an atxA pseudogene located on virulence plasmid pXO2. This amplification of signal domain-encoding genes is unique to BA2291 and B. anthracis, as proteins similar to the sensor domains of the other sporulation histidine kinases were not found to be encoded elsewhere in the genome and, to the best of our knowledge, such amplification is not known to occur in other organisms. The only exception would be the Bacillus cereus strain associated with an illness resembling inhalation anthrax, strain G9241, which carries a gene orthologue to the pXO1-118 and pXO2-61 genes on its virulence plasmid pBC218 (pBC218_0049, accession number NZ_AAEK01000004) (12). The presence of the pXO1-118 and pXO2-61 genes for these sensor domain proteins on the virulence plasmids of B. anthracis suggests a possible regulatory mechanism allowing the coordinate regulation of sporulation and virulence.
FIG. 1.
Amino acid sequence alignment of the B. anthracis sensor domains encoded by pXO1-118 and pXO2-61 and the BA2291 sensor domain (residues 1 to 161). Sequences were aligned by the ClustalW program. Asterisks indicate identical residues in all three sequences; colons denote conserved substitutions. Paired scores resulted in 34% identity between the pXO2-61 and BA2291 sensor domains, 29% identity between the pXO1-118 and BA2291 sensor domains, and 62% identity between the pXO1-118 and pXO2-61 sensor domains.
BA2291-dependent inhibition of B. anthracis and B. subtilis sporulation by overproduction of the sensor domains.
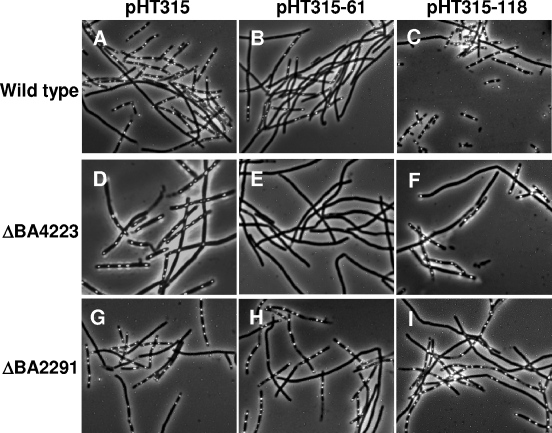
To determine if any regulatory effect was exerted by the virulence plasmid-encoded sensor domains on BA2291 in B. anthracis, each of the sensor domains was expressed from its own promoter on multicopy plasmid pHT315 (2) and introduced into several B. anthracis strains. The sporulation phenotypes of these strains were examined using phase-contrast microscopy of whole cells after 17 h of growth at 37°C in SM broth (Fig. 2). Expression of pXO2-61 (Fig. 2, pHT315-61) resulted in a marked decrease in sporulation in wild-type B. anthracis compared to that of the strain carrying the vector control pHT315 (Fig. 2A and B). The ability of B. anthracis carrying pHT315-61 to continue sporulating, albeit at a lower level, might be explained by the existence of seven putative sporulation sensor kinases active in this organism. A single deletion of any of these sporulation kinases results in only a minor reduction in sporulation, at least in laboratory media (6). However, when a deletion of the other major sporulation kinase in this organism, BA4223 (6), is combined with expression of pXO2-61 in B. anthracis, the inhibition of sporulation is complete (Fig. 2D and E). The inhibition of sporulation observed due to the presence of pXO2-61 is dependent on the presence of BA2291, because the level of sporulation in a B. anthracis ΔBA2291 strain carrying pHT315-61 was comparable to the one in the parental strain carrying pHT315 (Fig. 2G and H).
FIG. 2.
Sporulation phenotypes of B. anthracis parental 34F2, ΔBA4223, and ΔBA2291 strains harboring sensor domains encoded by pX01-118 (pHT315-118) and pXO2-61 (pHT315-61) expressed from their native promoters on multicopy plasmid pHT315. Cultures of each strain were grown in 5 ml of Schaeffer's sporulation medium (24) with the appropriate antibiotics for 17 h at 37°C with shaking.
The regulatory effect of pXO1-118 on sporulation in B. anthracis was less clear. Overexpression of pXO1-118 on pHT315 did not result in a significant decrease in sporulation in any of the B. anthracis strains tested, based on microscopic analysis (Fig. 2C, F, and I) or plate phenotypes on SM agar (data not shown). Because of the tendency of B. anthracis cells to remain in long chains rather than break into single units even after the initiation of the sporulation process, a reliable and reproducible quantitation of sporulation efficiency could not be carried out by the spore assay described in Materials and Methods.
To further explore a possible regulatory role for pXO1-118 and pXO2-61 in sporulation initiation, the effects of the sensor domains encoded by both virulence plasmids on the function of BA2291 were analyzed in the case of BA2291-dependent complementation of sporulation in B. subtilis. Each virulence plasmid-encoded sensor domain and its native promoter were cloned into the replicative vector pHT315 and transformed into B. subtilis sporulation sensor histidine kinase ΔkinA and ΔkinA ΔkinB mutants, respectively, carrying the gene encoding BA2291 integrated into the chromosome in a single copy. The sporulation phenotype of each strain was compared to that containing only the pHT315 vector by examining plate phenotypes on Schaeffer's sporulation agar (Fig. 3) and by carrying out sporulation assays in liquid cultures (Table 1).
FIG. 3.
Effects of overexpression of the sensor domains encoded by pX01-118 and pX02-61 on the sporulation phenotypes of B. subtilis wild-type, ΔkinA mutant, and ΔkinA ΔkinB mutant strains with (+2291) and without (−2291) the B. anthracis sporulation sensor kinase BA2291 integrated in a single copy on the chromosome. Strains were streaked on Schaeffer's sporulation medium agar (24) and incubated at 37°C for 48 h. Opaque sporulating strains appear white, and nonsporulating strains appear black/gray. The streak numbers correspond to the numbers in column 1 of Table 1.
TABLE 1.
Effects of pX01-118 and pX02-61 on sporulation in B. subtilisa
| No.b | Strain | Relevant genotype | Vector | Spores/ml | Viable cells/ml | % Sporulation |
|---|---|---|---|---|---|---|
| 1 | JH19190 | ΔkinA | pHT315 | 5.2 × 104 | 1.7 × 108 | 0.03 |
| 2 | JH19190 | ΔkinA | pHT315-61 | 1.6 × 105 | 3.1 × 108 | 0.05 |
| 3 | JH19190 | ΔkinA | pHT315-118 | 1.3 × 105 | 2.7 × 108 | 0.05 |
| 4 | JH19192 | ΔkinA amyE::B2291 | pHT315 | 1.4 × 108 | 5.0 × 108 | 28.0 |
| 5 | JH19192 | ΔkinA amyE::B2291 | pHT315-61 | 2.5 × 104 | 3.2 × 108 | 0.008 |
| 6 | JH19192 | ΔkinA amyE::B2291 | pHT315-118 | 1.0 × 108 | 4.0 × 108 | 25.0 |
| 7 | JH19191 | ΔkinA ΔkinB | pHT315 | 0 | 1.8 × 108 | 0 |
| 8 | JH19191 | ΔkinA ΔkinB | pHT315-61 | 0 | 2.8 × 108 | 0 |
| 9 | JH19191 | ΔkinA ΔkinB | pHT315-118 | 0 | 2.1 × 108 | 0 |
| 10 | JH19193 | ΔkinA ΔkinB amyE::B2291 | pHT315 | 5.5 × 107 | 2.8 × 108 | 19.0 |
| 11 | JH19193 | ΔkinA ΔkinB amyE::B2291 | pHT315-61 | 0 | 1.4 × 108 | 0 |
| 12 | JH19193 | ΔkinA ΔkinB amyE::B2291 | pHT315-118 | 7.7 × 107 | 3.5 × 108 | 22.6 |
| 13 | JH642 | Wild type | pHT315 | 1.4 × 108 | 4.8 × 108 | 29.2 |
| 14 | JH642 | Wild type | pHT315-61 | 1.1 × 108 | 4.0 × 108 | 27.5 |
| 15 | JH642 | Wild type | pHT315-118 | 1.2 × 108 | 3.9 × 108 | 30.7 |
| 16 | JH19169 | amyE::B2291 | pHT315 | 1.3 × 108 | 2.5 × 108 | 52.0 |
| 17 | JH19169 | amyE::B2291 | pHT315-61 | 6.8 × 107 | 4.0 × 108 | 17.0 |
| 18 | JH19169 | amyE::B2291 | pHT315-118 | 1.6 × 108 | 3.5 × 108 | 45.7 |
Strains were grown for 48 h at 37°C in SM plus erythromycin-lincomycin, and the spore assay was carried out as described in Materials and Methods. Values are representative of four independent experiments.
Numbers correspond to the streak numbers in Figure 3.
Introduction of either sensor domain into B. subtilis ΔkinA or ΔkinA ΔkinB mutants in the absence of BA2291 had no significant effect on sporulation compared to the vector-only control, as determined by the level of opacity within the streaks and isolated colonies on SM agar plates (Fig. 3, streaks 1, 2, 3, 7, 8, and 9). In B. subtilis, colony opacity increases with the level of sporulation; Spo0 mutant colonies are transparent. In contrast, when either sensor domain was introduced into the strains expressing BA2291, a significant decrease in BA2291-dependent sporulation, marked by a severe decrease in opacity, was observed (Fig. 3, streaks 4, 5, 6, 10, 11, and 12). This effect was much more severe in the presence of the sensor domain encoded by pXO2-61 than with that encoded by pXO1-118. However, in the presence of either sensor domain and BA2291, the level of sporulation was less than what was observed in the same strains in the absence of BA2291 (Fig. 3, compare streak 5 to streak 2, 6 to 3, 11 to 8, and 12 to 9). This indicates that not only was the BA2291-dependent complementation of sporulation previously observed inhibited, but the sporulation process induced by other sporulation kinases was actually blocked by the presence of the sensor domains in a BA2291-dependent manner.
The ability of the sensor domains to inhibit sporulation in B. subtilis in a BA2291-dependent manner was further demonstrated by examining the plate phenotypes of wild-type B. subtilis in the presence and absence of BA2291 and each sensor domain (Fig. 3). In the absence of BA2291, sporulation appears normal in strains in which either of the two sensor domains is expressed (Fig. 3, streaks 13, 14, and 15). However, in the presence of BA2291, sporulation is completely abolished when pXO2-61 is introduced (Fig. 3, streak 17), and sporulation is diminished with the introduction of pXO1-118 (Fig. 3, compare streak 18 to streak 16, in particular in the area with single colonies). Quantitation of sporulation efficiencies in liquid cultures essentially concurred with the visual analysis of agar plates (Table 1), except that the effect of pXO1-118 did not seem to be as detectable as it was when cells were grown on a solid surface. Perhaps growth in a liquid versus in a solid medium differentially affects the level of expression of pXO1-118, thus resulting in seemingly different phenotypes.
By examining the effect of each sensor domain on BA2291-dependent sporulation in B. subtilis rather than in B. anthracis, we were able to isolate the regulatory effects of both sensor domains on BA2291 independently from any additional regulatory networks that might exist in the native host.
The sensor domains convert the BA2291 kinase to an inhibitor of sporulation.
A mechanism by which the pXO1-118- and pXO2-61-encoded sensor domains regulate the activity of BA2291 to be either a contributor to or an inhibitor of sporulation is suggested by the observation that, while BA2291 in a single copy complements sporulation kinase-deficient mutants of B. subtilis, BA2291 expressed in multicopy completely abolishes the normally high levels of sporulation in wild-type B. subtilis (6). A possible explanation for this observation is that with a single copy, there is adequate signal available to activate BA2291 for autophosphorylation and subsequent phosphotransfer to the sporulation phosphorelay, resulting in sporulation. However, when BA2291 is present in multicopy, only a small portion of BA2291 in the cell is activated, due to insufficient levels of activating signal. The remaining portion of BA2291 that is not bound by activating signal is in a form that inhibits sporulation.
In vitro studies of purified BA2291 demonstrate that this histidine kinase does not autophosphorylate at a detectable level in vitro, yet it retains the ability to remove phosphoryl groups from Spo0F∼P that has been produced by phosphoryl group transfer from KinA∼P (the major sporulation kinase in B. subtilis) (7, 13) to Spo0F (Fig. 4A). We propose that the ability of BA2291 to inhibit sporulation is due to its ability to remove phosphoryl groups from the phosphorelay at the level of Spo0F and that it is this activity that predominates when BA2291 is not activated to autophosphorylate by activating signal, even when other sporulation kinases are activated by their own signals and feeding phosphoryl groups into the phosphorelay, as is the case in wild-type B. subtilis carrying multicopy BA2291. The BA2291-dependent inhibition of sporulation observed upon introduction of the virulence plasmid-encoded sensor domains is very similar to the inhibition of sporulation observed when BA2291 is present in wild-type B. subtilis in multicopy. We suggest that pXO1-118 and pXO2-61 interfere with the ability of BA2291 to perceive and/or transmit a signal for activation, which results in its conversion to an inhibitor of sporulation rather than a contributor to sporulation.
FIG. 4.
In vitro activity of the BA2291 histidine sensor kinase. (A) Autophosphorylation and phosphoryl transfer activity assays of BA2291 purified from E. coli were carried out as described in Materials and Methods. Autophosphorylation and phosphoryl transfer activities of purified B. anthracis BA2291 (5 μM) were compared to those observed for B. subtilis proteins KinA (0.2 μM) and Spo0F (2 μM) in the presence of γ-32P-labeled ATP at 0 and 60 min. The samples were run on 15% SDS-PAGE gels. (B) Schematic representation of the phosphorelay signal transduction system for sporulation initiation (7). Emphasized is the role of BA2291 in inducing sporulation (in the presence of activating signal) or inhibiting sporulation (in the absence of activating signal or in the presence of pXO1-118 and pXO2-61) by removing phosphoryl groups from Spo0F∼P.
DISCUSSION
We have identified a novel sensor domain family that includes three single-domain virulence plasmid-encoded proteins (those encoded by pXO1-118, pXO2-61, and pBC218_0049) and several multidomain histidine sensor kinases, orthologues of the B. anthracis BA2291 that is involved in sporulation initiation in the Bacillus cereus-B. anthracis-B. thuringiensis group of spore-forming organisms. There is a high level of amino acid sequence similarity between the pXO1-118- and pXO2-61-encoded proteins and the sensor domain of the sporulation histidine kinase BA2291 of B. anthracis (Fig. 1). Structural studies indicate that the plasmid-encoded sensor domains exist as homodimers and exhibit the same globin fold, characterized by a highly hydrophobic pocket suggestive of ligand-binding capabilities (G. Stranzl et al., unpublished data).
It is clear from the studies described in this report that the virulence plasmid-encoded sensor domains have a strong effect on the activity of sporulation sensor kinase BA2291. This effect results in the conversion of BA2291 from a normally functioning sporulation kinase that contributes phosphoryl groups to the sporulation phosphorelays of B. subtilis and B. anthracis to an enzyme that is able to inhibit sporulation. BA2291 becomes such a potent inhibitor of sporulation in the presence of pXO1-118 and pXO2-61 that it is able to abolish sporulation even in the presence of additional functional and active sporulation sensor kinases that can phosphorylate the Spo0F response regulator (6, 13). Deletion of the pXO1-118 gene does not result in a detectable sporulation phenotype as would be expected for a negative regulator. This is expected, given that only extreme sporulation defects are qualitatively and quantitatively detectable in B. anthracis and that the negative regulators of sporulation in B. subtilis (for example, KipI, Sda, Rap, or Spo0E phosphatases) give rise to often undetectable phenotypes when deleted (data not shown) (8, 20, 21, 27).
Because of the similarities among these sensor domains, there exist several possible mechanisms by which pXO1-118 and pXO2-61 might interrupt signaling to BA2291. It seems possible that heterodimers might form between monomers of either of the two virulence plasmid-encoded sensor domains and the sensor domain of BA2291. This would prevent normal homodimer formation between two BA2291 monomers, thus preventing the trans-autophosphorylation activity required for the input of phosphoryl groups into the sporulation phosphorelay upon binding by activating signal. Although this model is theoretically possible, it seems unlikely, due to the fact that the heterodimer proposed would still have to be able to interact appropriately with Spo0F in order to remove phosphoryl groups from the phosphorelay to inhibit the sporulation as observed. In addition, in pull-down assays in which overexpressed pXO1-118 or pXO2-61 was purified from B. anthracis, BA2291 failed to copurify with either protein (data not shown). This suggests that neither virulence plasmid-encoded sensor domain forms a strong heterodimer with BA2291 in vivo.
A more likely model is that the pXO1-118- and pXO2-61-encoded sensor domains competitively bind the same activating signal/receptor as the sensor domain of BA2291. In this manner, expression of pXO1-118 and pXO2-61 would result in the sequestering of BA2291 signal/receptor, resulting in the sporulation-inhibiting form of BA2291 (Fig. 4B).
Additional studies are required to understand the biochemical mechanism of pXO1-118 and pXO2-61 conversion of BA2291, but the fact that the BA2291 protein purified from E. coli is inactive as a kinase (presumably because of the lack of activating signal) has so far hampered our attempts to define a mechanism. However, it is clear that these virulence plasmid-encoded sensor domains regulate the activity of sporulation sensor kinase BA2291 and thus regulate sporulation. The fact that each virulence plasmid-encoded sensor domain is located within a pathogenicity island suggests that the regulation of the function of BA2291 by these domains may be the missing link in coordinating the onset of pathogenesis to the inhibition of sporulation required for pathogenesis. This is supported further by the observation that the trans-acting virulence gene regulator, AtxA, is also a regulator of pXO2-61 expression (4; Stranzl et al., unpublished). This illuminates a direct tie between inhibition of sporulation and toxin gene expression and adds to the increasingly complicated network of regulation between plasmid-encoded and chromosome-encoded functions in B. anthracis.
Acknowledgments
This study was supported in part by grant AI055860 from the National Institute of Allergy and Infectious Disease and grants GM019416 and GM055594 from the National Institute of General Medical Sciences, National Institutes of Health, United States Public Health Service. M.G. was supported in part by grant 2 PO4B o26 28 from the Polish State Committee for Scientific Research. Oligonucleotide synthesis and DNA sequencing costs were underwritten in part by the Stein Beneficial Trust.
We thank Robert Liddington for the gift of plasmid pXO2.
This article is manuscript number 18131-MEM from the Scripps Research Institute.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, L., A. Moir, and R. Manchee. 1998. The expression of the protective antigen of Bacillus anthracis in Bacillus subtilis. J. Appl. Microbiol. 84:741-746. [DOI] [PubMed] [Google Scholar]
- 4.Bourgogne, A., M. Drysdale, S. G. Hilsenbeck, S. N. Peterson, and T. M. Koehler. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brittingham, K. C., G. Ruthel, R. G. Panchal, C. L. Fuller, W. J. Ribot, T. A. Hoover, H. A. Young, A. O. Anderson, and S. Bavari. 2005. Dendritic cells endocytose Bacillus anthracis spores: implications for anthrax pathogenesis. J. Immunol. 174:5545-5552. [DOI] [PubMed] [Google Scholar]
- 6.Brunsing, R. L., C. La Clair, S. Tang, C. Chiang, L. E. Hancock, M. Perego, and J. A. Hoch. 2005. Characterization of sporulation histidine kinases of Bacillus anthracis. J. Bacteriol. 187:6972-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 8.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269-279. [DOI] [PubMed] [Google Scholar]
- 9.Gould, G. W. 1977. Recent advances in the understanding of resistance and dormancy in bacterial spores. J. Appl. Bacteriol. 42:297-309. [DOI] [PubMed] [Google Scholar]
- 10.Guidi-Rontani, C., and M. Mock. 2002. Macrophage interactions. Curr. Top. Microbiol. Immunol. 271:115-141. [DOI] [PubMed] [Google Scholar]
- 11.Guidi-Rontani, C., M. Weber-Levy, E. Labruyere, and M. Mock. 1999. Germination of Bacillus anthracis spores within alveolar macrophages. Mol. Microbiol. 31:9-17. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. D. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 14.Kang, T. J., M. J. Fenton, M. A. Weiner, S. Hibbs, S. Basu, L. Baillie, and A. S. Cross. 2005. Murine macrophages kill the vegetative form of Bacillus anthracis. Infect. Immun. 73:7495-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makino, S.-I., I. Uchida, N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 18.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perego, M., S. P. Cole, D. Burbulys, K. Trach, and J. A. Hoch. 1989. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J. Bacteriol. 171:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perego, M., C. G. Hanstein, K. M. Welsh, T. Djavakhishvili, P. Glaser, and J. A. Hoch. 1994. Multiple protein aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in Bacillus subtilis. Cell 79:1047-1055. [DOI] [PubMed] [Google Scholar]
- 21.Perego, M., and J. A. Hoch. 1987. Isolation and sequence of the spo0E gene: its role in initiation of sporulation in Bacillus subtilis. Mol. Microbiol. 1:125-132. [DOI] [PubMed] [Google Scholar]
- 22.Russell, A. D. 1990. Bacterial spores and chemical sporicidal agents. Clin. Microbiol. Rev. 3:99-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 26.Tzeng, Y.-L., and J. A. Hoch. 1997. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J. Mol. Biol. 272:200-212. [DOI] [PubMed] [Google Scholar]
- 27.Wang, L., R. Grau, M. Perego, and J. A. Hoch. 1997. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 11:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]