Abstract
Here we present evidence for a physiologically relevant light response mediated by the LOV domain-containing protein YtvA in the soil bacterium Bacillus subtilis. The loss and overproduction of YtvA abolish and enhance, respectively, the increase in σB-controlled ctc promoter activity at moderate light intensities. These effects were absent in the dark and in red light but present under blue-light illumination. Thus, activation of the general stress response in B. subtilis is modulated by blue light.
In Bacillus subtilis, the transcription factor σB controls a regulon composed of more than 200 genes (15) in response to energy (carbon, oxygen limitation) or environmental (acid, ethanol, heat, or salt) stress (8). The gene products of the σB regulon can be subdivided into a number of functional categories, including proteins involved in transcriptional regulation, cell protection (carotenoid synthases and catalases), influx and efflux (transporters and permeases), carbon metabolism, and macromolecular turnover (15). The transcription factor σB is regulated by a very complex signal transduction cascade in which several Ser/Thr kinase plus phosphatase couples play a key role. One of the proteins involved in this cascade is YtvA. Introduction of a null mutation of this protein into B. subtilis results in a twofold decrease in σB-regulated gene expression in response to salt, ethanol, or nutritional stress (1). YtvA is a 261-amino-acid polypeptide composed of an N-terminal LOV (light, oxygen, and voltage) domain and a C-terminal STAS (sulfate transporter and anti-sigma factor antagonist) domain (2, 13). The STAS domains have also been identified in other members of the family of regulators of the environmental signaling pathway that activates σB (2).
The most important members of the family of LOV domain-containing proteins from plants are the phototropins (5). They act as photoreceptors in higher plants to direct a number of physiological responses, such as phototropism, chloroplast relocation, and stomatal opening (6). Most of them contain two photoactive LOV domains at the N terminus which control the activity of a C-terminal serine/threonine kinase domain (9). Genes encoding LOV domain proteins have also been identified in green algae and bacteria (12). All LOV domains show strong mutual similarity and couple light sensing to diverse output domains, such as kinases, phosphodiesterases, transcription factors, and regulators of stress-responsive sigma factors (12). Interestingly, proteins containing the combination of a LOV domain and a STAS domain have also been found in a number of other gram-positive bacteria, including Oceanobacillus iheyensis, Listeria monocytogenes, and Listeria innocua (12).
The YtvA LOV domain contains a noncovalently bound flavin mononucleotide, which results in a yellow protein with absorption peaks at 375, 449, and 475 nm (13). Upon blue-light illumination, YtvA undergoes a photocycle that includes the formation of a signaling state with a thiol adduct between flavin mononucleotide C4 and Cys62 of the protein backbone (3, 13, 14). The rate of ground state recovery of YtvA is ∼4 × 10−4 s−1 (14), which indicates that a photosensory function of YtvA would saturate already at low light intensities.
The presence of both a LOV domain and a STAS domain in YtvA suggests that this protein could be a photoreceptor for light-regulated σB activation. However, no in vivo evidence has been presented to date for a light-induced physiological response that is regulated by YtvA or any of the other bacterial LOV proteins. Here we report the first evidence for a physiologically relevant light response in B. subtilis: blue-light-dependent activation of σB via the LOV domain protein YtvA.
To study a possible photoreceptor function of YtvA, we used transcriptional fusion strains of B. subtilis which report σB activity through β-galactosidase activity under the control of the ctc promoter (Pctc-lacZ) (1, 4). We subjected the wild-type (PB198) and ytvA null (PB565) reporter strains to salt stress in early logarithmic growth phase as described by Akbar et al. (1) in order to induce the σB response. First, we compared activities under these conditions in the dark and with moderate white light illumination of 50 microeinsteins m−2 s−1. Strains were cultured in TSB (30 g liter−1 tryptic soy broth) liquid medium supplemented with 0.5% glucose and 6 μg/ml chloramphenicol. Overnight cultures were diluted 1,000-fold in the same prewarmed medium in a final volume of 10 ml and incubated at 37°C and 250 rpm. Both strains were exposed to NaCl with a 0.3 M end concentration at an optical density at 600 nm (OD600) of 0.3. After 40 min, β-galactosidase activities were measured and expressed in Miller units as described previously (11). Light exposure of the salt-stressed wild-type strain led to higher levels of β-galactosidase activity (12.0 ± 0.6 Miller units) compared to the dark control (8.1 ± 0.4 Miller units). This activity was not significantly affected by light in the salt-stressed ytvA null mutant (6.4 ± 1.8 Miller units in the dark and 7.8 ± 0.4 Miller units in the light). These results indicate that YtvA mediates a light-dependent σB response.
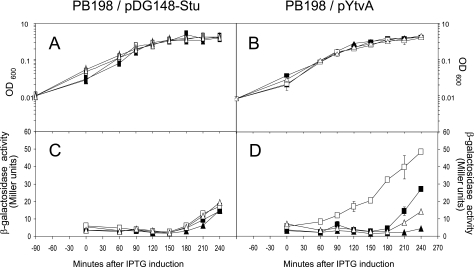
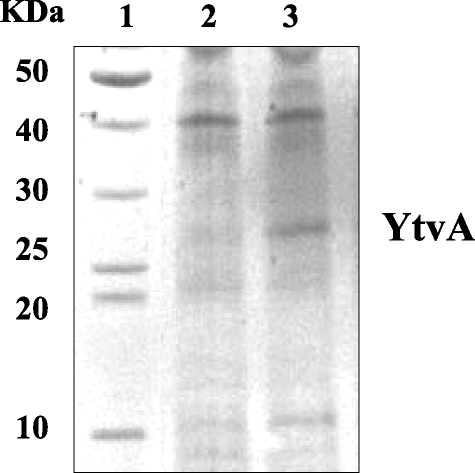
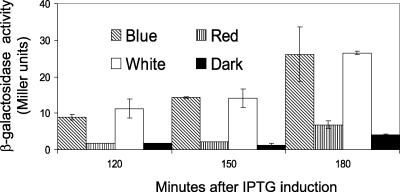
As the observed differences in β-galactosidase activity were relatively small and only clearly observable upon induction with salt stress, we checked the light-dependent control of σB activity in a strain that overproduces YtvA. We introduced the pYtvA vector, which contains ytvA under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac promoter, into PB198 (Table 1). The pYtvA plasmid was obtained by inserting the PCR product, coding for the full-length protein (the primers and the strain used for template DNA are described in Table 1), into the shuttle vector pDG148-Stu by ligation-independent cloning (10). The construct was verified by DNA sequencing; it showed the correct insertion and a 100% match with the ytvA gene sequence (identification no. 937254). B. subtilis strains PB198/pDG148-Stu and PB198/pYtvA were cultured in TSB glucose with 10 μg/ml kanamycin. Overnight cultures were diluted 100-fold in 16 100-ml Erlenmeyer flasks (eight cultures in duplicate; Fig. 1 contains more details). Half of the cultures were exposed to moderate white-light intensities as described above. The other half were covered with aluminum foil to prevent light exposure. During growth in the light and dark, the temperature was kept at 37°C. At an OD600 of 0.3, four illuminated and four dark cultures were induced with IPTG (1 mM final concentration) and cultured—together with the eight noninduced cultures—for another 4 h. β-Galactosidase activity was determined at 30-min intervals. As shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, an overproduced protein with a molecular mass close to that of YtvA (the calculated mass is 29,194.5 Da) was clearly observable 3 h after induction (Fig. 2). The growth rates of the two strains used were not substantially affected by the addition of IPTG and/or illumination (Fig. 1A and B). Approximately 180 min after induction, the β-galactosidase activity of each strain, under every growth condition tested, increased (Fig. 1C and D) because cells entered the stationary phase, which is known to activate σB (4, 17). Nevertheless, PB198/pYtvA β-galactosidase activities significantly increased in the logarithmic growth phase, from around 60 min after induction, but only in the presence of IPTG and light (Fig. 1D, open squares). This indicates that the overproduced YtvA protein is activated by light, resulting in an increase in the σB response. Minor but significant increases in β-galactosidase activity were observed as well from approximately 180 min onward in two other cultures (Fig. 1D). We attribute the increase in β-galactosidase levels with IPTG (filled squares) to the residual activity of YtvA in the dark. The small increase in the absence of IPTG and the presence of light (open triangles) probably resulted from leakage from the Pspac promoter. Relatively small differences in β-galactosidase activities between light and dark cultures were observed as well in cells that harbor the shuttle vector without the ytvA gene (Fig. 1C). We ascribe this small, light-induced rise in β-galactosidase activity to YtvA expressed from the chromosomal copy of the ytvA gene. In an additional experiment, we exposed cultures of the PB198/pYtvA strain to blue and red light with intensities of 30 microeinsteins m−2 s−1 under the same growth conditions as described for the experiment presented in Fig. 1. The results clearly show that blue and not red light induces σB activity (Fig. 3). This is in agreement with the blue-light-absorbing characteristics of YtvA (13).
TABLE 1.
Strains, plasmids, and primers used in this studya
| Strain, plasmid, or primer | Genotype, characteristics, or sequence | Source or reference |
|---|---|---|
| B. subtilis strains | ||
| 168 1A700 | trpC2 | Bacillus Genetic Stock Center |
| PB198 | amyE::Pctc-lacZ Cm trpC2 | 4 |
| PB565 | ytvAΔ1::ery amyE::Pctc-lacZ Cm trpC2 | 1 |
| Plasmids | ||
| pDG148-Stu | Kmr Apr Pspac | 10 |
| pYtvA | Kmr Apr Pspac::ytvA | This study |
| Primers | ||
| YTVAFW | 5′-AAGGAGGAAGCAGGTATGGCTAGTTTTCAATCATTTGGG-3′ | |
| YTVARV | 5′-GACACGCACGAGGTTACATAATCGGAAGCACTTTAAACG-3′ |
Strain 168 1A700 was used for the isolation of chromosomal template DNA, YTVAFW and YTVARV were used as forward and reverse primers, respectively; underlined sequences indicate the adaptor regions for ligation-independent cloning.
FIG. 1.
Effects of IPTG and light on growth and the σB reporter gene Pctc-lacZ of ytvA-overexpressing and control strains. In all panels, the x axis shows the time after addition of 1 mM IPTG. Cells were incubated in the presence of light (open symbols), in the dark (filled symbols), and in the presence (squares) or absence (triangles) of IPTG. Panels A and C show PB198/pDG148Stu (control). Panels B and D show PB198/pYtvA (ytvA overexpressing). In panels A and B, the y axis shows OD600 as a measurement of growth. Panels C and D show β-galactosidase activity (Miller units). Error bars indicate standard deviations calculated from two independent experiments, each with duplicate samples.
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (20%) of B. subtilis cells producing the YtvA protein (stained with Coomassie brilliant blue). Lane 1, molecular mass standards; lane 2, extracts from strain PB198/pYtvA in the absence of IPTG, lane 3, extracts from strain PB198/pYtvA in the presence of IPTG (1 mM final concentration), both harvested 3 h after induction.
FIG. 3.
Effect of blue, red, and white light on the β-galactosidase activity of ytvA-overexpressing strain PB198/pYtvA. At time zero, cells were induced with 1 mM (end concentration) IPTG. Light intensities were 50 microeinsteins m−2 s−1 for standard white-light illumination and 30 microeinsteins m−2 s−1 for red- and blue-light illumination. Error bars indicate standard deviations calculated from two independent experiments, each with duplicate samples.
In summary, here we report the first photoreception function for a bacterial LOV protein and also the first evidence of light activation of σB in Bacillus subtilis. Consequently, this function could be proposed for the other identified bacterial LOV-STAS proteins as well.
Interestingly, in B. subtilis and B. licheniformis a photoresponse has been described that gives rise to a light-induced delay in sporulation (16). Sporulation in B. subtilis is controlled by a pathway that regulates the activity of the sporulation transcription factor σF. The transcription factors σB and σF are controlled by proteins that are paralogous between the two pathways, and under certain conditions cross talk between these pathways has been observed (5). A fascinating extension of the role of YtvA as a photoreceptor for the light regulation of sporulation is currently under investigation in our laboratory.
ADDENDUM
Consistent with the findings presented here, recent work by Gaidenko et al. shows that Cys62, a residue critical for the photochemical reactions displayed by LOV domain proteins, is important for the positive regulatory role of YtvA (7).
Acknowledgments
This work was supported by the Council for Earth and Life Science of The Netherlands Organization for Scientific Research (NWO).
We are very grateful to Alex Ter Beek for initial work on this project and to Chester Price for kindly donating the PB198 and PB565 B. subtilis strains.
REFERENCES
- 1.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 2000. The STAS domain—a link between anion transporters and antisigma-factor antagonists. Curr. Biol. 10:R53-R55. [DOI] [PubMed] [Google Scholar]
- 3.Bednarz, T., A. Losi, W. Gartner, P. Hegemann, and J. Heberle. 2004. Functional variations among LOV domains as revealed by FT-IR difference spectroscopy. Photochem. Photobiol. Sci. 3:575-579. [DOI] [PubMed] [Google Scholar]
- 4.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor B by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carniol, K., T.-J. Kim, C. W. Price, and R. Losick. 2004. Insulation of the σF regulatory system in Bacillus subtilis. J. Bacteriol. 186:4390-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie, J. M., T. E. Swartz, R. A. Bogomolni, and W. R. Briggs. 2002. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32:205-219. [DOI] [PubMed] [Google Scholar]
- 7.Gaidenko, T. A., T.-J. Kim, A. L. Weigel, M. S. Brody, and C. W. Price. 2006. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188:6387-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecker, M., W. Schumann, and U. Voelker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 9.Huala, E., P. W. Oeller, E. Liscum, I. S. Han, E. Larsen, and W. R. Briggs. 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278:2120-2123. [DOI] [PubMed] [Google Scholar]
- 10.Joseph, P., J. R. Fantino, M. L. Herbaud, and F. Denizot. 2001. Rapid orientated cloning in a shuttle vector allowing modulated gene expression in Bacillus subtilis. FEMS Microbiol. Lett. 205:91-97. [DOI] [PubMed] [Google Scholar]
- 11.Kenney, T. J., and C. P. Moran, Jr. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Losi, A. 2004. The bacterial counterparts of plant phototropins. Photochem. Photobiol. Sci. 3:566-574. [DOI] [PubMed] [Google Scholar]
- 13.Losi, A., E. Polverini, B. Quest, and W. Gartner. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Losi, A., B. Quest, and W. Gartner. 2003. Listening to the blue: the time-resolved thermodynamics of the bacterial blue-light receptor YtvA and its isolated LOV domain. Photochem. Photobiol. Sci. 2:759-766. [DOI] [PubMed] [Google Scholar]
- 15.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, K. C. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 16.Propst-Ricciuti, C., and L. B. Lubin. 1976. Light-induced inhibition of sporulation in Bacillus licheniformis. J. Bacteriol. 128:506-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]