Abstract
AdpA in the A-factor regulatory cascade in Streptomyces griseus activates a number of genes required for secondary metabolism and morphological differentiation, forming an AdpA regulon. The Streptomyces subtilisin inhibitor (SSI) gene, sgiA, in S. griseus was transcribed in response to AdpA, showing that sgiA is a member of the AdpA regulon. AdpA bound a single site upstream of the sgiA promoter at approximately position −70 with respect to its transcriptional start point. Mutational analysis of the AdpA-binding site showed that the AdpA-binding site was essential for transcriptional activation. Mutants in which sgiA was disrupted had higher trypsin, chymotrypsin, metalloendopeptidase, and total protease activities than the wild-type strain, which showed that SgiA modulated the activities of these extracellularly produced proteases. Because a number of genes encoding chymotrypsins, trypsins, and metalloendopeptidases, most of which are SSI-sensitive proteases, are also under the control of AdpA, the A-factor regulatory cascade was thought to play a crucial role in modulating the extracellular protease activities by triggering simultaneous production of the proteases and their inhibitor at a specific timing during growth. Mutants in which sgiA was disrupted grew normally and formed aerial hyphae and spores with the same time course as the wild-type strain. However, exogenous addition of purified SgiA to substrate mycelium grown on agar medium resulted in a delay in aerial mycelium formation, indicating that SgiA is involved in aerial hypha formation in conjunction with proteases.
In Streptomyces griseus, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) acts as a chemical signaling molecule or a microbial hormone that triggers secondary metabolism and cell differentiation (5, 28, 30). We studied the A-factor signaling cascade that leads to production of almost all the secondary metabolites produced by this organism and to formation of aerial mycelium and spores. The maximum concentration of A-factor that is gradually accumulated in a growth-dependent manner, about 25 ng/ml (100 nM), occurs at or near the middle of the exponential growth phase (1). When the concentration of A-factor reaches a critical level, this molecule binds ArpA, which is bound to the promoter of adpA, and removes ArpA from the target DNA, thus inducing transcription of adpA (29). The transcriptional factor AdpA activates a number of genes required for secondary metabolism and morphological differentiation, forming an AdpA regulon. The members of the AdpA regulon include a variety of genes responsible for secondary metabolite formation and morphological development (5, 28, 30).
We have identified several members of the AdpA regulon for morphogenesis. These genes include adsA encoding an extracytoplasmic function sigma factor of RNA polymerase essential for aerial mycelium formation (41), ssgA encoding a small acidic protein essential for spore septum formation (42), amfR encoding a regulatory protein essential for aerial mycelium formation (43), and sgmA encoding a metalloendopeptidase that affects aerial mycelium formation (11). In addition, two trypsins (SprT and SprU) (9) and three chymotrypsins (SprA, SprB, and SprD) (37), all of which are serine proteases that are produced extracellularly from the late exponential growth stage to the stationary growth stage, are also controlled by AdpA. Because serine proteases have been reported to be involved in aerial mycelium formation in several Streptomyces spp. (14-16), we disrupted the five AdpA-dependent protease genes in S. griseus to determine the possible effects on morphogenesis. Contrary to our expectations, however, no detectable morphological changes were observed; even a ΔsprT ΔsprU double mutant or a ΔsprA ΔsprB ΔsprD triple mutant grew normally and developed aerial hyphae and spores with the same time course as the wild-type strain (9, 37). These findings ended our plan to study a possible link via extracellular proteases between A-factor and morphogenesis.
We then examined the Streptomyces subtilisin inhibitor (SSI), which is a proteinaceous inhibitor that inhibits serine proteases, including trypsins and chymotrypsins. Although the biochemistry of SSIs has been extensively studied (24, 32, 36), little is known about their physiological roles in the organisms that produce them. We searched the genome sequence of S. griseus (unpublished data) for an open reading frame (ORF) encoding a protein that is similar to the SSI family members. A single ORF, designated sgiA, was found. An unexpected finding was that sgiA is a member of the AdpA regulon; its transcription completely depends on AdpA. During these studies, Kim et al. (12) reported that the SSI gene in Streptomyces coelicolor A3(2) is a target of AdpA. These workers also showed that disruption of the SSI gene has no detectable effect on the morphogenesis of S. coelicolor A3(2).
In this paper, we describe (i) the discovery of sgiA as an SSI gene in S. griseus, (ii) a transcriptional analysis of sgiA, (iii) the dependence of sgiA transcription on AdpA, and (iv) the involvement of SgiA in aerial hypha formation. Because disruption of sgiA resulted in no detectable effects on morphogenesis of S. griseus like the effects found for S. coelicolor A3(2) (10, 12), we purified SgiA and added it to substrate mycelium grown on agar medium. SgiA caused a delay in the formation of aerial mycelium, as did pefablocSC, a serine protease inhibitor (17). These results show that the AdpA-dependent, SSI-sensitive proteases and SSI play a role in aerial mycelium formation, perhaps in the hydrolysis of proteins in substrate hyphae for reuse in the formation of aerial hyphae.
MATERIALS AND METHODS
General recombinant DNA studies.
All the bacterial strains, plasmids, and media used have been described previously (9, 11). The strategies used for gene disruption, alteration of the AdpA-binding sites by PCR, a gel mobility shift assay, and S1 nuclease mapping have also been described previously (9). Low-copy-number plasmid pKUM20 was described previously (43).
S1 nuclease mapping.
Total RNA was isolated with ISOGEN (Nippon Gene) from cells grown on cellophane on the surface of YMPD agar. Hybridization probes were prepared by performing PCR with a pair of 32P-labeled and nonlabeled primers. The PCR primers used for low- and high-resolution S1 mapping were sF and sR (Table 1). Primer sR was labeled at the 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase before PCR. The same 32P probe was also used for transcriptional assays of the native and mutated sgiA promoters on pKUM20 in S. griseus mutant ΔsgiA.
TABLE 1.
Primers used in this study
| Primer | Position | Sequence (5′ to 3′) |
|---|---|---|
| F1 | −343 to −324 | CGGGATTCGGAATCGTGTCC |
| R1 | 104 to 85 | GTGGCGAATCCACCGAGCAG |
| F2 | 58 to 77 | TTCGCGACCGTCACGGCGAC |
| R2 | 484 to 465 | CGGTGGGTCAGAAGGCGAAC |
| F3 | −340 to −321 | GATTCGGAATCGTGTCCTCC |
| R3 | −111 to −130 | CGTGAGGCGTCCAGCATATG |
| F4 | −183 to −164 | CCGTGTATCCGCAGGTCAGG |
| R4 | 26 to 7 | GCTCCTTGGTCGAATGCCTG |
| F5 | −43 to −24 | CCGTCGGCGTGCGCTTACTT |
| sF | −127 to −108 | ATGCTGGACGCCTCACGCCG |
| sR | 76 to 57 | TCGCCGTGACGGTCGCGAAG |
| dLF | 463 to 491 | CCCAAGCTTCACGTGGTGTTCGCCTTCTGACCCACCGGGGCGCG |
| dLR | 2377 to 2348 | CCGGAATTCGACCATGGACCCATGGGCAGAGAATCCCCC |
| dRF | −1833 to −1814 | CTTGCCGATGAGTTCGACGG |
| dRR | 36 to 16 | CCCAAGCTTCATGGGTGAGGCTCCTTGGTC |
| pF | 121 to 147 | GGAATTCCATATGGCCATGGCACCGGCGGGGGCCCCGAGCCTCTAC |
| pR | 474 to 455 | CGGGATCCGAGAAGGCGAACACGGCATTGC |
| cF | −756 to −737 | AAGTCGGAACGGGACAGGGC |
| cR | 774 to 755 | CCGGAATTCTGACGCAGGAGTCCTCCGAC |
| mR | 104 to 85 | CGGGGTACCGTGGCGAATCCACCGAGCAG |
| mAF | −71 to −48 | GGAATTCGAACGCCCGCCCGCGGCTGGAAG |
| mAR | −77 to −97 | GGAATTCACCGAATGGTCATTCGGGCAG |
| R-20mer | ATGACCATGATTACGCCAAG |
Gel mobility shift assay.
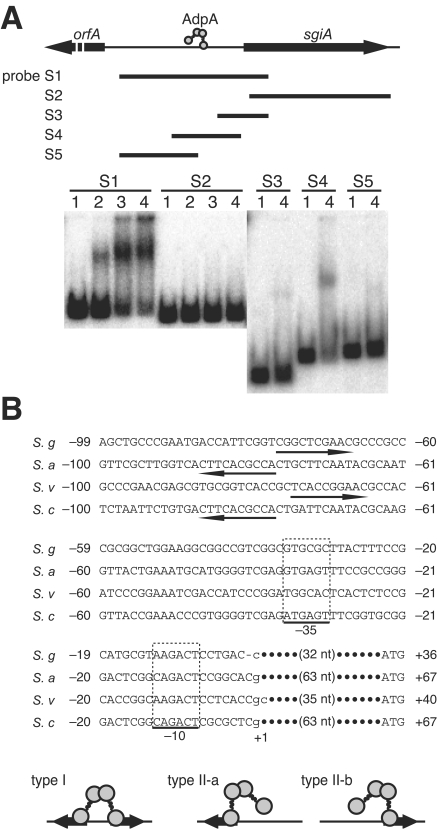
The DNA fragments used for 32P-labeled probes were amplified by PCR and 32P labeled with T4 polynucleotide kinase. Various regions upstream and in the coding sequence of sgiA were used as 32P-labeled probes. Table 1 shows the primer sequences used for preparing these probes. Five probes, S1 to S5, were prepared as follows: F1 and R1 were used for probe S1; F2 and R2 were used for probe S2; F5 and R1 were used for probe S3; F4 and R4 were used for probe S4; and F3 and R3 were used for probe S5.
Alteration of the AdpA-binding sequence by PCR.
A mutation was introduced into the AdpA-binding site of sgiA by PCR. A 1.5-kb fragment (positions −756 to 774) was amplified by performing PCR with primers cF and cR with an EcoRI site. The amplified fragment was trimmed with EcoRI and SalI (at position −726 in the amplified fragment) and cloned between the EcoRI and SalI sites of pUC19, generating pUC-sgiA, and the results were checked by nucleotide sequencing. The HindIII (in the pUC19 sequence)-EcoRI fragment was inserted between the HindIII and EcoRI sites of pKUM20, generating pW. The CGGCTC sequence in the AdpA-binding site was changed to the EcoRI cleavage sequence GAATTC by PCR using pUC-sgiA as the template (see Fig. 4A). The region upstream of the AdpA-binding site was amplified with primers mAR (containing an EcoRI sequence at the 5′ end) and R-20mer (corresponding to the pUC19 sequence) and was cut with SalI and EcoRI. The region downstream of the AdpA-binding site was amplified with primers mAF (containing an EcoRI sequence at the 5′ end) and mR (containing a KpnI sequence at the 5′ end) and was cut with EcoRI and KpnI. The SalI-EcoRI fragment and the EcoRI-KpnI fragment were inserted between the SalI and KpnI sites of pUC19 by three-fragment ligation, generating pUC-pre-mt, and the results were checked by nucleotide sequencing. The SalI-SphI fragment from pUC-pre-mt and the SphI-EcoRI fragment from pUC-sgiA were inserted between the SalI and EcoRI sites of pUC19 by three-fragment ligation, generating pUC-mt. The HindIII (in the pUC19 sequence)-EcoRI (mutation site) fragment from pUC-mt was inserted between the HindIII and EcoRI sites of pKUM20, generating pKU-pre-mt. pKU-pre-mt was digested with EcoRI, treated with calf intestine alkaline phosphatase, and ligated with the EcoRI fragment (containing the sgiA coding region) from pUC-mt, resulting in pM.
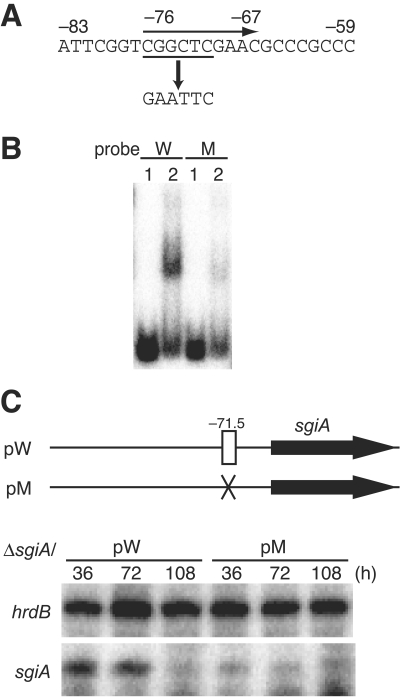
FIG. 4.
Importance of the AdpA-binding sequence for transcriptional activation of sgiA by AdpA. (A) A mutation was introduced into the AdpA-binding sequence by PCR to generate an EcoRI recognition sequence. (B) Gel mobility shift assay for determination of AdpA binding to the mutated sequence. The 32P-labeled probe (positions −183 to 26) containing the intact AdpA-binding sequence (W) yielded a distinct shifted signal, whereas a similar probe containing the EcoRI mutation (M) did not. The amounts of AdpA used were 0 μg (lane 1) and 0.8 μg (lane 2). (C) Transcriptional analysis of the promoter containing the EcoRI mutation in the AdpA-binding site. pW, as a control, contained the sgiA coding region and the intact AdpA-binding site on pKUM20, and pM contained the mutated AdpA-binding site. The distance between the center of the AdpA-binding site and the transcriptional start point of sgiA is 71.5 bp. RNA was prepared from cells of mutant ΔsgiA harboring pW or pM that were grown at 28°C for the times indicated on YMPD agar containing 50 μg/ml of thiostrepton. Almost no transcription from the sgiA promoter with the mutated AdpA-binding site was observed, whereas distinct sgiA transcripts from the sgiA promoter with the intact AdpA-binding site were detected at 36 and 72 h.
Gene disruption.
For disruption of sgiA on the chromosome, the sgiA sequence was replaced by the neomycin resistance gene aphII. Briefly, a 1.9-kb sequence upstream of the sgiA coding sequence and a 1.9-kb sequence downstream of the sgiA coding sequence, together with aphII, were assembled in pUC19 (see Fig. 5A). This mutagenic plasmid was linearized by digestion with DraI and introduced by transformation into S. griseus IFO13350, and neomycin (10 μg/ml)-resistant colonies were isolated. Correct disruption was checked by Southern hybridization with the sgiA sequence (positions 463 to 2377, prepared with primers dLF and dLR) and the aphII sequence as 32P-labeled probes against the chromosomal DNA digested with EcoRI plus PvuII.
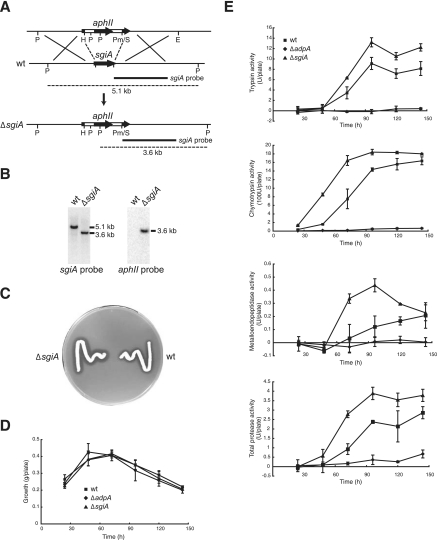
FIG. 5.
Disruption of the chromosomal sgiA gene and protease activities in the sgiA disruptant. (A) Schematic diagram of disruption of sgiA on the chromosome of S. griseus IFO13350. Restriction enzymes: E, EcoRI; H, HindIII; P, PvuII; Pm, PmaCI; S, SalI. (B) Southern hybridization to check the correct disruption of sgiA with the probes indicated against the EcoRI/PvuII-digested chromosome. (C) Strains grown at 28°C for 48 h on Bennett medium containing 0.7% skim milk. (D) Growth of the S. griseus strains expressed as cell mass. (E) Trypsin, chymotrypsin, metalloendopeptidase, and total protease activities produced extracellularly by the strains grown on agar medium, determined using artificial chromogenic substrates. The values are means ± standard deviations (n = 3). wt, wild type.
Extracellular protease assays.
The extracellular protease activity of mutant ΔsgiA was assayed by using synthetic substrates, as described previously (9). S. griseus strains were grown for various times as lawns on cellophane on the surface of YMPD agar. After the cellophane and the cells were removed, an agar layer (5 by 5 cm) was cut out and homogenized by passage through a 50-ml plastic syringe in 10 ml of buffer A (100 mM Tris-HCl [pH 8.0], 10 mM CaCl2). The resulting suspension was centrifuged at 14,000 × g at 4°C for 10 min in order to remove agar debris, and the supernatant was used as a crude enzyme. The reaction mixture used for the chymotrypsin activity assay, which consisted of 980 μl of buffer A, 10 μl of 30 mM N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (Sigma), and 10 μl of the enzyme solution, was incubated at 37°C for 15 min. The reaction was stopped by adding 400 μl of stop solution (30% acetic acid in dioxane). The chymotrypsin activity was measured spectrophotometrically at 405 nm by determining the release of p-nitroanilides due to the hydrolysis of the artificial chromogenic substrate. One unit of hydrolytic activity was defined as the amount of enzyme that resulted in a 0.1 U increase in absorbance per min under the conditions described above. The reaction mixture used for the analysis of trypsin activity consisted of 890 μl of buffer B (50 mM Tris-HCl [pH 8.0], 20 mM CaCl2), 10 μl of 50 mM N-benzoylarginine p-nitroanilide (Sigma), and 100 μl of the enzyme solution, and the trypsin activity was assayed like the chymotrypsin activity. For the assay of total protease activity, the reaction mixture consisting of 125 μl of buffer C (50 mM potassium phosphate [pH 7.0], 2% azocasein) and 75 μl of the enzyme solution was incubated at 37°C for 1 h. The reaction was stopped by adding 600 μl of 10% trichloroacetic acid. The mixture was centrifuged at 8,000 × g for 3 min. To a portion (600 μl) of the supernatant, 700 μl of 1 M NaOH was added, and the azo form released was measured spectrophotometrically at 440 nm. For the assay of metalloendopeptidase activity, 1.25 μmol of EDTA was added to the reaction mixture, and the assay was performed like the assays described above. The reaction mixture was incubated for 12 h. The metalloendopeptidase activity was calculated by subtracting the activity measured in the presence of EDTA from the activity measured in the absence of EDTA.
Production and purification of SgiA.
The probable pro form plus mature sequence (Ala-30 to Phe-147) of SgiA was expressed as a fusion to the PelB leader sequence at the N terminus and to a histidine tag at the C terminus, as follows. The sgiA sequence encoding Ala-30 to Phe-147 was amplified by PCR with primers pF (containing EcoRI, NdeI, and NcoI sites) and pR (containing a BamHI site). The sgiA coding sequence was excised as an EcoRI-BamHI fragment and placed between the EcoRI and BamHI sites of pUC19, and the results were checked by nucleotide sequencing. The NcoI-BamHI fragment was excised from this plasmid and placed between the NcoI and BamHI sites of pET26b, resulting in pET26-sgiA. Escherichia coli BL21(DE3) harboring pET26-sgiA was cultured at 26.5°C for 3 h in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). SgiA with a His tag at the C terminus was purified from the soluble fraction with an Ni spin column (QIAGEN) used according to the manual of the manufacturer. The SgiA preparation was dialyzed against 10 mM Tris-HCl (pH 7.5).
The trypsin inhibitory activity of the purified SgiA protein was determined by incubation (at 37°C for 15 min) of a reaction mixture consisting of 940 μl of buffer B, 40 μl of a SgiA sample (containing 29 to 1,160 pmol), 10 μl of 50 mM N-benzoylarginine p-nitroanilide, and 10 μl (168 pmol) of bovine trypsin (Sigma). For assays of chymotrypsin inhibitory activity, the reaction mixture contained 940 μl of buffer A, 40 μl of a SgiA sample (containing 1.45 to 58 pmol), 10 μl of 30 mM N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide, and 10 μl (4 pmol) of bovine chymotrypsin (Washington Biochemical Corp.).
Nucleotide sequence accession number.
The nucleotide sequence of sgiA has been deposited in the DDBJ, EMBL, and GenBank DNA databases under accession no. AB259170.
RESULTS
SSI (sgiA) gene in S. griseus.
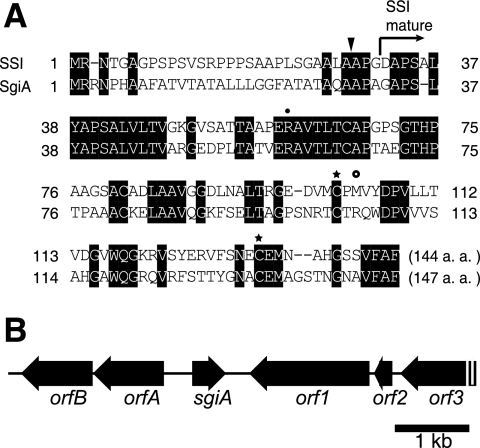
We searched the genome sequence of S. griseus IFO13350 (unpublished data) for ORFs encoding an SSI-like protein, although Kuramoto et al. (21) reported the absence of any gene homologous to an SSI gene on the basis of DNA hybridization. A single ORF, designated sgiA, that encoded a protein exhibiting end-to-end similarity to members of the SSI family was found. Figure 1A shows an alignment of the amino acid sequences of SgiA and the SSI from Streptomyces albogriseolus S-3253 (7, 27). The N-terminal amino acid of the SSI from S. albogriseolus is Asp-32 (27). This SSI is probably cleaved by a signal peptidase between Ala-28 and Ala-29, as predicted by the Signal P program (http://www.cbs.dtu.dk/services/SignalP/), and matures due to a second cleavage between Gly-31 and Asp-32. Because the amino acid sequences that include the processing sites of the two SSIs are similar, we assume that SgiA is probably cleaved by a signal peptidase between Ala-29 and Ala-30 and matures due to cleavage between Ala-32 and Gly-33. The two cysteine residues (Cys-103 and Cys-133) which form a disulfide bridge to maintain conformational rigidity around the reactive site of the inhibitor (18) and Arg-60, which is important for maintaining the tertiary structure by forming a salt bridge with the C-terminal carboxylate (31), are all conserved in SgiA. The P1 site, the center of the reactive site (19), is Met in the SSI of S. albogriseolus and Arg in SgiA. Site-directed replacement of the Met by Lys or Arg yielded a mutant SSI that exhibited potent inhibition of trypsins (19, 24). Consistent with this, as described below, SgiA exhibited potent trypsin inhibitory activity but no chymotrypsin inhibitory activity in vitro.
FIG. 1.
Amino acid sequence of SgiA (A) and gene organization in the neighborhood of sgiA (B). (A) Alignment of the amino acid sequences of SSI from S. albogriseolus S-3253 and SgiA from S. griseus. The Arg residue indicated by a solid circle is required to maintain the tertiary structure by formation of a salt bridge with the C-terminal carboxylate. The Cys residues indicated by stars are also required for conformational rigidity around the reactive site of the inhibitor by formation of a disulfide bridge. The amino acid indicated by an open circle represents the P1 site (33). The N-terminal site of the mature form of SSI is indicated by an arrow. A probable cleavage site for a signal peptidase is indicated by a solid triangle. a.a., amino acids. (B) Positions and directions of open reading frames predicted by the S. griseus genome sequence are indicated by arrows. The predicted ORFs are as follows: orfA, ABC transporter; orfB, iron ion transporter; orf1, putative export protein; orf2, ribosomal protein; orf3, hypothetical protein.
The gene organization in the neighborhood of sgiA is shown in Fig. 1B. sgiA is located between the genes encoding an ABC transporter (orfA) and a putative export protein (orf1). Because the direction of sgiA is opposite the direction of orfA and orf1, sgiA is not cotranscribed with the neighboring genes. The gene organization in the region around the SSI gene (SCO0762) in S. coelicolor A3(2) (10, 12) is totally different from that in S. griseus. SCO0762 is located between the genes encoding a functionally unknown protein and an oxidoreductase. The gene organizations in the neighboring regions of three ORFs encoding probable SSIs in Streptomyces avermitilis (SAV7486, SAV2156, and SAV4204) (6) are also totally different from those in S. griseus and S. coelicolor A3(2).
Dependence of sgiA transcription on AdpA.
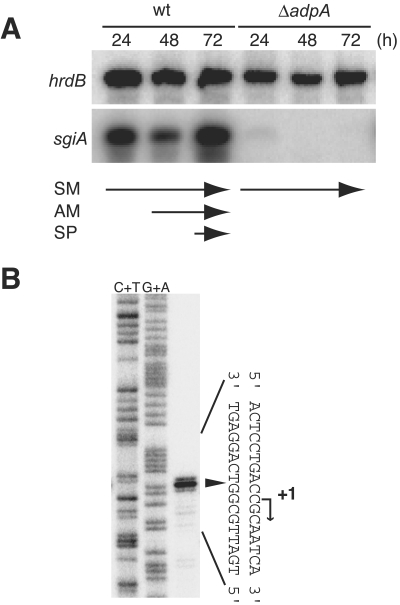
We analyzed transcription of sgiA by S1 nuclease mapping by using RNA prepared from cells grown on cellophane on the surface of agar (Fig. 2A). Under these culture conditions, S. griseus grew as substrate mycelium at 24 h, as a mixture of substrate mycelium and aerial mycelium at 48 h, and as a mixture of aerial mycelium and spores at 72 h. The hrdB gene, which is transcribed throughout growth, was used as an internal control to check the integrity and amount of mRNA used. We found that sgiA was actively transcribed throughout growth. The transcriptional start point was determined to be 33 nucleotides upstream of the translational start codon, as determined by high-resolution S1 mapping (Fig. 2B). We propose that the potential −35 and −10 sequences are 5′-GTGCGC-3′ and 5′-AAGACT-3′, respectively, with a space of 17 nucleotides between them (Fig. 3B).
FIG. 2.
Transcriptional analysis of sgiA. (A) Time course of sgiA transcription determined by low-resolution S1 mapping with RNA prepared from cells grown at 28°C for the times indicated on cellophane on the surface of YMPD medium. As an internal control, transcription of hrdB (constitutive) was also determined. SM, substrate mycelium; AM, aerial mycelium; SP, spore; wt, wild type. Mutant ΔadpA grew as substrate mycelium throughout growth. (B) Transcriptional start point of sgiA determined by high-resolution S1 mapping. RNA prepared from the wild-type strain grown for 24 h on YMPD medium was used. The arrowhead indicates the position of the S1-protected fragment. The 5′ terminus of the mRNA was assigned to position +1, because the fragments generated by the chemical sequencing reactions migrated 1.5 nucleotides further than the corresponding fragments generated by S1 digestion of the DNA-RNA hybrids migrated (0.5 residue from the presence of the 3′-terminal phosphate group and 1 residue from elimination of the 3′-terminal nucleotide).
FIG. 3.
Binding of AdpA to the region upstream of the sgiA promoter. (A) Gel mobility shift assays with AdpA and five 32P-labeled probes, S1 to S5. The amounts of AdpA used were 0.2 μg (lane 2), 0.4 μg (lane 3), and 0.8 μg (lane 4). Lane 1 was a control lane in which no AdpA was present. (B) Nucleotide sequence including the −35 and −10 elements of the sgiA promoter and a consensus AdpA-binding sequence. The consensus AdpA-binding sequences, from the 5′ end to the 3′ end, are indicated by arrows. The transcriptional start point, determined by S1 mapping (Fig. 2B), is indicated by +1. The alignment includes the SSI genes from S. griseus (S. g), S. albogriseolus S-3253 (S. a), S. venezuelae (S. v), and S. coelicolor A3(2) (S. c). Schematic diagrams of AdpA binding to two types of binding sites (types I and II) are also shown.
An unexpected finding was that the transcription of sgiA was under the control of AdpA (Fig. 2A). In S. griseus mutant ΔadpA, almost no sgiA transcription occurred at any stage of growth. A similar finding concerning transcriptional control of the SSI gene by AdpA in S. coelicolor A3(2) has recently been reported by Kim et al. (12). As described below, a single AdpA-binding site at nucleotide −71 was present. These data suggested that there is transcriptional control of sgiA by AdpA.
Detection of an AdpA-binding site upstream of sgiA.
Because sgiA was found to be controlled by AdpA, we determined whether AdpA binds regions upstream of sgiA by a gel mobility shift assay. AdpA activates target genes by binding various positions with respect to the transcriptional start point (for example, more than 200 bp upstream and 25 bp downstream from the transcriptional start point) (44). In addition, some target genes contain multiple AdpA-binding sites. Furthermore, a consensus AdpA-binding sequence is rather relaxed (see below). Therefore, we designed five 32P-labeled probes for various regions and used them for gel mobility shift assays. The AdpA protein used was purified from E. coli harboring pET-adpA and had a histidine tag at its C-terminal end with the structure AdpA-Leu-Glu-His6 (41). Two of the five probes tested, S1 (nucleotide positions −343 to 104 with respect to the transcriptional start point) and S4 (positions −183 to 26), gave a single retarded signal (Fig. 3A).
In the sequence of probe S4 there was a potential AdpA-binding sequence (consensus sequence, 5′-TGGCSNGWWY-3′, where S = G or C, W = A or T, Y = T or C, and N = any nucleotide) (44) (Fig. 3B). The gel mobility shift assay with S4 showed that AdpA bound to this probe, but similar assays with probes S2, S3, and S5 showed that there was no AdpA-binding site (Fig. 3A). Some AdpA-binding sites contain two consensus sequences as an inverted repeat (type I) (Fig. 3B), and others contain a single AdpA-binding sequence (type II). A dimer of AdpA binds both types of sites (44). The AdpA-binding site for sgiA is a type II site, to which AdpA probably binds by anchoring the DNA via the two DNA-binding motifs in one subunit of the AdpA dimer (44). Most target genes activated by AdpA contain one AdpA-binding site at positions −40 to −50 with the same orientation of the recognition sequence (5′ to 3′) and of transcription. The AdpA dimer bound to this site is predicted to recruit RNA polymerase to initiate transcription. In fact, AdpA facilitates RNA polymerase activity to form an open complex that is competent for transcriptional initiation at the promoters of strR (38), ssgA (44), and adsA (44). The distance of the AdpA-binding site from the transcriptional start point and its direction with respect to the direction of transcription suggest that an AdpA dimer binds this site by anchoring the DNA via the DNA-binding domain in the subunit close to the RNA polymerase on the promoter (type II-b). The same topology can be applied to the AdpA binding to the SSI gene of Streptomyces venezuelae (39). In S. albogriseolus S-3253 (35) and S. coelicolor A3(2) (12), on the other hand, AdpA is assumed to bind the SSI gene in a type II-a manner.
Alteration of the AdpA-binding sequence.
To determine the importance of the AdpA-binding sequence, we introduced a mutation into the AdpA-binding sequence of sgiA by site-directed mutagenesis. The effects of the mutation on the promoter activity were examined to determine whether AdpA directly controls the transcription of sgiA. The six nucleotides in the consensus AdpA-binding sequence for sgiA were changed to an EcoRI recognition sequence consisting of six nucleotides (Fig. 4A). The 32P-labeled probe (positions −183 to 26) containing the EcoRI mutation and the sgiA promoter gave almost no retarded signal, whereas a similar probe containing the intact AdpA-binding sequence gave a distinct signal (Fig. 4B). These data showed that the consensus sequence from position −76 to position −67 actually contributed to the AdpA-binding site.
We constructed pW, containing a 1.5-kb fragment, which contained the sgiA promoter together with its upstream region as far as position −726 and the complete sgiA-coding sequence, with the low-copy-number plasmid pKUM20 (Fig. 4C). We also constructed the similar pM plasmid, in which the AdpA-binding site was replaced by the EcoRI mutation. Because the 32P-labeled end of the probe used for S1 mapping was deleted from the chromosome in mutant ΔsgiA (see below), the transcript detected in this mutant should have originated from the sgiA promoter on the plasmids. The sgiA transcript in mutant ΔsgiA harboring pW was detected at 36 and 72 h, whereas almost no sgiA transcripts were detected in mutant ΔsgiA harboring pM. Therefore, AdpA bound at position −71 directly controls sgiA transcription. The transcriptional activity of sgiA in pW was greatly reduced compared to that on the chromosome (Fig. 2A); the reason for this is unclear.
Disruption of the chromosomal sgiA gene.
We replaced most of sgiA with aphII to determine the possible role of sgiA in morphological development (Fig. 5A). The correct disruption of sgiA was checked by Southern hybridization with appropriate probes, as shown in Fig. 5B.
The extracellular trypsin, chymotrypsin, metalloendopeptidase, and total protease activities of mutant ΔsgiA grown on YMPD agar were compared to the activities of the wild-type strain and mutant ΔadpA (Fig. 5E). Mutant ΔadpA was used as a negative control because most of the trypsin, chymotrypsin, and metalloendopeptidase genes are under the control of AdpA (9, 11, 37). The growth of these three strains was almost the same (Fig. 5D). At 50 h and thereafter, mutant ΔsgiA exhibited higher trypsin, chymotrypsin, metalloendopeptidase, and total protease activities than the wild-type strain exhibited, showing that SgiA plays a direct or indirect role in modulating these protease activities in vivo.
Proteases have been suggested to be important for morphological differentiation, particularly in aerial mycelium formation (14-16). We expected that higher activities of the SgiA-sensitive proteases caused by the sgiA mutation might affect morphological differentiation. Mutant ΔsgiA was grown on various media, such as YMPD, R2YE, Trypto-Soya, and minimal media, as a lawn by spreading a lump of mycelium and as a colony by inoculating a lump of mycelium or spores with a toothpick. The carbon source (glucose) in these media was also changed to mannitol and glycerol. However, no apparent phenotypic changes were observed in the mutant; mutant ΔsgiA grew normally and formed aerial hyphae and spores with the same time course as the wild-type strain.
When bacteria grow on skim milk-containing medium, they hydrolyze it and form a clear zone. S. griseus also produced a clear zone on skim milk-containing medium (Fig. 5C). Because mutant ΔsgiA formed a larger clear zone than the wild-type strain formed, some of the skim milk-hydrolyzing proteases were modulated by SgiA. It should be noted that the trypsins and chymotrypsins, which are produced in response to AdpA, are probably not responsible for the formation of the clear zone. ΔsprABD and ΔsprTU mutants of S. griseus produced clear zones that were the same size as the clear zone produced by the wild-type strain (37), although there may have been some interregulation among proteases and compensatory changes in different proteases.
We repeatedly attempted to complement the defect in protease inhibitory activity of mutant ΔsgiA or to overexpress sgiA in the wild-type strain by introducing sgiA on a low-copy-number plasmid (pKUM20). Although mutant ΔsgiA transformants harboring the plasmids that were designed were isolated, the defect in protease inhibitory activity of the transformants was not restored. In addition, no increase in the protease inhibitory activity of the wild-type strain harboring the plasmids that were designed was observed. As shown in Fig. 4C, the transcription of sgiA on a plasmid was greatly reduced. We observed a similar failure to complement a mutation in the SSI gene in S. coelicolor A3(2) or to overexpress the SSI gene (10).
Exogenous supply of SgiA to growing mycelium.
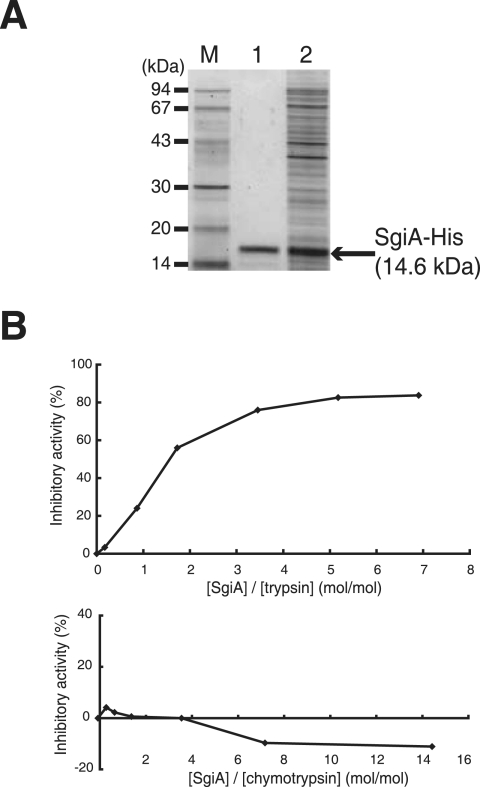
We next purified SgiA from recombinant E. coli cells and applied it to growing mycelium to determine whether there was a possible effect on morphogenesis. For this purpose, we constructed pET26-sgiA that expressed a 16.9-kDa fusion protein consisting of the PelB leader sequence, Met (as a linker), the pro form plus the mature sequence of SgiA (from Ala-30 to Phe-147), and a His tag. SSIs are reported to show inhibitory activity also in their pro forms (34). The SgiA protein was produced in the soluble fraction of E. coli harboring pET26-sgiA and was purified using His-bind resin (Fig. 6A). The size of the purified protein was calculated to be 15 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. N-terminal amino acid sequencing of the purified protein indicated that its N terminus was Met, which showed that a signal peptidase in E. coli cleaved the primary product between the PelB leader sequence and the Met linker, generating the protein Met-(Ala-30 to Phe-147) (calculated molecular mass, 14.6 kDa).
FIG. 6.
Purification of His-tagged SgiA from E. coli. (A) E. coli harboring pET26b-sgiA was grown in the presence of IPTG, and the His-tagged SgiA protein was purified with His-bind resin. The soluble fraction (lane 2) and the purified sample (lane 1), together with molecular size markers (lane M), were electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel. (B) Trypsin inhibitory activity of the His-tagged SgiA protein. Saturated inhibition occurred when [SgiA]/[trypsin] was 2.5 ([SgiA] is the concentration of the SgiA monomer, although SgiA acts as a homodimer).
We checked the protease inhibitory activity of the purified SgiA protein by using commercially available proteases from mammals (Fig. 6B). SgiA was expected to inhibit trypsin activity because the P1 amino acid is Arg, as described above. Saturated trypsin inhibitory activity of the SgiA sample was observed when [SgiA]/[trypsin] was 2.5. SSI inhibits subtilisin BPN′ when [SSI]/[subtilisin BPN′] is 1, because a dimer of SSI inhibits protease activity by forming a complex with two molecules of a protease monomer (8, 20). Therefore, we concluded that SgiA strongly inhibits even bovine trypsin, despite the great difference in the amino acid sequence and thus the overall conformation between bovine trypsin and trypsin-like proteases in Streptomyces. On the other hand, SgiA exhibited no bovine chymotrypsin inhibitory activity, which also reflected a great difference in amino acid sequence between bovine chymotrypsin and chymotrypsin-like proteases in Streptomyces. The high trypsin, chymotrypsin, and total protease activities in mutant ΔsgiA (Fig. 5E), however, suggested that SgiA inhibited multiple serine proteases to various extents, thus lowering the overall protease activity in vivo.
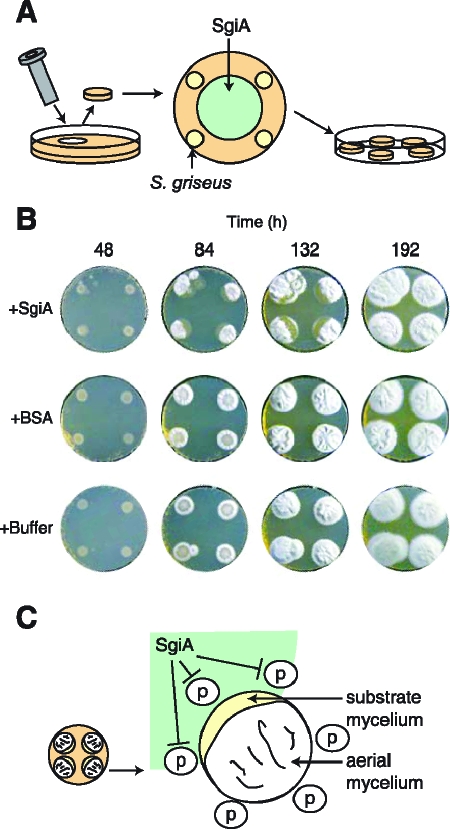
We supplied various amounts of the purified SgiA protein to growing mycelium at various timings to examine the possible effect of exogenously supplied SgiA on morphological development. An agar plug (diameter, 2.1 cm; height, 5 mm) was prepared from R2YE medium in a petri dish with a cork borer, on which S. griseus IFO13350 was inoculated at four spots with a toothpick (Fig. 7A). After 12 h of incubation at 28°C, a SgiA preparation (3.7 mg, 0.25 μmol) in Tris-HCl buffer was applied so that the edges of the colonies and the SgiA-containing spot did not overlap. As controls, equal volumes of the same buffer and the buffer containing bovine serum albumin (BSA) (3.7 mg) were applied similarly. Careful examination of the morphological development of the colonies revealed a delay in aerial hypha formation in a crescent-shaped region of a colony when SgiA was applied (Fig. 7B and C). No such delay was observed when BSA or the buffer was applied. During prolonged cultivation, the substrate mycelia in the inhibited region inoculated close to SgiA formed aerial hyphae and spores that were as abundant as the aerial hyphae and spores that were formed by substrate mycelia in the other region. The delay in aerial hypha formation caused by SgiA was also observed when colonies were formed on YMPD medium. Perhaps the delay in aerial hypha formation resulted from the inhibition of protein hydrolysis in the substrate mycelium by SgiA, which resulted in an insufficient supply of amino acids as nutrients for generation of aerial hyphae. During development of a Streptomyces colony, aerial hyphae emerge from the surface layer by reuse of material assimilated into the substrate hyphae in the underlying layer, such as DNA, proteins, and storage compounds (3, 22, 23, 40).
FIG. 7.
Inhibition of aerial hypha formation by SgiA exogenously applied to growing substrate mycelium. (A) Schematic diagram of inoculation of S. griseus on an R2YE agar plug (diameter, 2.1 cm; height, 5 mm) supplemented with purified SgiA. After S. griseus IFO13350 was inoculated at four spots with a toothpick and grown at 28°C for 12 h, the SgiA preparation was applied, as shown. (B) Morphogenesis of S. griseus cultures supplemented with SgiA (0.25 μmol, 3.7 mg), BSA (3.7 mg), and Tris-HCl buffer (pH 7.5). (C) Diagram of the crescent-like zone where no aerial hyphae are formed. This shape of inhibition zones was observed for S. griseus grown for 84 and 132 h in the presence of SgiA. p, proteases excreted from the substrate mycelium.
The crescent-like shape of the zone of aerial mycelium inhibition by SgiA and the eventual recovery of aerial growth (Fig. 7C) are consistent with the idea that the purpose of the protease activity is to recycle proteins from the substrate mycelium to provide nutrients for aerial growth. According to this model, nutrients could be supplied to the area of inhibition by diffusion from areas where protease activity has not been inhibited.
DISCUSSION
SgiA as a member of the AdpA regulon.
The present study demonstrated that AdpA directly triggers the transcription of sgiA, because (i) there was little sgiA transcription in the adpA mutant, (ii) AdpA bound the upstream activation site in front of its promoter, and (iii) a mutation in the upstream activation site severely impaired transcription of sgiA. SgiA begins to be produced at or just before the decision point when A-factor at a critical concentration switches the cell physiology from vegetative growth to differentiation conditions. The decision point is in the middle of the exponential growth phase (25), at which point the A-factor concentration reaches a critical level (1). Therefore, SgiA functions during and after the second exponential growth phase after the decision point.
The sgiA ortholog genes in S. albogriseolus (35), S. venezuelae (39), and S. coelicolor A3(2) (12) contain a sequence matching the AdpA-binding site at positions −70 to −80 (Fig. 3B). Their directions and distances with respect to the −35 and −10 promoter elements suggest that type II-a binding occurs for the sgiA promoters of S. albogriseolus and S. coelicolor A3(2) and that type II-b binding occurs for the sgiA promoters of S. griseus and S. venezuelae. An important implication of these findings is that the SSI genes that may be involved in morphological development in Streptomyces spp. are invariably under the control of AdpA, although the control of adpA differs in different strains (4).
Simultaneous control of extracellular protease activities by SSI and A-factor via AdpA.
We previously showed that two extracellular trypsin-type proteases, SprT and SprU (9), three chymotrypsin-type proteases, SprA, SprB, and SprD (37), and a metalloendopeptidase, SgmA (11), are under strict control of AdpA. Therefore, these proteases function during and after the second exponential growth phase after the decision point. Some of these proteases are presumably inhibited by SgiA to various extents. This means that the production of these proteases and their inhibitor protein are simultaneously triggered by A-factor. In addition, they localize extracellularly because they are released from the membrane by cleavage of their signal sequences, followed by a second cleavage by themselves or by some other protease for maturation.
What is the role of SSI in Streptomyces?
Many hydrolytic enzymes required for the degradation of cytoplasmic contents, such as proteases, nucleases, and lipases, are thought to be required during aerial mycelium formation, because the aerial mycelium reuses material that is first assimilated into the substrate mycelium (3, 22, 23, 40). Furthermore, several observations have suggested that there is a possible relationship between serine proteases and aerial mycelium formation. Kim et al. (14) and Nicieza et al. (26) observed that a trypsin inhibitor eliminated or impaired aerial mycelium formation in several Streptomyces spp. Kim and Hong (17) observed that addition of a serine protease inhibitor to S. griseus delayed aerial mycelium formation by 1 to 2 days. Kim and Lee (15, 16) concluded that a trypsin-like protein was involved in the lysis of substrate hyphae in Streptomyces exfoliatus by studying the trypsin in combination with its inhibitor, leupeptin, and a leupeptin-inactivating enzyme. In addition, Taguchi et al. (36) reported that in a mutant of S. albogriseolus deficient in SSI there was a marked decrease in sporulation ability. All these observations suggest, but still do not prove unambiguously, that there is a possible link between morphogenesis, proteases, and protease inhibitors.
When we think of the involvement of proteases in aerial mycelium formation and sporulation by lysing mycelium, the absence of morphological changes in a ΔsprT ΔsprU double mutant deficient in trypsin activity (9) or in a ΔsprA ΔsprB ΔsprD triple mutant deficient in chymotrypsin activity (37) can be explained by compensation by the remaining proteases for the defect in protease activity to the level required for normal morphogenesis. Conversely, an increase in protease activity in mutant ΔsgiA is not enough to cause obvious changes in morphogenesis or growth. A mutation in the SSI gene in S. coelicolor A3(2) does not result in any detectable morphological changes (10, 12). It is possible that on artificial agar medium in a petri dish under laboratory conditions minute regulation of morphogenesis by proteases and SSIs in Streptomyces is undetectable. However, exogenous supply of SgiA to growing substrate mycelium apparently delayed aerial hypha formation. A crescent-like area in which no aerial hyphae developed (Fig. 7) showed that SgiA inhibited aerial hypha formation. The shape of the inhibition area also suggested that amino acids that were nutrients diffusing from the lysed substrate hyphae into the surface layer of the SgiA-inhibited substrate hyphae permitted development of aerial hyphae. Thus, the importance of the SgiA-sensitive proteases in morphological development is apparent. However, the simultaneous control of the proteases and their inhibitor, SSI, by AdpA is a mystery to be evaluated in the future.
Kim et al. (12, 13) proposed a model for a cascade linking bldA (encoding the only tRNA for translation of the UUA leucine codon) to mycelial lysis in S. coelicolor A3(2) on the basis of the finding that the SSI gene is controlled by bldA via the TTA-containing adpA gene. Although the bld cascade in S. griseus is not well understood, bldA was shown to affect aerial mycelium formation in this species (2) and adpA contains a TTA leucine codon (29). A clear difference in activation of adpA between S. griseus and S. coelicolor A3(2) is that A-factor acts as a master switch in the former organism. S. griseus has accreted an extra signaling step involving A-factor/ArpA (4).
Acknowledgments
J. Kato was supported by the Japan Society for the Promotion of Science. This work was supported by Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from Monkasho and by the BioDesign Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan.
REFERENCES
- 1.Ando, N., K. Ueda, and S. Horinouchi. 1997. A Streptomyces griseus gene (sgaA) suppresses the growth disturbance caused by high osmolality and a high concentration of A-factor during early growth. Microbiology 143:2715-2723. [DOI] [PubMed] [Google Scholar]
- 2.Babcock, M. J., and K. E. Kendrick. 1988. Cloning of DNA involved in sporulation of Streptomyces griseus. J. Bacteriol. 170:2802-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braña, A. F., C. Méndez, L. A. Díaz, M. B. Manzanal, and C. Hardisson. 1986. Glycogen and trehalose accumulation during colony development in Streptomyces antibioticus. J. Gen. Microbiol. 132:1319-1326. [DOI] [PubMed] [Google Scholar]
- 4.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 5.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045-d2057. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 7.Ikenaka, T., S. Odani, M. Sakai, Y. Nabeshima, S. Sato, and S. Murao. 1974. Amino acid sequence of an alkaline proteinase inhibitor (Streptomyces subtilisin inhibitor) from Streptomyces albogriseolus S-3253. J. Biochem. 76:1191-1209. [DOI] [PubMed] [Google Scholar]
- 8.Inouye, K., B. Tonomura, K. Hiromi, S. Sato, and S. Murao. 1977. The stoichiometry of inhibition and binding of a protein proteinase inhibitor from Streptomyces (Streptomyces subtilisin inhibitor) against subtilisin BPN′. J. Biochem. 82:961-967. [DOI] [PubMed] [Google Scholar]
- 9.Kato, J., W.-J. Chi, Y. Ohnishi, S.-K. Hong, and S. Horinouchi. 2005. Transcriptional control by A-factor of two trypsin genes in Streptomyces griseus. J. Bacteriol. 187:286-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato, J., S. Hirano, Y. Ohnishi, and S. Horinouchi. 2005. The Streptomyces subtilisin inhibitor (SSI) gene in Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem. 69:1624-1629. [DOI] [PubMed] [Google Scholar]
- 11.Kato, J., A. Suzuki, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2002. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6016-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, D.-W., K. Chater, K.-J. Lee, and A. Hesketh. 2005. Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. J. Bacteriol. 187:2957-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, D. W., S. G. Kang, I. S. Kim, B. K. Lee, Y. T. Rho, and K. J. Lee. 2006. Proteases and protease inhibitors produced in streptomycetes and their roles in morphological differentiation. J. Microbiol. Biotechnol. 16:5-14. [Google Scholar]
- 14.Kim, I. S., S. G. Kang, and K. J. Lee. 1995. Physiological importance of trypsin-like protease during morphological differentiation of streptomycetes. J. Microbiol. 33:315-321. [Google Scholar]
- 15.Kim, I. S., and K. J. Lee. 1995. Physiological roles of leupeptin and extracellular proteases in mycelium development of Streptomyces exfoliatus SMF13. Microbiology 141:1017-1025. [DOI] [PubMed] [Google Scholar]
- 16.Kim, I. S., and K. J. Lee. 1996. Trypsin-like protease of Streptomyces exfoliatus SMF13, a potential agent in mycelial differentiation. Microbiology 142:1797-1806. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J.-M., and S.-K. Hong. 2000. Streptomyces griseus HH1, an A-factor deficient mutant, produces diminished level of trypsin and increased level of metalloproteases. J. Microbiol. 38:160-168. [Google Scholar]
- 18.Kojima, S., I. Kumagai, and K. Miura. 1993. Requirement for a disulfide bridge near the reactive site of protease inhibitor SSI (Streptomyces subtilisin inhibitor) for its inhibitory action. J. Mol. Biol. 230:395-399. [DOI] [PubMed] [Google Scholar]
- 19.Kojima, S., S. Obata, I. Kumagai, and K. Miura. 1990. Alteration of the specificity of the Streptomyces subtilisin inhibitor by gene engineering. Bio/Technology 8:449-452. [DOI] [PubMed] [Google Scholar]
- 20.Kojima, S., M. Terabe, S. Taguchi, H. Momose, and K. Miura. 1994. Primary structure and inhibitory properties of a proteinase inhibitor produced by Streptomyces cacaoi. Biochim. Biophys. Acta 1207:120-125. [DOI] [PubMed] [Google Scholar]
- 21.Kuramoto, A., A. Lezhava, S. Taguchi, H. Momose, and H. Kinashi. 1996. The location and deletion of the genes which code for SSI-like protease inhibitors in Streptomyces species. FEMS Microbiol. Lett. 139:37-42. [DOI] [PubMed] [Google Scholar]
- 22.Méndez, C., A. F. Braña, M. B. Manzanal, and C. Hardisson. 1985. Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol. 31:446-450. [DOI] [PubMed] [Google Scholar]
- 23.Miguélez, E. M., C. Hardisson, and M. B. Manzanal. 1999. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J. Cell Biol. 145:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura, K., I. Kumagai, S. Obata, S. Kojima, and S. Taguchi. 1988. Partial alteration of a protein Streptomyces subtilisin inhibitor by site-directed mutagenesis. Proc. Jpn. Acad. 64:147-149. [Google Scholar]
- 25.Neumann, T., W. Piepersberg, and J. Distler. 1996. Decision phase regulation of streptomycin production in Streptomyces griseus. Microbiology 142:1953-1963. [Google Scholar]
- 26.Nicieza, R. G., J. Huergo, B. A. Connolly, and J. Sanchez. 1999. Purification, characterization, and role of nucleases and serine proteases in Streptomyces differentiation. Analogies with the biochemical processes described in late steps of eukaryotic apoptosis. J. Biol. Chem. 274:20366-20375. [DOI] [PubMed] [Google Scholar]
- 27.Obata, S., S. Taguchi, I. Kumagai, and K. Miura. 1989. Molecular cloning and nucleotide sequence determination of gene encoding Streptomyces subtilisin inhibitor (SSI). J. Biochem. 105:367-371. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi, Y., and S. Horinouchi. 2004. The A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces. Biofilms 1:319-328. [DOI] [PubMed] [Google Scholar]
- 29.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi, Y., H. Yamazaki, J. Kato, A. Tomono, and S. Horinouchi. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69:431-439. [DOI] [PubMed] [Google Scholar]
- 31.Sakai, M., S. Odani, and T. Ikenaka. 1980. Importance of the carboxyl-terminal four amino acid residues in the inhibitory activity of Streptomyces subtilisin inhibitor (with a revision of its carboxyl-terminal sequence). J. Biochem. 87:891-898. [DOI] [PubMed] [Google Scholar]
- 32.Taguchi, S., S. Kojima, M. Terabe, Y. Kumazawa, H. Kohriyama, M. Suzuki, K. Miura, and H. Momose. 1997. Molecular phylogenetic characterization of Streptomyces protease inhibitor family. J. Mol. Evol. 44:542-551. [DOI] [PubMed] [Google Scholar]
- 33.Taguchi, S., S. Kojima, M. Terabe, K. Miura, and H. Momose. 1994. Comparative studies on the primary structures and inhibitory properties of subtilisin-trypsin inhibitors from Streptomyces. Eur. J. Biochem. 220:911-918. [DOI] [PubMed] [Google Scholar]
- 34.Taguchi, S., I. Kumagai, and K. Miura. 1990. Comparison of secretory expression in Escherichia coli and Streptomyces of Streptomyces subtilisin inhibitor (SSI) gene. Biochim. Biophys. Acta 1049:278-285. [DOI] [PubMed] [Google Scholar]
- 35.Taguchi, S., K. Nishiyama, I. Kumagai, and K. Miura. 1989. Analysis of transcriptional control regions in the Streptomyces subtilisin-inhibitor-encoding gene. Gene 84:279-286. [DOI] [PubMed] [Google Scholar]
- 36.Taguchi, S., A. Odaka, Y. Watanabe, and H. Momose. 1995. Molecular characterization of a gene encoding extracellular serine protease isolated from a subtilisin inhibitor-deficient mutant of Streptomyces albogriseolus S-3253. Appl. Environ. Microbiol. 61:180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomono, A., Y. Tsai, Y. Ohnishi, and S. Horinouchi. 2005. Three chymotrypsin genes are members of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. J. Bacteriol. 187:6341-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomono, A., Y. Tsai, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2005. Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 187:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Mellaert, L., E. Lammertyn, S. Schacht, P. Proost, J. Van Damme, B. Wroblowski, J. Anne, T. Scarcez, E. Sablon, J. Raeymaeckers, and A. Van Broekhoven. 1998. Molecular characterization of a novel subtilisin inhibitor protein produced by Streptomyces venezuelae CBS762.70. DNA Seq. 9:19-30. [DOI] [PubMed] [Google Scholar]
- 40.Wildermuth, H. 1970. Development and organization of the aerial mycelium in Streptomyces coelicolor. J. Gen. Microbiol. 60:43-50. [DOI] [PubMed] [Google Scholar]
- 41.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2003. Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamazaki, H., Y. Takano, Y. Ohnishi, and S. Horinouchi. 2003. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 50:1173-1187. [DOI] [PubMed] [Google Scholar]
- 44.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555-572. [DOI] [PubMed] [Google Scholar]