Abstract
The penultimate step in the biosynthesis of riboflavin (vitamin B2) involves the condensation of 3,4-dihydroxy-2-butanone 4-phosphate with 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione, which is catalyzed by 6,7-dimethyl-8-ribityllumazine synthase (lumazine synthase). Pathogenic Brucella species adapted to an intracellular lifestyle have two genes involved in riboflavin synthesis, ribH1 and ribH2, which are located on different chromosomes. The ribH2 gene was shown previously to specify a lumazine synthase (type II lumazine synthase) with an unusual decameric structure and a very high Km for 3,4-dihydroxy-2-butanone 4-phosphate. Moreover, the protein was found to be an immunodominant Brucella antigen and was able to generate strong humoral as well as cellular immunity against Brucella abortus in mice. We have now cloned and expressed the ribH1 gene, which is located inside a small riboflavin operon, together with two other putative riboflavin biosynthesis genes and the nusB gene, specifying an antitermination factor. The RibH1 protein (type I lumazine synthase) is a homopentamer catalyzing the formation of 6,7-dimethyl-8-ribityllumazine at a rate of 18 nmol mg−1 min−1. Sequence comparison of lumazine synthases from archaea, bacteria, plants, and fungi suggests a family of proteins comprising archaeal lumazine and riboflavin synthases, type I lumazine synthases, and the eubacterial type II lumazine synthases.
Vitamin B2 (riboflavin) (compound 6 [Fig. 1]) is the precursor of flavin mononucleotide and flavin adenine dinucleotide, essential cofactors for a wide variety of redox enzymes. Moreover, they are involved in numerous other physiological processes involving light sensing, bioluminescence, circadian time keeping, and DNA repair (for a review, see reference 39). The vitamin is biosynthesized by plants, fungi, and certain microorganisms but must be obtained from dietary sources and/or the intestinal flora by animals.
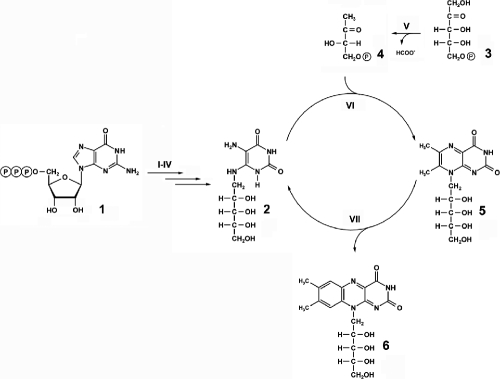
FIG. 1.
Biosynthesis of riboflavin in eubacteria. I, GTP cyclohydrolase II; II, 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate deaminase; III, 5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate reductase; IV, hypothetical phosphatase; V, 3,4-dihydroxy-2-butanone 4-phosphate synthase; VI, 6,7-dimethyl-8-ribityllumazine synthase; VII, riboflavin synthase; 1, GTP; 2, 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione; 3, ribulose 5′-phosphate; 4, 3,4-dihydroxy-2-butanone 4-phosphate; 5, 6,7-dimethyl-8-ribityllumazine; 6, riboflavin.
The pathways of riboflavin biosynthesis in microorganisms and plants have been reviewed recently (9, 10). The final steps are catalyzed by 6,7-dimethyl-8-ribityllumazine synthase (lumazine synthase [compound VI]) and riboflavin synthase (compound VII). More specifically, lumazine synthase catalyzes the condensation of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (substrate 2) with 3,4-dihydroxy-2-butanone 4-phosphate (substrate 4), resulting in the pteridine derivative, substrate 5 (Fig. 1).
Lumazine synthases from a variety of eubacteria (including Escherichia coli, Bacillus subtilis, Mycobacterium tuberculosis, and the hyperthermophile Aquifex aeolicus), archaea (Methanococcus jannaschii), fungi (Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Magnaporthe grisea), and a plant (spinach) have been studied in some detail (11, 16, 22, 32, 34, 35, 40-42, 44, 56, 57). The enzymes from fungi and from M. tuberculosis are C5-symmetric homopentamers, and the lumazine synthases of plants, most eubacteria, and archaea are 532-symmetric, hollow capsids, which are best described as dodecamers of pentamers. The subunit folds of these enzymes and the topology of the pentamer moieties are closely similar. The topologically equivalent active sites (5 in the case of the pentameric enzymes and 60 in the case of the icosahedral enzymes) are invariably located at interfaces between adjacent subunits in the pentamer moieties. Recently, it was found that pentameric riboflavin synthases of archaea are closely related to 6,7-dimethyl-8-ribityllumazine synthases (15, 25, 45).
Brucellosis is a disease of humans and livestock that is caused by closely related Brucella species adapted to intracellular life within the cells of a variety of mammals; the main pathogenic species for domestic animals are Brucella abortus, Brucella melitensis, and Brucella suis. Goldbaum and coworkers have shown that an 18-kDa B. abortus antigen with sequence similarity to lumazine synthases is a serological marker of active disease in human brucellosis patients (17, 19). Immunization with the protein has been shown to induce both cellular and humoral immune responses in mice. Moreover, the generation of protective immunity has also been observed in this model (2). Hence, the 18-kDa protein is of considerable immunologic interest and has been suggested to be a general carrier for the engineering of subunit vaccines (36). In preliminary enzymatic studies, this protein was shown to catalyze the formation of 6,7-dimethyl-8-ribityllumazine (compound 5), albeit at a low rate. Recently, the catalytic properties of this protein have been analyzed in closer detail (31). The three-dimensional structure of the 18-kDa antigen has been studied in considerable detail by Goldbaum and coworkers, and the protein has been reported to be a homodecamer which is best described as a D5-symmetric dimer of pentamers (31, 58). The 18-kDa antigen of B. abortus is the first lumazine synthase that has been reported to exhibit this quaternary arrangement.
Brucella spp. are known to share a genome topology characterized by two circular chromosomes with approximate sizes of 2.1 MDa (chromosome I) and 1.2 MDa (chromosome II). The recently published B. abortus genome sequence indicates that the 18-kDa antigen is specified by the ribH2 gene, located on chromosome II of B. abortus. Moreover, a second gene (designated ribH1) with sequence similarity to lumazine synthase was found on chromosome I.
This paper describes the identification, cloning, and expression of the ribH1 gene of B. abortus and provides a biochemical characterization of the encoded type I lumazine synthase. Based on this study, we suggest that the decameric protein encoded by the ribH2 gene be designated type II lumazine synthase. We also describe the genomic organization of the ribH1 and ribH2 genes and provide a phylogenetic analysis of lumazine synthases and related pentameric riboflavin synthases derived from different organisms.
MATERIALS AND METHODS
Materials.
5-Amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (substrate 2) and 6,7-dimethyl-8-ribityllumazine (substrate 5) were synthesized according to published procedures (1, 49). Recombinant 3,4-dihydroxy-2-butanone 4-phosphate synthase of E. coli was used for preparation of 3,4-dihydroxy-2-butanone 4-phosphate (substrate 4) (46).
Cloning and expression of the ribH1 gene of B. abortus.
The ribH1 gene was amplified by PCR using B. abortus chromosomal DNA as the template and the oligonucleotides RibH1-Ndel-Vo 5′-ATAATAATACATATGGAGTTTCTCATGTCCAAGCAC-3′ and RibH1-HindIII-Hi 5′-TATTATTATAAGCTTAGGCTCCGAATTTTTTGCGCAGGC-3′ as primers. The amplicon was digested with NdeI and HindIII. The resulting fragment was purified with an agarose gel extraction kit (QIAGEN, Hilden, Germany) and was then ligated to the plasmid pT7-7 (51), which had been treated with the same restriction enzymes. The ligation mixture was transformed into E. coli strain XL1-Blue (3), resulting in the recombinant strain XL-1-pT7-7-BARibH1. The reisolated plasmid was sequenced by the method of Sanger (47) (GATC Biotech, Konstanz, Germany) and was transformed into E. coli BL21(DE3) competent cells (50) (Stratagene, La Jolla, CA), resulting in the recombinant E. coli strain BL21(DE3)-pT7-7-BARibH1.
Bacterial culture.
E. coli BL21(DE3)-pT7-7-BARibH1 was grown to an optical density (at 600 nm) of 1.0 in LB medium containing 100 μg of ampicillin per ml at 37°C with shaking (150 rpm). An aliquot (5 ml) of this culture was diluted into 500 ml of medium, and incubation with shaking was continued to an optical density of 1.0. Isopropyl-β-thiogalactoside was added to a final concentration of 1 mM, and the suspension was incubated for 4 h at 37°C with shaking (150 rpm).
Purification of type I lumazine synthase of B. abortus.
Frozen bacterial cell mass was thawed in 50 mM potassium phosphate, pH 7.0. The suspension was ultrasonically treated and centrifuged. The supernatant was loaded on top of a Q-Sepharose column (1.8 by 25 cm) that had been equilibrated with 50 mM potassium phosphate, pH 7.0. The column was developed with a linear gradient of 0.05 to 1.0 M potassium phosphate, pH 7.0. Fractions were combined and concentrated by ultrafiltration. The solution was loaded on top of a Superdex-200 column (2.6 by 60 cm), which was developed with 100 mM potassium phosphate, pH 7.0. Fractions were combined and concentrated by ultrafiltration.
Assay of lumazine synthase activity.
Steady-state kinetic experiments were performed as described previously (12). Corrected initial rates were fitted to the Hill equation (equation 1) or equation 2 (see Results), which represents a classical substrate inhibition model, with the program package Origin or the program DynaFit, version 3.28.024 (33).
Analytical ultracentrifugation.
Experiments were performed with an analytical ultracentrifuge, Optima XL-A, from Beckman Instruments (Palo Alto, CA), equipped with absorbance and interference optics. Aluminum double-sector cells equipped with quartz windows were used throughout. The protein concentration was monitored photometrically at 280 nm. For boundary sedimentation experiments, a solution containing 100 mM potassium phosphate, pH 5.0, 300 mM sodium chloride, and 1.2 mg of protein per ml was centrifuged at 59,000 rpm and 20°C. Sedimentation equilibrium experiments were performed with a solution containing 100 mM potassium phosphate, pH 5.0, 300 mM sodium chloride, and 0.4 mg of protein per ml. Samples were centrifuged at 10,000 rpm and 4°C for 72 h. The partial specific volume was estimated from the amino acid composition, resulting in a value of 0.733 ml g−1 (37).
Phylogenetic analysis.
Sequences used for the phylogenetic analysis of lumazine synthase proteins were obtained from the HMM library, the genome assignment server Superfamily 1.69, and the NCBI database. The amino acid sequences were aligned with the ClustalX (version 1.81) program for multiple sequence alignment (53). Phylogenetic analysis of this alignment was inferred with the maximum-likelihood heuristic algorithm implemented by PHYML, version 2.4.4 (21), under the JTT substitution model (26). The reliability of tree nodes was analyzed by generating 1,000 bootstrap trees (8).
RESULTS
Genomic organization of B. abortus lumazine synthase genes.
The sequences of the complete genomes of B. abortus, B. suis, and B. melitensis were recently published (4, 23, 43) and show that the 18-kDa lumazine synthase antigen (type II lumazine synthase) is specified by the ribH2 gene, which is located on chromosome II. In all Brucella species analyzed, open reading frames (ORFs) with similarity to formate dehydrogenase (BMEII0588, BAb2-0534) and a sugar binding periplasmic precursor (BMEII0590) were found, and ORFs with unknown function surround the ribH2 gene (Fig. 2). This group of genes does not appear to have a common transcriptional control, as judged by analysis of the ORF orientation and promoter localization. Moreover, analysis of the 5′ untranslated region suggests that the ribH2 gene is under the regulatory control of a putative RFN element that is believed to sense the flavin mononucleotide concentration (54, 55).
FIG. 2.
Topology of riboflavin biosynthesis operons. Sequences were derived from fully sequenced genomes available from GenBank. Orientations of the genes involved in riboflavin biosynthesis or adjacent to those genes are given as arrows (not drawn to scale). Organisms (accession numbers) are as follows: Brucella meliloti 16 M chromosome I (NC 003317), Brucella suis 1330 chromosome I (NC 004310), Brucella abortus biovar 1 strain 9-941 chromosome I (NC 006932), Sinorhizobium meliloti 1021 (NC_003047), Mesorhizobium loti MAFF303099 (NC 002678), Bartonella henselae strain Houston-1 (NC 005956), Rhodopseudomonas palustris CGA009 (NC 005296), Bradyrhizobium japonicum USDA 110 (NC 004463), Caulobacter crescentus CB15 (NC_002696), Xanthomonas axonopodis pv. citri strain 306 (NC 003919), Xanthomonas oryzae KACC10331 (NC 006834), Xylella fastidiosa Temecula1 (NC_004556), Pseudomonas syringae pv. tomato strain DC3000 (NC_004578), Pseudomonas putida KT2440 (NC 002947), Methylococcus capsulatus strain Bath (NC 002977), Acinetobacter sp. ADP1 (NC 005966), Escherichia coli K-12 (NC_000913), Shigella sonnei Ss046 (NC 007384), Shigella flexneri 2a strain 301 (NC_004337), Salmonella enterica subsp. enterica serovar Choleraesuis strain SC-B67 (NC_006905), Yersinia pestis KIM (NC_004088), Photorhabdus luminescens subsp. laumondii TTO1 (NC 005126), Haemophilus influenzae 86-028NP (NC_007146), Geobacter sulfurreducens PCA (NC 002939), Aquifex aeolicus VF5 (NC 000918), Helicobacter pylori J99 (NC_000921), Candidatus (Blochmannia) floridanus (NC_005061), Buchnera aphidicola strain Sg (Schizaphis graminum) (NC_004061), Caulobacter crescentus CB15 (NC_002696), Ralstonia solanacearum GMI1000 (NC 003295), Chromobacterium violaceum ATCC 12472 (NC 005085), Azoarcus sp. EbN1 (NC 006513), Nitrosomonas europaea ATCC 19718 (NC 004757), Bordetella bronchiseptica RB50 (NC 002927), B. meliloti 16 M chromosome II (NC 003318), Brucella suis 1330 chromosome II (NC 004311), B. abortus biovar 1 strain 9-941 chromosome II (NC 006933), M. loti MAFF303099 (NC 002678), S. meliloti 1021 (NC_003047), R. palustris CGA009 (NC 005296), B. japonicum USDA 110 (NC 004463), P. syringae pv. tomato strain DC3000 (NC_004578).
Surprisingly, closer study of the three sequenced Brucella genomes revealed the presence of a second gene locus with sequence similarity to lumazine synthase genes. This gene is located on chromosome I and appears to be part of an operon-like gene arrangement (Fig. 2). This operon comprises four ORFs with sequence similarity to ribD (specifying a putative bifunctional deaminase/reductase), ribH (specifying a putative lumazine synthase), ribB (specifying a putative riboflavin synthase), and nusB (specifying a putative antitermination factor). The operon is preceded by a gene of unknown function, possibly a transcriptional regulator, with sequence similarity to the ytcG gene of B. subtilis. The predicted roles of these gene products in the riboflavin biosynthesis pathway are illustrated in Fig. 1. The B. abortus operon as a whole has similarity to the rib operons of various other eubacteria, most notably to an E. coli operon that has been studied in some detail (42, 46, 52).
Figure 2 shows the genomic localizations of ribH genes from a variety of organisms. Besides Brucella, other organisms (Mesorhizobium loti, Sinorhizobium meliloti, Rhodopseudomonas palustris, Bradyrhizobium japonicum, and Pseudomonas syringae) also have two different ORFs with sequence similarity to lumazine synthase (designated ribH1 and ribH2). This analysis could not detect any organism presenting only the ribH2 gene; moreover, the ribH2 gene was never found within an operon, with the exception of in Caulobacter crescentus. In the C. crescentus genome, we found a rib operon with a lumazine synthase resembling the ribH2 gene, whereas a gene with similarity to the ribH1 genes is located at a different site on the same chromosome; this putative ribH1 gene is probably not part of a larger transcription unit. Nine organisms, including Brucella, have shortened rib operons without ribA genes (bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase); isolated ribA genes are located elsewhere on chromosome I. Interestingly, Pseudomonas syringae harbors a ribA gene within a rib operon and another gene coding for an additional monofunctional GTP cyclohydrolase II. In Sinorhizobium meliloti, the genes ribD/ribB and ribH/nusB are separated by three ORFs with unknown functions and opposite directions spanning 2,555 bp. Buchnera aphidicola is the only organism in this study that has separate genes for deaminase and reductase (ribD1 and ribD2), though arranged in tandem.
In many cases, the genes in the respective operons overlap by a few base pairs. Notably, in almost all operons (32 of 33 analyzed genomes), the nusB gene appears downstream of the rib genes, in most cases followed by thiL, which specifies thiamine monophosphate kinase (Fig. 2). The rib genes are invariably oriented in the same direction as nusB and thiL, although the functional relevance of this association is still unknown.
The amino acid sequence identity between the type I and type II lumazine synthases of B. abortus is 21% (Fig. 3). Interestingly, both lumazine synthases have higher sequence similarities with other lumazine synthases, e.g., B. subtilis (type I, 33%; type II, 24%) and E. coli (type I, 37%; type II, 29%) than with each other.
FIG. 3.
Sequence alignment of lumazine synthases from eubacteria and yeast and an archaeal riboflavin synthase. BabRibH2, type II lumazine synthase of B. abortus (P61711); BabRibH1, type I lumazine synthase of B. abortus (Q57DY1); BsuLS, lumazine synthase of Bacillus subtilis (P11998); AaeLS, lumazine synthase of Aquifex aeolicus (O66529); SpoLS, lumazine synthase of Schizosaccharomyces pombe (Q9UUB1); MjaRS, riboflavin synthase of Methanocaldococcus jannaschii (Q58584). Identical residues are printed in white on black, and similar residues are shaded in gray.
Quaternary structure of the type I lumazine synthase of B. abortus.
For a detailed characterization, the ribH1 gene of B. abortus was cloned into an expression plasmid under the control of a T7 RNA polymerase promoter and lacZ operator. In a recombinant E. coli strain, the expression construct directed the formation of a 17-kDa protein, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). The recombinant protein was purified to apparent homogeneity by anion-exchange chromatography, followed by gel permeation chromatography. Edman degradation of the N terminus of the recombinant protein resulted in the sequence MEFLMSKHEADA, in agreement with the translated ORF (Fig. 3), thus confirming the integrity of the protein sequence at the N terminus.
The type I lumazine synthase of B. abortus sediments at an apparent velocity of 5.9S at 20°C (Fig. 4A). In comparison, it should be noted that pentameric lumazine synthases from the yeasts S. cerevisiae and S. pombe have similar apparent sedimentation coefficients, of 5.5S and 5.0S, respectively (11, 42). Sedimentation equilibrium experiments indicated a molecular mass of 88 kDa, with an ideal monodisperse model used for calculation (Fig. 4B). The calculated subunit molecular mass of 17,599 Da indicates a pentameric mass of 88 kDa, in excellent agreement with the experimental data.
FIG. 4.
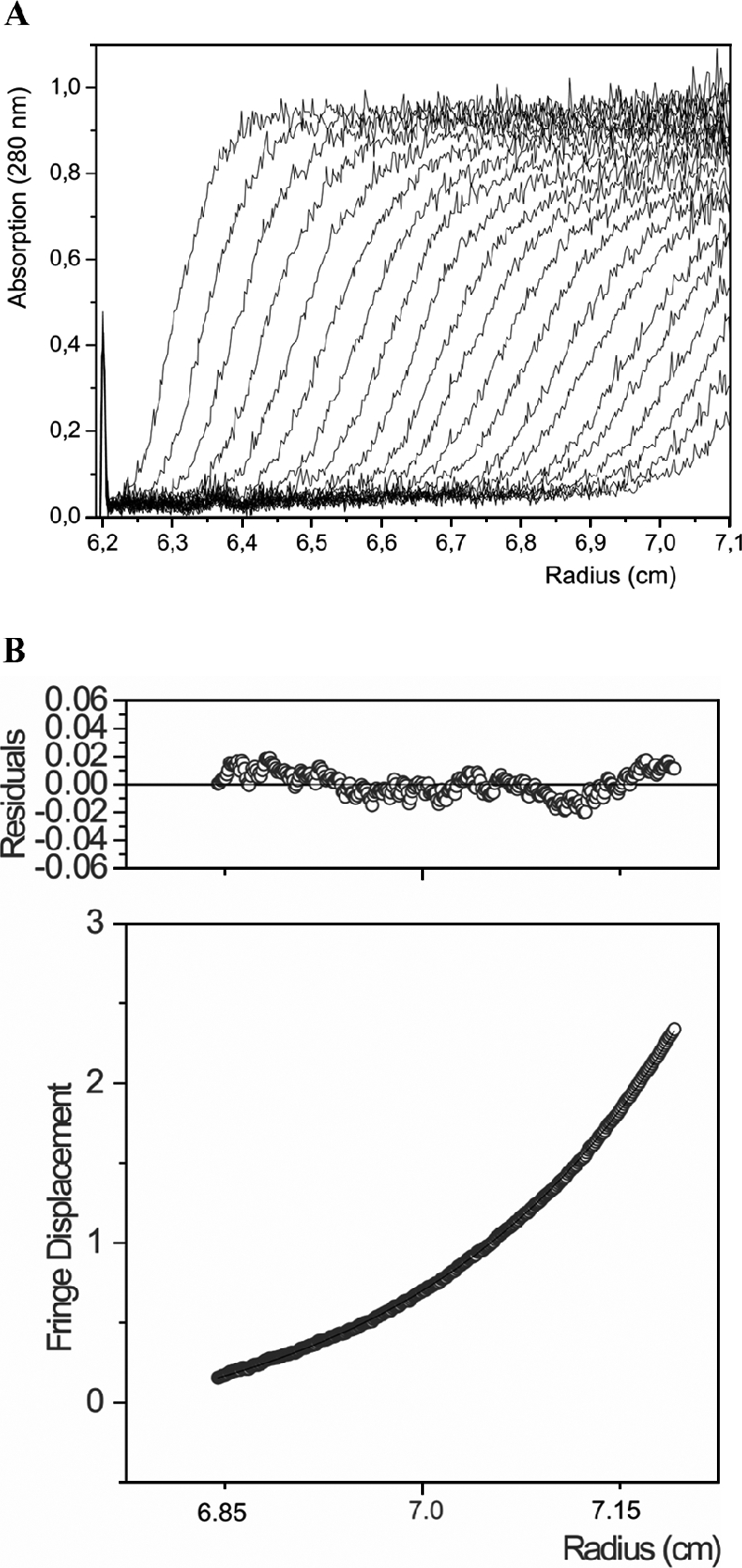
Analytical ultracentrifugation analysis of the type I lumazine synthase of B. abortus. (A) For boundary sedimentation centrifugation analysis, a solution containing 100 mM potassium phosphate, pH 5.0, 300 mM sodium chloride, and 1.2 mg of protein per ml was centrifuged at 59,000 rpm and 20°C for 3 h. The protein concentration was monitored photometrically (280 nm) at intervals of 5 min. (B) For sedimentation equilibrium centrifugation analysis, a solution containing 100 mM potassium phosphate, pH 5.0, 300 mM sodium chloride, and 0.4 mg of protein per ml was centrifuged at 10,000 rpm and 4°C for 72 h.
Functional characterization of the type I lumazine synthase of B. abortus.
Enzymatic studies show that the type I lumazine synthase catalyzes the formation of 6,7-dimethyl-8-ribityllumazine (substrate 5) at a rate of about 18 ± 2 nmol mg−1 min−1 at 37°C (Table 1). Notably, this value is about 1 order of magnitude below the catalytic rates of lumazine synthases from other mesophilic microorganisms and from spinach (Table 1). The kinetic data for the substrate, 3,4-dihydroxy-2-butanone 4-phosphate (substrate 4), were fitted to the Hill equation (equation 1),
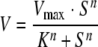 |
(1) |
where V is the reaction velocity, Vmax is the maximum reaction velocity, S is the substrate concentration, K is the reaction constant, and n is the Hill coefficient, resulting in a K value of 125 ± 10 μM for substrate 4.
TABLE 1.
Properties of lumazine synthases
| Origin |
Km (μM)a
|
Vmax at 37°C (nmol mg−1 min−1) | Sedimentation velocity (S) | Source or reference | |
|---|---|---|---|---|---|
| Substrate 4 | Substrate 2 | ||||
| B. subtilis | 55 ± 5 | 9.0 ± 1 | 242 ± 6 | 26.5 | 29 |
| E. coli | 62 | 4.2 | 197 | 26.8 | 42 |
| S. cerevisiae | 90 | 4.0 | 257 | 5.5 | 42 |
| S. pombe | 67 | 5.0 | 217 | 5.0 | 11 |
| S. oleracea | 26 ± 3 | 20.0 ± 2 | 275 | 27 | |
| B. abortus | |||||
| Type I lumazine synthase | 125 ± 10* | 90 ± 16 | 18 ± 2 | 5.9 | This study |
| Type II lumazine synthase | 4,000 | 10 | 20 ± 3 | 31 | |
Substrate 4, 3,4-dihydroxy-2-butanone 4-phosphate; substrate 2, 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione. *, Hill coefficient (n = 2 ± 0.3).
Steady-state kinetic experiments showed a decreasing reaction velocity at high concentrations of the substrate 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione. The data could be best approximated with the model represented by equation 2, indicating a Km of 90 ± 16 μM and an inhibition constant of 370 ± 70 μM (Table 1):
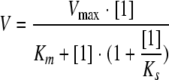 |
(2) |
where V is the reaction velocity, Vmax is the maximum reaction velocity, S is the concentration of substrate 2, Km is the Michaelis-Menten constant, and KS is the substrate inhibition constant.
As shown in Table 1, the affinity for 3,4-dihydroxy-2-butanone 4-phosphate (substrate 4) is in the same range as the values reported for other lumazine synthases. In contrast, the Km value for substrate 2 is about 10 times higher than that observed for other orthologs.
Table 2 summarizes the genomic localization and the structural and biochemical characterization of B. abortus type I and type II lumazine synthases.
TABLE 2.
Summary of genomic, structural, and biochemical knowledge about B. abortus lumazine synthases
Structure-based phylogenetic analysis of lumazine synthases.
We conducted a phylogenetic analysis comparing atypical, duplicated genes of archaea and bacteria with structurally characterized pentameric and icosahedral lumazine synthases. From this analysis, depicted in Fig. 5, a completely new picture emerges, showing that in addition to eubacterial type I lumazine synthases and archaeal lumazine synthases, this folding gave origin to at least two new evolutionarily related functions: the archaeal riboflavin synthases, which are lumazine synthase-like riboflavin synthases with no detectable lumazine synthase activity (13, 15, 45), and the eubacterial type II lumazine synthases.
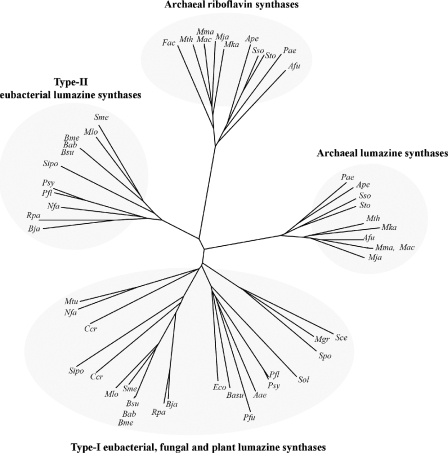
FIG. 5.
Phylogenetic analysis of the quaternary arrangements of lumazine synthases or related pentameric riboflavin synthases derived from different organisms, showing a distribution into four clearly defined branches: (i) type II lumazine synthases (decameric), (ii) archaeal pentameric riboflavin synthases, (iii) archaeal lumazine synthases, and (iv) type I eubacterial, fungal, and plant lumazine synthases. The corresponding organisms are indicated as follows (references describing the structures of the enzymes involved in this study are given in brackets): Aae, A. aeolicus (lumazine synthase [56, 57]); Afu, Archaeoglobus fulgidus; Ape, Aeropyrum pernix; Bab, B. abortus; Basu, B. subtilis (lumazine synthase [34, 35]); Bja, Bradyrhizobium japonicum; Bme, B. melitensis; Bsu, B. suis (RibH2 protein [58]); Ccr, Caulobacter crescentus; Eco, E. coli (lumazine synthase [42]); Fac, Ferroplasma acidarmanus; Mac, Methanosarcina acetivorans; Mgr, M. grisea (lumazine synthase [44]); Mja, Methanococcus jannaschii (riboflavin synthase [15, 45], lumazine synthase [22]); Mka, Methanopyrus kandleri; Mlo, M. loti; Mma, Methanosarcina mazei; Mth, Methanobacterium thermoautotrophicum; Mtu, M. tuberculosis (lumazine synthase [41]); Nfa, Nocardia farcinica; Pae, Pyrobaculum aerophilum; Pfl, Pseudomonas fluorescens; Pfu, Pyrococcus furiosus; Psy, P. syringae; Rpa, R. palustris; Sce, S. cerevisiae (lumazine synthase [40]); Sipo, Silicibacter pomeroyi; Sme, S. meliloti; Sol, S. oleracea (lumazine synthase [44]); Spo, S. pombe (lumazine synthase [16, 32]); Sso, Sulfolobus solfataricus; Sto, Sulfolobus tokodaii.
Eubacteria that harbor a ribH2 gene have a type I lumazine synthase of apparent pentameric arrangement, with the exception of that of Pseudomonas syringae, which is clustered with the icosahedral lumazine synthases. In addition, pentameric type I lumazine synthases from some of the α-Proteobacteria (Brucella, Rhizobium, and Rhodobacter) diverge from pentameric fungal and yeast lumazine synthases (S. cerevisiae, S. pombe, and M. grisea).
DISCUSSION
Genomic analysis of Brucella identified two related ORFs (ribH1 and ribH2) coding for two proteins designated type I and type II lumazine synthases, respectively. The similarity between the type I and type II lumazine synthases, which are located on two different chromosomes, is quite low (Fig. 3). Hence, we must assume that either the separation of both lumazine synthase genes occurred very early in evolution or one of the ribH genes was acquired prior to α-proteobacterial speciation by lateral gene transfer. At this time, there is no experimental evidence that would permit a decision.
Recent studies have shown the existence of a family of pentameric lumazine synthase-like riboflavin synthases (Fig. 5 and 6). These enzymes have been found exclusively in archaea, which have also been shown to be devoid of riboflavin synthases of the trimeric eubacterium/yeast/plant type. The cavity harboring the active site of the pentameric riboflavin synthase of M. jannaschii is similar to that of the lumazine synthases. The binding mode of the acceptor lumazine molecule in pentameric riboflavin synthase closely resembles the pyrimidinedione-binding site of the lumazine synthases (Fig. 6). Notably, these archaeal riboflavin synthases are evolutionarily old and have no detectable lumazine synthase activity (13, 15, 45). Completely sequenced archaeal genomes typically comprise sets of two similar genes, coding for a lumazine synthase-like riboflavin synthase and a regular lumazine synthase. Interestingly, present-day lumazine synthases have retained the capacity to bind riboflavin. For example, the lumazine synthase of the yeast S. pombe is yellow colored due to the presence of a tightly bound riboflavin (11).
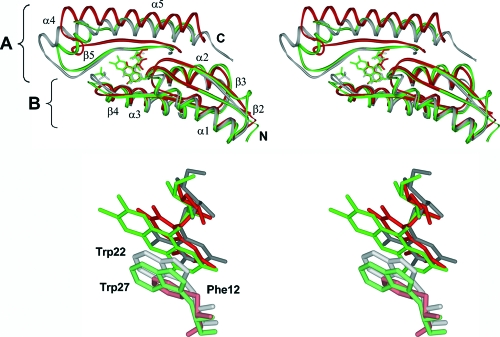
FIG. 6.
Structural comparison of type I and type II lumazine synthases and archaeal riboflavin synthase. (Top) Structural superposition of riboflavin synthase from M. jannaschii (MjaRS; PDB entry code 2B99 [45]), type I lumazine synthase from S. pombe (SpoLS; PDB entry code 1KYZ [16]), and type II lumazine synthase from B. abortus (BabRibH2; PDB entry code 1T13 [31]). The active sites are formed by two adjacent monomers of SpoLS with bound riboflavin (green; residues E17 to D112 and S113 to L158), BabRibH2 with bound 5-nitro-6-(d-ribitylamino)-2,4(1H,3H)-pyrimidinedione (gray; residues S12 to E106 and T107 to L156), and MjaRS with bound 6,7-dioxo-8-ribityllumazine, resembling the acceptor lumazine molecule (red; residues T2 to M90 and T91 to Y135). Secondary structure element labeling refers to SpoLS. The five topologically equivalent active sites of pentameric lumazine synthases are located at the interfaces between adjacent monomers of the pentamer; two of them (A and B) are shown. Ligands are drawn in the respective colors. (Bottom) Enlarged ligand-binding sites of all three enzymes, with Trp27 from SpoLS, Trp22 from BabRibH2, and Phe12 from MjaRS.
The situation in the archaea is comparable to that in the eubacteria, with type I and type II lumazine synthases as we describe in the present work (Fig. 5). However, there is no evidence that either type I or type II lumazine synthase has any riboflavin synthase activity (results not shown).
It should be noted that the eubacterial type I lumazine synthases that have been reported in the literature have low catalytic activities, in the range of about 200 to 300 nmol mg−1 min−1 when assayed near the optimum growth temperature of the cognate species (Table 1). By comparison with these data, the activity of the B. abortus type I lumazine synthase is more than 10-fold lower.
The generally low catalytic activity of lumazine synthases does not result from a particularly large free energy barrier of the reaction catalyzed. Quite to the contrary, the condensation of 3,4-dihydroxy-2-butanone 4-phosphate (substrate 4) and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (substrate 2) is characterized by such a low energy barrier that it can proceed at an appreciable rate at room temperature in aqueous solution and at neutral pH (30). Thus, it is mandatory that blank assays without enzyme be run in order to correct for the contribution of the uncatalyzed reaction in lumazine synthase activity measurements (30). In the case of the lumazine synthase of B. abortus, with its particularly low catalytic rate, correction for the contribution of the uncatalyzed reaction becomes most important.
The low catalytic activities of the lumazine synthases are in no way unique among the enzymes of the riboflavin pathway. All activity values for the entire riboflavin pathways in E. coli, B. subtilis, and yeast are in the range of nanomoles per milligram per minute (5-7, 11, 12, 14, 20, 24, 28, 38, 46, 48). Since, at least in the case of lumazine synthase and riboflavin synthase, the inherent free energy barriers of the catalyzed reactions cannot be the reason for these low rates, we must assume, for lack of other arguments, that the selective pressure controlling this pathway favors the evolution of catalysts with low reaction rates. In fact, riboflavin is required in only small amounts, and excess production would unnecessarily deplete the precursor pools. Still, it remains an open question why the catalytic activity of the B. abortus type I enzyme is at the lower end of all documented lumazine synthases.
An earlier study showed that the decameric arrangement of type II B. abortus lumazine synthase is related to a very high Km for 3,4-dihydroxy-2-butanone 4-phosphate (substrate 4) (31). The in vivo concentration of 3,4-dihydroxy-2-butanone 4-phosphate is unknown. However, unless we assume that it is in the same numerical range as the Km of the type II enzyme, we must assume that the bulk of the 6,7-dimethyl-8-ribityllumazine (substrate 6) would be generated by the enzyme with the lower Km value, i.e., the type I lumazine synthase, whereas the type II enzyme could at best supply a minor amount of the overall riboflavin production.
Thus, the question of which selective pressures could have prevented the loss of the ribH2 gene (in case both lumazine synthase genes are evolutionarily old in Brucella-related organisms) or could have favored its more recent acquisition by horizontal gene transfer is still open. Interestingly, the type II lumazine synthase is an immunodominant antigen of B. abortus, and there is unpublished evidence that links this protein to Brucella virulence, suggesting that the type II lumazine synthase has evolved for a new, as-yet-unknown function.
Acknowledgments
This work was supported by a Howard Hughes Medical Institute international grant (to F.A.G.) and by a grant from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) República Argentina. F.A.G. is a member of the research career of CONICET, and V.Z. and S.K. are recipients of a fellowship from CONICET. This work was supported by the Fonds der Chemischen Industrie and the Hans-Fischer-Gesellschaft eV. M.F. and F.A.G. acknowledge support for exchange visits between laboratories by the BMBF and SECyT (project ARG 04/Z06).
We acknowledge R. Ugalde, D. Comerci, and Ines Marchesini for genomic B. abortus DNA and for early genomic analysis and Diana Posadas for helping us in the phylogenetic analysis.
REFERENCES
- 1.Bacher, A. 1986. Heavy riboflavin synthase from Bacillus subtilis. Methods Enzymol. 122:192-199. [DOI] [PubMed] [Google Scholar]
- 2.Berguer, P. M., J. Mundinano, I. Piazzon, and F. A. Goldbaum. 2006. A polymeric bacterial protein activates dendritic cells via TLR4. J. Immunol. 176:2366-2372. [DOI] [PubMed] [Google Scholar]
- 3.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strains with beta-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 4.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberhardt, S., S. Korn, F. Lottspeich, and A. Bacher. 1997. Biosynthesis of riboflavin: an unusual riboflavin synthase of Methanobacterium thermoautotrophicum. J. Bacteriol. 179:2938-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberhardt, S., G. Richter, W. Gimbel, T. Werner, and A. Bacher. 1996. Cloning, sequencing, mapping and hyperexpression of the ribC gene coding for riboflavin synthase of Escherichia coli. Eur. J. Biochem. 242:712-719. [DOI] [PubMed] [Google Scholar]
- 7.Echt, S., S. Bauer, S. Steinbacher, R. Huber, A. Bacher, and M. Fischer. 2004. Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans. J. Mol. Biol. 341:1085-1096. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1987. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 12:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, M., and A. Bacher. 2005. Biosynthesis of flavocoenzymes. Nat. Prod. Rep. 22:324-350. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, M., and A. Bacher. 2006. Biosynthesis of Vitamin B2 in Plants. Physiol. Plant. 126:304-318. [Google Scholar]
- 11.Fischer, M., I. Haase, R. Feicht, G. Richter, S. Gerhardt, J. P. Changeux, R. Huber, and A. Bacher. 2002. Biosynthesis of riboflavin: 6,7-dimethyl-8-ribityllumazine synthase of Schizosaccharomyces pombe. Eur. J. Biochem. 269:519-526. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, M., I. Haase, K. Kis, W. Meining, R. Ladenstein, M. Cushman, N. Schramek, R. Huber, and A. Bacher. 2003. Enzyme catalysis via control of activation entropy: site-directed mutagenesis of 6,7-dimethyl-8-ribityllumazine synthase. J. Mol. Biol. 326:783-793. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, M., W. Romisch, B. Illarionov, W. Eisenreich, and A. Bacher. 2005. Structures and reaction mechanisms of riboflavin synthases of eubacterial and archaeal origin. Biochem. Soc. Trans. 33:780-784. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, M., W. Romisch, S. Saller, B. Illarionov, G. Richter, F. Rohdich, W. Eisenreich, and A. Bacher. 2004. Evolution of vitamin B2 biosynthesis: structural and functional similarity between pyrimidine deaminases of eubacterial and plant origin. J. Biol. Chem. 279:36299-36308. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, M., A. K. Schott, W. Romisch, A. Ramsperger, M. Augustin, A. Fidler, A. Bacher, G. Richter, R. Huber, and W. Eisenreich. 2004. Evolution of vitamin B2 biosynthesis. A novel class of riboflavin synthase in Archaea. J. Mol. Biol. 343:267-278. [DOI] [PubMed] [Google Scholar]
- 16.Gerhardt, S., I. Haase, S. Steinbacher, J. T. Kaiser, M. Cushman, A. Bacher, R. Huber, and M. Fischer. 2002. The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase. J. Mol. Biol. 318:1317-1329. [DOI] [PubMed] [Google Scholar]
- 17.Goldbaum, F. A., J. Leoni, J. C. Wallach, and C. A. Fossati. 1993. Characterization of an 18-kilodalton Brucella cytoplasmic protein which appears to be a serological marker of active infection of both human and bovine brucellosis. J. Clin. Microbiol. 31:2141-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldbaum, F. A., I. Polikarpov, A. A. Cauerhff, C. A. Velikovsky, B. C. Braden, and R. J. Poljak. 1998. Crystallization and preliminary x-ray diffraction analysis of the lumazine synthase from Brucella abortus. J. Struct. Biol. 123:175-178. [DOI] [PubMed] [Google Scholar]
- 19.Goldbaum, F. A., C. A. Velikovsky, P. C. Baldi, S. Mortl, A. Bacher, and C. A. Fossati. 1999. The 18-kDa cytoplasmic protein of Brucella species—an antigen useful for diagnosis—is a lumazine synthase. J. Med. Microbiol. 48:833-839. [DOI] [PubMed] [Google Scholar]
- 20.Graupner, M., H. Xu, and R. H. White. 2002. The pyrimidine nucleotide reductase step in riboflavin and F420 biosynthesis in Archaea proceeds by the eukaryotic route to riboflavin. J. Bacteriol. 184:1952-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 22.Haase, I., S. Mortl, P. Kohler, A. Bacher, and M. Fischer. 2003. Biosynthesis of riboflavin in archaea. 6,7-Dimethyl-8-ribityllumazine synthase of Methanococcus jannaschii. Eur. J. Biochem. 270:1025-1032. [DOI] [PubMed] [Google Scholar]
- 23.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 187:2715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey, R. A., and G. W. Plaut. 1966. Riboflavin synthetase from yeast. Properties of complexes of the enzyme with lumazine derivatives and riboflavin. J. Biol. Chem. 241:2120-2136. [PubMed] [Google Scholar]
- 25.Illarionov, B., W. Eisenreich, N. Schramek, A. Bacher, and M. Fischer. 2005. Biosynthesis of vitamin B2: diastereomeric reaction intermediates of archaeal and non-archaeal riboflavin synthases. J. Biol. Chem. 280:28541-28546. [DOI] [PubMed] [Google Scholar]
- 26.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 27.Jordan, D. B., K. O. Bacot, T. J. Carlson, M. Kessel, and P. V. Viitanen. 1999. Plant riboflavin biosynthesis. Cloning, chloroplast localization, expression, purification, and partial characterization of spinach lumazine synthase. J. Biol. Chem. 274:22114-22121. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser, J., N. Schramek, S. Eberhardt, S. Puttmer, M. Schuster, and A. Bacher. 2002. Biosynthesis of vitamin B2. Eur. J. Biochem. 269:5264-5270. [DOI] [PubMed] [Google Scholar]
- 29.Kis, K., and A. Bacher. 1995. Substrate channeling in the lumazine synthase/riboflavin synthase complex of Bacillus subtilis. J. Biol. Chem. 270:16788-16795. [DOI] [PubMed] [Google Scholar]
- 30.Kis, K., K. Kugelbrey, and A. Bacher. 2001. Biosynthesis of riboflavin. The reaction catalyzed by 6,7-dimethyl-8-ribityllumazine synthase can proceed without enzymatic catalysis under physiological conditions. J. Org. Chem. 66:2555-2559. [DOI] [PubMed] [Google Scholar]
- 31.Klinke, S., V. Zylberman, D. R. Vega, B. G. Guimaraes, B. C. Braden, and F. A. Goldbaum. 2005. Crystallographic studies on decameric Brucella spp. Lumazine synthase: a novel quaternary arrangement evolved for a new function? J. Mol. Biol. 353:124-137. [DOI] [PubMed] [Google Scholar]
- 32.Koch, M., C. Breithaupt, S. GerhardtHaase, S. Weber, M. Cushman, R. Huber, A. Bacher, and M. Fischer. 2004. Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase. Eur. J. Biochem. 271:3208-3214. [DOI] [PubMed] [Google Scholar]
- 33.Kuzmic, P. 1996. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237:260-273. [DOI] [PubMed] [Google Scholar]
- 34.Ladenstein, R., K. Ritsert, R. Huber, G. Richter, and A. Bacher. 1994. The lumazine synthase/riboflavin synthase complex of Bacillus subtilis. X-ray structure analysis of hollow reconstituted beta-subunit capsids. Eur. J. Biochem. 223:1007-1017. [DOI] [PubMed] [Google Scholar]
- 35.Ladenstein, R., M. Schneider, R. Huber, H. D. Bartunik, K. Wilson, K. Schott, and A. Bacher. 1988. Heavy riboflavin synthase from Bacillus subtilis. Crystal structure analysis of the icosahedral beta 60 capsid at 3.3 A resolution. J. Mol. Biol. 203:1045-1070. [DOI] [PubMed] [Google Scholar]
- 36.Laplagne, D. A., V. Zylberman, N. Ainciart, M. W. Steward, E. Sciutto, C. A. Fossati, and F. A. Goldbaum. 2004. Engineering of a polymeric bacterial protein as a scaffold for the multiple display of peptides. Proteins 57:820-828. [DOI] [PubMed] [Google Scholar]
- 37.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 38.Liao, D. I., P. V. Viitanen, and D. B. Jordan. 2000. Cloning, expression, purification and crystallization of dihydroxybutanone phosphate synthase from Magnaporthe grisea. Acta Crystallogr. D 56:1495-1497. [DOI] [PubMed] [Google Scholar]
- 39.Massey, V. 2000. The chemical and biological versatility of riboflavin. Biochem. Soc. Trans. 28:283-296. [PubMed] [Google Scholar]
- 40.Meining, W., S. Mortl, M. Fischer, M. Cushman, A. Bacher, and R. Ladenstein. 2000. The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue. J. Mol. Biol. 299:181-197. [DOI] [PubMed] [Google Scholar]
- 41.Morgunova, E., W. Meining, B. Illarionov, I. Haase, G. Jin, A. Bacher, M. Cushman, M. Fischer, and R. Ladenstein. 2005. Crystal structure of lumazine synthase from Mycobacterium tuberculosis as a target for rational drug design: binding mode of a new class of purinetrione inhibitors. Biochemistry 44:2746-2758. [DOI] [PubMed] [Google Scholar]
- 42.Mortl, S., M. Fischer, G. Richter, J. Tack, S. Weinkauf, and A. Bacher. 1996. Biosynthesis of riboflavin. Lumazine synthase of Escherichia coli. J. Biol. Chem. 271:33201-33207. [DOI] [PubMed] [Google Scholar]
- 43.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persson, K., G. Schneider, D. B. Jordan, P. V. Viitanen, and T. Sandalova. 1999. Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly. Protein Sci. 8:2355-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramsperger, A., M. Augustin, A. K. Schott, S. Gerhardt, T. Krojer, W. Eisenreich, B. Illarionov, M. Cushman, A. Bacher, R. Huber, and M. Fischer. 2006. Crystal structure of an archaeal pentameric riboflavin synthase in complex with a substrate analog inhibitor: stereochemical implications. J. Biol. Chem. 281:1224-1232. [DOI] [PubMed] [Google Scholar]
- 46.Richter, G., R. Volk, C. Krieger, H. W. Lahm, U. Rothlisberger, and A. Bacher. 1992. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J. Bacteriol. 174:4050-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheuring, J., M. Fischer, M. Cushman, J. Lee, A. Bacher, and H. Oschkinat. 1996. NMR analysis of site-specific ligand binding in oligomeric proteins. Dynamic studies on the interaction of riboflavin synthase with trifluoromethyl-substituted intermediates. Biochemistry 35:9637-9646. [DOI] [PubMed] [Google Scholar]
- 49.Sedlmaier, H., F. Muller, P. J. Keller, and A. Bacher. 1987. Enzymatic synthesis of riboflavin and FMN specifically labeled with 13C in the xylene ring. Z. Naturforsch. C 42:425-429. [DOI] [PubMed] [Google Scholar]
- 50.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 51.Tabor, S., and C. C. Richardson. 1990. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Effect of pyrophosphorolysis and metal ions. J. Biol. Chem. 265:8322-8328. [PubMed] [Google Scholar]
- 52.Taura, T., C. Ueguchi, K. Shiba, and K. Ito. 1992. Insertional disruption of the nusB (ssyB) gene leads to cold-sensitive growth of Escherichia coli and suppression of the secY24 mutation. Mol. Gen. Genet. 234:429-432. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkler, W. C., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 99:15908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, X., W. Meining, M. Cushman, I. Haase, M. Fischer, A. Bacher, and R. Ladenstein. 2003. A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus. J. Mol. Biol. 328:167-182. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, X., W. Meining, M. Fischer, A. Bacher, and R. Ladenstein. 2001. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 306:1099-1114. [DOI] [PubMed] [Google Scholar]
- 58.Zylberman, V., P. O. Craig, S. Klinke, B. C. Braden, A. Cauerhff, and F. A. Goldbaum. 2004. High order quaternary arrangement confers increased structural stability to Brucella sp. lumazine synthase. J. Biol. Chem. 279:8093-8101. [DOI] [PubMed] [Google Scholar]