Abstract
Sinorhizobium meliloti uses proline betaine (PB) as an osmoprotectant when osmotically stressed and as an energy source in low-osmolarity environments. To fulfill this dual function, two separate PB transporters, BetS and Hut, that contribute to PB uptake at high and low osmolarity, respectively, have been previously identified. Here, we characterized a novel transport system that mediates the uptake of PB at both high and low osmolarities. Sequence analysis of Tn5-luxAB chromosomal insertions from several PB-inducible mutants has revealed the presence of a four-gene locus encoding the components of an ABC transporter, Prb, which belongs to the oligopeptide permease (Opp) family. Surprisingly, prb mutants were impaired in their ability to transport PB, and oligopeptides were not shown to be competitors for PB uptake. Further analysis of Prb specificity has shown its ability to take up other quaternary ammonium compounds such as choline and, to a lesser extent, glycine betaine. Interestingly, salt stress and PB were found to control prb expression in a positive and synergistic way and to increase Prb transport activity. At low osmolarity, Prb is largely implicated in PB uptake by stationary-phase cells, likely to provide PB as a source of carbon and nitrogen. Furthermore, at high osmolarity, the analysis of prb and betS single and double mutants demonstrated that Prb, together with BetS, is a key system for protection by PB.
Bacteria have to cope with variations in their environment and particularly with changes in the external osmolarity. They protect themselves against high external osmolarity by accumulating osmoprotective organic compounds in their cytoplasm, the so-called compatible solutes (10). These usually nonionic, highly water-soluble molecules can be accumulated to very high levels in the cytoplasm of stressed cells without interfering with vital cellular functions. They correspond to a limited number of compounds including sugars (trehalose), free amino acids (e.g., glutamate and proline), and quaternary ammonium compounds (e.g., glycine betaine, proline betaine [PB], γ-butyrobetaine, and carnitine) that bacteria accumulate from de novo synthesis or from externally provided osmoprotectants such as choline, the precursor of glycine betaine (15, 36). Many bacteria are equipped with systems that facilitate the efficient transport of osmoprotectants under stressed conditions, and several of these osmoregulated systems have been identified previously (64). Among these systems, mainly ATP-binding cassette (ABC) transporters and secondary transporters have been characterized at the molecular level. All the ABC transporters reported until now in nonhalophilic bacteria are related to the ProU system from Escherichia coli (58), including OpuA, OpuB, and OpuC of Bacillus subtilis (29, 30), OusB of Erwinia chrysanthemi (14), and BusA of Lactococcus lactis (8).
In a few bacteria such as Sinorhizobium meliloti, the root symbiont of Medicago sativa (alfalfa), quaternary ammonium transporters also have an important role at low osmolarity. In fact, this bacterium has the capacity to use compatible solutes such as glycine betaine and PB as carbon and nitrogen sources in addition to osmoprotection (5, 25). PB, also known as N,N-dimethylproline or stachydrine, is available in the rhizosphere, where it is secreted by germinating seedlings of many Medicago species nodulated by S. meliloti (47). The molecule is converted in proline after two steps of demethylation, with the first step of demethylation depending on stc2 and stc4 genes organized in a cluster on the megaplasmid pSyma and with the second step requiring the stcD gene located on the megaplasmid pSymb (11, 25, 46). PB catabolism is active at low osmolarity but reduced at high osmolarity, whereas PB uptake is increased. As a consequence, PB is accumulated in the cytoplasm, contributing to the reestablishment of turgor pressure (24). The expression of stc2, stc4, and stcD genes is enhanced in the presence of PB in the growth medium (25, 46), and PB has also been shown to induce the expression of nodulation (nod) genes via the activation of the NodD2 transcriptional regulator (45). Thus, more likely, the PB uptake activity also contributes to PB-mediated gene regulation in S. meliloti.
Two systems involved in the uptake of PB in S. meliloti have previously been characterized. Hut is a ProU-like ABC transporter with a high affinity for histidine, proline, and PB. Its activity is controlled at the transcriptional level by histidine but not by salt stress, and Hut contributes to the use of histidine as a nitrogen source (6). The BetS system is a Na+-coupled secondary transporter, with a high affinity for glycine betaine and PB. This system is activated posttranslationally by osmotic stress and plays a crucial role in the rapid response to osmotic upshock (7). However, a betS mutant is still able to use PB as an osmoprotectant, suggesting the existence of another PB transport system(s). In this study, we described the characterization of a new ABC transporter, called Prb, involved in the uptake of PB. Prb is homologous to an oligopeptide permease and not to ProU, while it also transports choline and glycine betaine. We also demonstrated that Prb is active at low and high osmolarities and is required for a maximal adaptation to high-salinity conditions, since a prb betS mutant does not use PB as an osmoprotectant.
MATERIALS AND METHODS
Chemicals.
Proline betaine was purchased from Extrasynthèse (Genay, France). Choline, glycine betaine, proline, Gly-Gly, Gly-His, Ala-Pro, Gly-Pro-Arg-Pro, Tyr-Gly-Gly-Phe-Leu (leucine enkephaline), and N,N-dimethylglycine were purchased from Sigma-Aldrich (Saint Quentin Fallavier, France). N-Methylproline was synthesized from the sodium salt of proline, as described previously (44). γ-Butyrobetaine and dimethylsulfonioacetate (DMSA) were prepared as described previously (3). Radioactive [U-14C]PB (4.6 GBq/mmol) was obtained from the Commissariat à l'Energie Atomique (Gif-sur-Yvette, France), and [methyl-14C]choline (2.04 GBq/mmol) was purchased from Amersham Corp. (Little Chalfont, United Kingdom). [methyl-14C]glycine betaine was prepared from [methyl-14C]choline as previously described (44).
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. meliloti strains were maintained at 30°C in Luria-Bertani (LB) medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2. For luciferase, transport, and osmoprotection assays, S. meliloti strains were grown in MCAA semisynthetic medium containing 0.1% sodium malate as the carbon source and 0.1% Casamino Acids (technical) as the nitrogen source (57). For physiological analysis of the role of Prb and Hut, cells were grown in M9 minimal medium (37) supplemented with 0.2% mannitol or 0.2% PB as a carbon source. The osmolarity of the medium was increased by the addition of NaCl, as indicated. E. coli strains were grown in LB broth at 37°C. Antibiotics were used in E. coli (or S. meliloti) cultures at the following concentrations: kanamycin, 25 μg/ml; spectinomycin, 200 μg/ml; chloramphenicol, 20 μg/ml; streptomycin, 200 μg/ml; neomycin, 200 μg/ml. Bacterial growth was monitored spectrophotometrically at 600 nm, and protein concentrations were determined by the method of Bradford (9), with bovine serum albumin as a standard.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or transposon | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| Rm1021 | SU47 str-21 Smr | 37 |
| Rm5000 | SU47 rif-5 Rifr | 20 |
| Rm1021-S1 (S1) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| Rm1021-S2 (S2) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| Rm1021-S3 (S3) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| Rm1021-S4 (S4) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| Rm1021-S5 (S5) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| Rm1021-S6 (S6) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| Rm1021-S7 (S7) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| Rm1021-S8 (S8) | Rm1021::Tn5 pRL1063a Smr Neor | 46 |
| RmUNA369 | Rm1021 betS::Ω Smr Spcr | This work |
| RmUNA370 | S7 betS::Ω Smr Spcr Neor | This work |
| RmHY220 | Rm5000 hutX::Ω | 6 |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 53 |
| MT616 | MT607 pRK600 | 21 |
| Plasmids | ||
| pRK600 | ColE1 replicon with RK2 transfer region; Cmr | 21 |
| pH 45-Ω | pBR322 derivative with interposon Ω; Ampr Smr Spcr | 49 |
| pSup 202 | ColE1 replicon; Mob+ Tcr Ampr Cmr | 56 |
| pSBI39 | Derivative of pSup202 with an S. meliloti 3.4-kb insert containing the betS gene | 7 |
| pSM28 | pSBI39 betS::Ω (KpnI insertion) | This work |
| pSIF7 | 10-kb EcoRI fragment of Rm1021-S7 containing prbB::Tn5 pR1063a; Kmr | This work |
| pSIF5 | 4.3-kb EcoRI fragment of Rm1021-S5 containing prbB::Tn5pR1063a; Kmr | This work |
| pSIF3 | 4.3-kb EcoRI fragment of Rm1021-S3 containing prbC::Tn5 pR1063a; Kmr | This work |
| Transposon | ||
| Tn5pRL1063a | Tn5 derivative; promoterless luxAB; oriV Kmr | 63 |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Neor, neomycin resistance; Rifr, rifampicin resistance; Spcr, spectinomycin resistance; Smr, streptomycin resistance; Tcr, tetracycline resistance.
DNA manipulations and sequence analysis.
Genomic and plasmidic DNA extractions, digestions by restriction enzymes, ligations, and transformations were performed by using standard methods (53). To recover S. meliloti genomic sequences contiguous with the inserted transposon, genomic DNA from each Rm1021::Tn5 pRL1063a mutant was digested by EcoRI, which does not cut within the transposon, ligated, and transformed into DH5α cells where oriV in pRL1063a maintained the DNA as a plasmid. For each mutant strain, plasmid DNA was purified, and the DNA sequence of the S. meliloti fragments was determined using unique outward primers corresponding to the left and right ends of Tn5 pRL1063a as described previously (38). Nucleotide sequence determination was performed by MWG-Biotech (Ebersberg, Germany) by using the fluorescent ABI dye-labeled deoxy terminator method. DNA and protein sequences were analyzed by using BLAST protocols (2).
Construction of betS mutants.
betS mutants were constructed by using the Ω interposon encoding spectinomycin resistance. pSBI39, which carries the entire betS gene (7), was linearized by KpnI, which cuts inside the betS coding sequence, blunt ended, and ligated with an SmaI-digested Ω interposon. The resulting plasmid was transferred by conjugation into strains Rm1021 and S7 by using triparental spot matings with E. coli MT616 as a helper strain (21). The Ω insertions were finally recombined into the S. meliloti genome by the plasmid incompatibility technique (52) as previously described (43). Genetic exchange of the wild-type betS gene by the betS-Ω insertion was confirmed by Southern hybridization analysis.
Measurement of luciferase activity.
Cells from 500 μl of bacterial culture were harvested, resuspended in fresh MCAA medium to obtain a final A600 of 1, mixed with 2 μl of n-decyl aldehyde (Sigma, St. Louis, Mo.), and vortexed for 30 s. Light emission was measured for 60 s with a luminometer (CHEM-GLOW photometer model J4-7441; American Instrument Company, Md.). The assays were performed on two independent cultures in triplicates. Bioluminescence is expressed as arbitrary defined relative light units.
Transport assays.
S. meliloti cells were harvested at an A600 of 0.4 to 1.2, washed twice in the medium used for the culture, and resuspended at a final A600 of 0.5. All assays were carried out at 30°C with 1 ml of the cell suspension. Uptake of radioactive substrate (100,000 dpm) was determined after a 2-min incubation period by rapid filtration through GF/F glass microfiber filters (Whatman), which were rinsed with 3 ml of the same medium, as previously described (19). The radioactivity remaining on the filters was determined with a liquid scintillation spectrometer (model LS6000SC; Beckman Instruments, Villepinte, France). For competition experiments, unlabeled compounds were added at a final concentration of 4 mM to a 40 μM [14C]PB solution.
Periplasmic protein extraction and binding assays.
S. meliloti strain Rm1021 was grown to an A600 of 1.5 in MCAA medium supplemented with 0.3 M NaCl and 100 μM PB. Cells were collected by centrifugation (10,000 × g for 10 min at 20°C) and resuspended in 10 mM Tris-HCl (pH 7.5). Periplasmic proteins were released by cold osmotic shock according to a method described previously by Neu and Heppel (42). The activity of one periplasmic marker enzyme, cyclic phosphodiesterase, and the activity of one typical cytoplasmic marker, malate deshydrogenase, were measured to determine the amount of marker enzyme activities released, as described previously (35). Periplasmic proteins were concentrated by ultrafiltration according to a standard procedure (35). Binding assays with 150 μg of periplasmic proteins were performed using ammonium sulfate precipitation, as described previously (19). Measurements were done in triplicates from two independent periplasmic preparations.
RESULTS
Identification of prb, a new PB-inducible locus of S. meliloti.
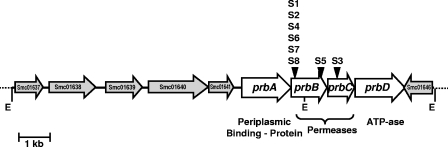
Twelve Tn5-luxAB insertions mutants (S1 to S12) containing PB-inducible genes have been isolated from S. meliloti Rm1021 (46). The only two mutants that could not use PB as a source of carbon and nitrogen were characterized further and were shown to be inactivated in the putA gene, encoding proline dehydrogenase (strain S11), and in the stcD gene, which is involved in PB catabolism (strain S10) (46). In order to identify other PB-inducible loci, we cloned the tagged genes in other mutants in E. coli, and their DNA sequences were determined and compared to the genomic sequence of S. meliloti strain Rm1021 (http://sequence.toulouse.inra.fr/meliloti/html). Eight insertions were located in the same region of the chromosome that we called prb (prb for proline betaine-inducible transporter), which represents a cluster of four open reading frames (ORFs) oriented in the same direction, designated prbA (Smc01642), prbB (Smc01643), prbC (Smc01644), and prbD (Smc01645) (Fig. 1). Seven insertions (S1, S2, S4, S5, S6, S7, and S8) were localized in prbB, with strains S1, S2, S4, S6, S7, and S8 corresponding to a single site of insertion. The transposon insertion in strain S3 was located in the third gene of the locus, prbC. At the 5′ extremity of the locus, a cluster of four genes involved in the metabolism of acyl coenzyme A (CoA) was found directly upstream of the prb locus, separated from prbA by a 317-bp-long sequence. At the 3′ extremity, a 795-bp-long ORF (Smc01646) of unknown function was found on the reverse strand, 29 bp downstream of prbD. Inside the prb locus, a strong translational coupling probably exists between all the genes, as supported by (i) the identity of the last base of the TGA stop codon of prbA and the first base of the ATG initiation codon of prbB, (ii) the overlapping of four bases in the ATGA sequence of prbB and prbC, and (iii) the overlapping of eight bases in the ATGCCTGA sequence of prbC and prbD.
FIG. 1.
Genetic organization of the chromosomal prb locus region of S. meliloti strain Rm1021. EcoRI sites used for subcloning are indicated (E), and locations of the Tn5-luxAB insertions are indicated by an arrowhead. Open reading frames are as follows: Smc01637, unknown function; Smc01638, hydroxyacyl-CoA dehydrogenase; Smc01639, acyl-CoA dehydrogenase; Smc01640, acyl-CoA synthetase; Smc01641, enoyl-CoA hydratase; Smc01646, unknown function.
This locus resembles an ABC transporter operon, with a periplasmic binding protein (PrbA), two inner membrane components (PrbB and PrbC), and a peripheral ATPase (PrbD). The ORF corresponding to prbA (Smc01642) is reported to encode a hydrophilic protein of 538 amino acid residues according to the S. meliloti website. However, sequence analysis (http://www.cbs.dtu.dk/services/SignalP/) of the NH2-terminal part of the potential encoded protein revealed the presence of a characteristic signal peptide for secretion in the periplasm if the first amino acid of the encoded protein is the Met residue located 75 bp downstream of the predicted one. Thus, we estimated that the prbA-encoded peptide is 513 amino acid residues long and corresponds to the unprocessed form of the periplasmic protein PrbA. PrbB and PrbC (319 and 275 amino acids, respectively) are highly hydrophobic and contain six and five putative transmembrane helices, respectively, typical of integral membrane proteins. The last gene, prbD, encodes a 543-amino-acid-residue protein, which is likely the ATPase of the transporter. This protein contains two fused ABC domains presenting 33% sequence identity, and each of them contains the Walker A and Walker B sites, the Q loop, and the switch domain, forming the nucleotide-binding site (54, 55). The conserved signature motif (LSGGQ) unique to the ABC transporters (54, 55) is present in the carboxyl-terminal ABC domain, while a very close sequence (VSGGQ) is present in the amino-terminal ABC domain.
Comparison of the Prb protein sequences with protein sequences in the data libraries revealed that the highest amino acid similarity was obtained with the components from systems of the peptide ABC transporter superfamily (Table 2). These transporters are known for their broad substrate specificities and have been shown to be involved in the uptake of peptides and nonpeptide molecules such as nickel or sugars (16).
TABLE 2.
Sequence identities and similaritiesa between components of Prb and selected transporters of the oligopeptide transport system family in gram-negative bacteria
| Transporter (bacterium) | % Identity (% similarity)
|
Substrate(s) | Reference | |||
|---|---|---|---|---|---|---|
| PrbA | PrbB | PrbC | PrbDb | |||
| Dpp (E. coli) | 22 (46) | 27 (44) | 28 (53) | 33 (56)/26 (49) | Dipeptides, 5-aminolevulinic acid | 1, 61 |
| 34 (59)/39 (62) | 61 | |||||
| Opp (Salmonella enterica serovar Typhimurium) | 17 (41) | 23 (44) | 25 (48) | 32 (57)/32 (54) | Oligopeptides | 27 |
| 37 (56)/40 (63) | ||||||
| Dpp (Rhizobium leguminosarum) | 21 (41) | 28 (49) | 31 (55) | 32 (54)/30 (48) | Dipeptides, 5-aminolevulinic acid | 13 |
| 36 (58)/38 (63) | ||||||
| Agp (S. meliloti) | 20 (38) | 23 (47) | 28 (53) | 36 (60) | α-Galactosides | 23 |
| Nik (E. coli) | 19 (41) | 22 (43) | 28 (53) | 31 (50)/26 (49) | Nickel | 41 |
| 31 (49)/36 (60) | ||||||
| Acc (Agrobacterium tumefaciens) | 20 (40) | 28 (50) | 35 (55) | 28 (49)/34 (52) | Agrocinopine | 26 |
| 33 (55)/35 (56) | ||||||
The percentages of identity and similarity (in parentheses) were determined with Clustal W.
Each half containing the ABC domain of PrbD (amino-terminal domain/carboxy-terminal domain) was compared to the first encoded (first line) and the second encoded (second line) ATPase of the transporter. For AgpD, which is the unique ATPase encoded by Agp and which contains two ABC cassette domains, comparison with PrbD concerns the entire proteins.
PB and NaCl inducibility of prb gene expression.
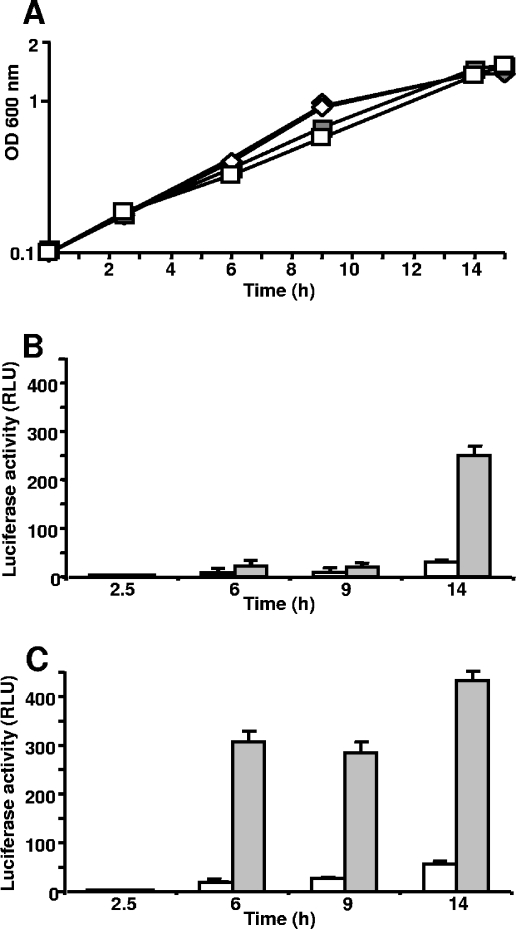
As a first step to characterize Prb function, the potential induction of prb gene expression was analyzed under different growth conditions. The PB inducibility of Tn5-luxAB mutants, and particularly of the prb mutants, was initially detected in cells grown at low osmolarity (46). Since PB has an important role as an osmoprotectant in S. meliloti, PB inducibility of prb gene expression was determined in cells grown at high osmolarity and was compared to that of cells grown at low osmolarity. The prb expression level was measured by monitoring the luciferase activity in strain S7, a prbB::luxAB insertion mutant. This strain was grown in MCAA semisynthetic medium with or without PB (100 μM) in the presence or absence of 0.3 M NaCl, a salt concentration that is not too restrictive for growth (Fig. 2A). In the absence of NaCl, PB induction of the luxAB reporter gene was significant only at stationary phase, and the activity was eightfold higher in induced cultures than in uninduced cultures after 14 h of growth (Fig. 2B). The addition of NaCl led to a much earlier and stronger PB induction, since after only 6 h of growth in MCAA medium containing NaCl and PB, the cell luciferase activity was more than 30-fold higher than the activity measured in MCAA medium (Fig. 2C); activity was even higher in cells entering stationary phase, after 14 h. The positive effect of NaCl without PB in the growth medium on prbB expression was also measurable from 6 h to 14 h, with about a twofold increase compared to the level in MCAA medium (Fig. 2B and C). These results showed that prb gene expression is controlled by both PB and NaCl, with a synergistic effect between the two compounds. Since the higher level of expression was observed at the beginning of the stationary phase under all tested conditions, the growth phase also plays a role in the control of prb gene expression. Moreover, the PB inducing effect could not be attributed to catabolite derivatives of PB, as neither N-methylproline nor proline induced the expression of prb (data not shown). Similarly, no induction by choline or glycine betaine could be detected, whereas these compounds are also transported by Prb (see below). Furthermore, despite the homology between Prb and oligopeptide transporters, no induction was observed after growth in the presence of a dipeptide (Gly-Gly) and oligopeptides of various sizes (Gly-Pro-Arg-Pro and Tyr-Gly-Gly-Phe-Leu), strengthening the specificity of the PB inducing effect on prb gene expression.
FIG. 2.
prb gene expression is induced by both PB and NaCl. Cells of strain S7 (prbB::Tn5-luxAB fusion) were grown overnight in MCAA medium and then diluted in the same medium or in MCAA medium with 0.3 M NaCl in the presence (gray symbols and gray bars) or absence (white symbols and white bars) of PB. (A) Growth curves in MCAA medium (⋄ and ♦) and MCAA medium with NaCl (□ and ▪). (B and C) Luciferase activity at various times during the growth cycle in MCAA medium and MCAA with NaCl. Results are means of duplicates from three independent experiments. The error bars represent the standard deviations from the means. OD, optical density; RLU, relative light units.
Prb contribution to PB uptake at low osmolarity.
Assuming that PB inducibility of prb genes indicates that PB is a potential substrate for the Prb transport system, the contribution of Prb to PB uptake was investigated. Transport experiments using radiolabeled PB at 40 μM were performed with wild-type and S7 cells grown in low-osmolarity MCAA medium in the presence or absence of PB (100 μΜ). PB uptake was measured using cells grown to mid-exponential phase and cells grown to stationary phase. As shown in Table 3, wild-type and mutant strains collected at the exponential phase presented similar PB uptake activities, which were not enhanced by the presence of PB. This result is in full agreement with the low level of prb gene induction by PB at this growth stage, as shown previously. On the contrary, in stationary-phase cells, the addition of PB was followed by a 2.6-fold increase in PB uptake activity in the wild-type strain, but such stimulation was not observed in the mutant strain. These results showed that PB uptake activity can be stimulated by the presence of PB in a low-osmolarity growth medium and that such increased activity is Prb dependent.
TABLE 3.
A prb mutant is affected in the uptake of proline betaine at low osmolaritya
| Strain | Growth with PB | PB uptake activityb (nmol/min/mg of protein) ± SD
|
|
|---|---|---|---|
| Exponential phase | Stationary phase | ||
| Rm1021 | No | 0.56 ± 0.10 | 0.70 ± 0.10 |
| Yes | 0.53 ± 0.05 | 1.82 ± 0.12 | |
| S7 | No | 0.51 ± 0.05 | 0.69 ± 0.03 |
| Yes | 0.39 ± 0.05 | 0.78 ± 0.03 | |
Cells were grown in MCAA medium with or without 100 μM PB and harvested at mid-log phase (A600 of 0.4) or early stationary phase (A600 of 1.5).
Uptake measurements of PB were done at a final substrate concentration of 40 μM. Values are means from duplicates of three independent cultures ± standard deviations.
To investigate the physiological role of Prb in cells maintained at low osmolarity, growth experiments were performed using minimal medium containing PB as the unique carbon and nitrogen source. No difference in the growth kinetics between the wild-type and the prb mutant could be observed, which probably indicates that another PB transport system(s) compensates for the Prb deficiency. Hut was a good candidate, but the growth of hut mutant strains could not be distinguished from that of the wild-type strain, unless histidine (100 μM) was present as a hut inducer in the growth medium. Thus, in addition to Prb, it is more likely that another transporter is involved in the utilization of PB as a carbon and nitrogen source.
Prb contribution to PB uptake at high osmolarity.
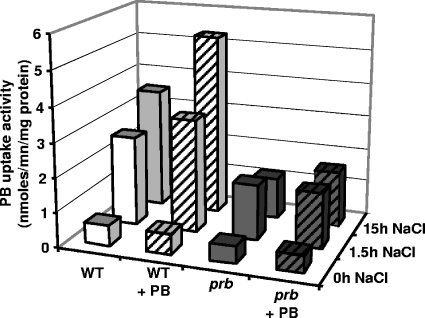
PB transport assays were also performed using cells that were exposed to NaCl in MCAA medium with 0.3 M NaCl during 1.5 h and 15 h in the presence (100 μM) or absence of PB (Fig. 3). In the wild-type strain, the PB uptake activity increased in response to the time of exposure to NaCl. Interestingly, this NaCl stimulation was enhanced by the presence of PB in the growth medium: after 15 h, the activity increased from 3.5 to 5.3 nmol/min/mg of protein when PB was added. This higher level corresponds to about a 10-fold increase in PB uptake activity compared to the activity in cells grown in MCAA medium (0.6 nmol/min/mg of protein) (Table 2). In the mutant strain, stimulation of uptake by NaCl was much lower, with a maximal activity of 1.6 nmol/min/mg of protein. Consequently, after 15 h of growth in the presence of both NaCl and PB, PB uptake activity in the prb mutant represented only one-third of the activity in wild-type cells grown under the same conditions. These results demonstrated that Prb is the major PB uptake system functioning under prolonged conditions of high salinity and that the enhancement of PB uptake activity by the presence of PB is essentially due to Prb stimulation.
FIG. 3.
A prb mutant is affected in the uptake of PB at high osmolarity. Cells of strains Rm1021 (wild type [WT]) and S7 (prb) grown to mid-log phase in MCAA medium were subjected to a 1.5-h NaCl treatment (final concentration of 0.3 M) in the presence or absence of 100 μM PB or diluted in MCAA medium with 0.3 M NaCl supplemented or not supplemented with 100 μM PB and grown for 15 h to mid-log phase. Uptake measurements of PB were done at a final concentration of 40 μM. Results are means of duplicates from three independent experiments, and standard deviations did not exceed 15%.
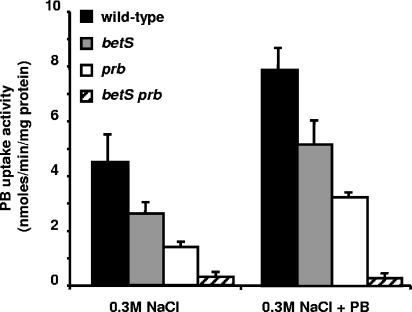
Since both Prb and BetS systems are involved in the uptake of PB at high osmolarity (7), a betS prb double mutant should be severely impaired in PB uptake. Thus, a betS inactivated allele was introduced into strains Rm1021 and S7, resulting in strains UNA369 and UNA370 (see Materials and Methods). PB uptake activities of prb and betS single and double mutants (strains S7, RmUNA369, and RmUNA370, respectively) in cells grown at high osmolarity during 18 h in the presence or the absence of PB were then determined. Figure 4 shows that PB uptake activity was nearly abolished in the betS prb double mutant, indicating that only Prb and BetS significantly contributed to PB uptake at a high osmolarity. Moreover, PB uptake activity in betS and prb single mutants increased following the addition of PB to MCAA medium, suggesting that BetS activity, in addition to Prb activity, was stimulated by PB.
FIG. 4.
PB uptake activity is severely impaired in a prb betS double mutant. Cells were grown for 18 h in MCAA medium containing 0.3 M NaCl or 0.3 M NaCl and 100 μM PB. Assays for the uptake of PB were done at a final concentration of 40 μM using cells from strains Rm1021 (wild type), RmUNA369 (betS), S7 (prb), and RmUNA370 (prb betS). The results are means of duplicates from three independent cultures; standard deviations from the means are also shown.
Evidence for PB-binding activity in periplasmic fractions.
In gram-negative bacteria, high-affinity ABC transporters are dependent upon at least one periplasmic substrate binding protein, and Prb-mediated uptake of PB more likely depends on the existence of such a component. However, no PB-binding activity has previously been detected in periplasmic fractions of cells grown at low or high osmolarity (24). Since the results presented above have shown that Prb activity is strongly increased in the presence of both NaCl and PB, binding assays were performed with periplasmic fractions extracted from cells grown in MCAA medium containing NaCl (0.3 M) and PB (100 μM). In such fractions, a PB-binding activity of 35 pmol/mg of protein was detected in the presence of a free-ligand concentration of 80 μM. In extracts from cells grown at low osmolarity or in the presence of 0.3 M NaCl without PB, binding activity was barely measurable. These results demonstrated the existence of a PB binding protein(s) among periplasmic fractions from S. meliloti but with a level of activity that was rather low.
Prb is involved in the uptake of choline and glycine betaine.
In E. coli and B. subtilis, the different systems involved in PB uptake also transport other betaines or related compounds (31). Similarly, in S. meliloti, BetS is a high-affinity transporter for PB and also for glycine betaine (7). In addition, in the wild-type strain, uptake of PB is inhibited by other quaternary ammonium compounds (24). In a first approach to determine potential substrates for Prb, different molecules were used in competition experiments conducted with strain UNA369 (betS mutant). Uptake of 40 μM PB in the presence of a 100-fold molar excess of unlabeled competitors was measured in cells grown in MCAA medium containing 0.3 M NaCl (Table 4). Trigonelline, which strongly inhibited PB uptake in the wild-type strain (24), poorly competed in the betS mutant, suggesting that it is a substrate for BetS and not for Prb. In contrast, several other compounds (glycine betaine, γ-butyrobetaine, choline, N,N-dimethylglycine, N-methylproline, and DMSA) effectively inhibited the uptake of PB in the betS mutant. Among these compounds, only choline and glycine betaine are available in a radioactive form. Thus, in order to precisely define the substrate of Prb, transport assays with radiolabeled choline and glycine betaine were performed using prb+ strains and prb mutants grown at high osmolarity (0.3 M NaCl). Choline (40 μM) uptake activity was significantly affected in the prb mutant, and the presence of PB in the growth medium strongly increased the uptake activity in the wild-type strain (Table 5). This suggested that stimulation of Prb by PB likewise plays an important role in the uptake of choline at high osmolarity. Conversely, the transport of glycine betaine (40 μM) was neither significantly stimulated by the addition of PB nor affected in the single prb mutant. However, it was more affected in the prb betS mutant than in the single betS mutant, showing that Prb also contributes to glycine betaine uptake but at a lower affinity than BetS. Finally, because of the rather high level of homology between Prb and oligopeptide transporters, peptides of different sizes (Ala-Pro, Gly-His, Gly-Pro-Arg-Pro, and Tyr-Gly-Gly-Phe-Leu) were used in competition experiments using the wild-type strain. None of them significantly inhibited PB uptake in cells grown in the presence or absence of NaCl (data not shown), suggesting that oligopeptide uptake is not mediated by Prb.
TABLE 4.
Various compounds inhibit proline betaine uptake in a betS mutanta
| Competitor | % Inhibition of PB uptakeb with unlabeled competitor |
|---|---|
| Glycine betaine | 94 |
| Trigonelline | 9 |
| γ-Butyrobetaine | 53 |
| Choline | 70 |
| N,N-Dimethylglycine | 40 |
| N-Methylproline | 55 |
| DMSA | 84 |
Cells of RmUNA369 (betS::Ω) were grown to mid-log phase in MCAA medium with 0.3 M NaCl.
Uptake assays were performed with 40 μM PB and different competitors added at a 100-fold molar excess. The results are expressed as the percentage of inhibition of PB uptake and are means of duplicates with standard deviations of less than 5%. The control value without competitor was 2.2 nmol of PB transported per min per mg of protein.
TABLE 5.
A prb mutant is affected in the uptake of cholinea
| Strain | Uptake activity (nmol/min/mg of protein)b ± SD
|
|||
|---|---|---|---|---|
| Choline
|
Glycine betaine
|
|||
| A | B | A | B | |
| Rm1021 (wild type) | 0.8 ± 0.1 | 6.8 ± 0.4 | 6.7 ± 0.4 | 7.3 ± 1.0 |
| S7 (prb) | 0.5 ± 0.1 | 0.3 ± 0.1 | 7.7 ± 0.5 | 6.3 ± 0.4 |
| UNA369 (betS) | ND | ND | 2.5 ± 0.1 | 3.9 ± 0.8 |
| UNA370 (prb betS) | ND | ND | 1.5 ± 0.1 | 1.6 ± 0.2 |
Cells grown in MCAA medium with 0.3 M NaCl in the absence (A) or the presence (B) of 100 mM PB were harvested at mid-log phase.
Uptake assays of choline and glycine betaine were done at a final substrate concentration of 40 μM. Values are means from duplicates of three independent cultures ± standard deviations. ND, not determined.
Prb plays a significant role in osmoprotection by PB and glycine betaine.
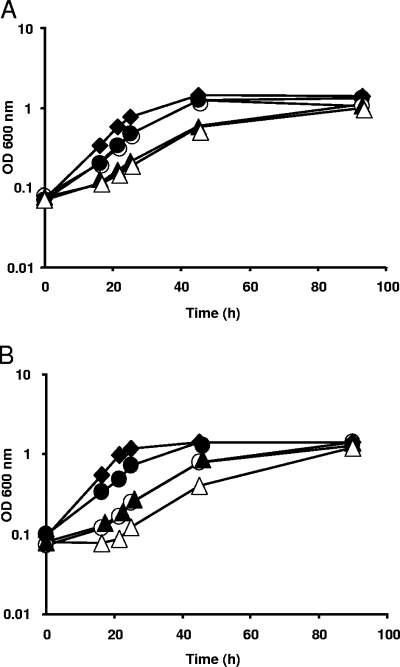
Since Prb is involved in the transport of PB, choline, and glycine betaine in cells grown at high osmolarity, it could significantly contribute to osmoprotection. Therefore, the effect of prb inactivation was investigated by growing the cells at high osmolarity (0.6 M NaCl) in the absence or presence of PB, choline, or glycine betaine. No difference in osmoprotection by choline (7 mM) could be observed in a prb mutant (strain S7) compared to the wild-type strain, indicating that another choline uptake activity compensated for the prb deficiency (data not shown). This result corroborates the previous identification of a choline uptake activity that is inducible by choline and fully active at high osmolarity (48). When PB (1 mM) was added, growth inhibition was much more efficiently alleviated in the wild-type strain than in betS and prb mutants (Fig. 5A). It is noteworthy that PB was no longer used as an osmoprotectant by the betS prb double mutant. These results offer evidence that Prb is necessary for maximal osmoprotection by PB and that beside Prb and BetS, no other transport system is required for adaptation to salinity stress through PB utilization. Osmoprotection by glycine betaine was also less efficient in the prb mutant than in the wild-type strain but was clearly much more efficient than that in the betS mutant (Fig. 5B). This result is consistent with the moderate contribution of Prb to glycine betaine uptake, as indicated by the results of transport assays. The remaining capacity of the betS prb double mutant to use glycine betaine for osmoprotection also indicates the existence of another system or other systems for glycine betaine transport in S. meliloti.
FIG. 5.
Inactivation of prb has a strong negative effect on osmoprotection by PB and glycine betaine. Cells were grown in MCAA medium with 0.6 M NaCl in the presence of 1 mM PB (A) or 1 mM glycine betaine (GB) (B). Strains used were Rm1021 (wild-type) (⧫), S7 (prb) (•), RmUNA369 (betS) (○), and RmUNA370 (prbB betS) (▴). Growth of RmUNA370 in MCAA medium with 0.6 M NaCl (without osmoprotectant) is also shown (▵). The results are means of three independent cultures, and standard deviations did not exceed 5%. OD, optical density.
DISCUSSION
Proline betaine, a quaternary ammonium compound released in the rhizosphere by germinating alfalfa seeds (47), functions as an osmoprotectant, an energy substrate, and an inducer of nodulation genes in S. meliloti. For the first physiological processes, this bacterium has evolved several types of PB uptake systems, two of which are the Hut (6) and the BetS (7) transporter systems. In this study, we have localized Tn5-luxAB insertions in genes encoding components of a novel ABC transporter, called Prb, also involved in the uptake of PB, which differs from Hut and BetS by its inducibility by both PB and salt stress. The general organization of the prb locus is similar to that of ABC transporter operons, and each component of the Prb system shows significant similarity with components of the peptide ABC transporters (Opp-like transporters) of various bacteria. These systems are known to transport a large variety of substrates, such as peptides of various lengths, used as carbon and nitrogen sources or as pheromones (32, 34) but also heme precursor (13, 61), nickel (41), and sugar and modified sugar molecules (23, 26). In S. meliloti, 27 of the 176 predicted ABC transporters in the whole genome belong to the peptide transport system subfamily (4, 12, 22). However, their substrate specificities have not been characterized so far, except in the case of Agp system, which transports α-galactosides (23). It has been suggested that some Opp-like transporters from S. meliloti could be involved in osmotic adaptation, since several genes from opp-like operons are among the salt-induced genes identified using a microarray approach (51). The presence of prbB and prbD among these genes and our present data demonstrating the contribution of Prb in PB, choline, and glycine betaine uptake at high osmolarity strongly support this suggestion. It is noteworthy that in spite of the homology between Prb and Opp proteins, Prb is unlikely to be involved in the uptake of peptides, since peptides of 2 to 5 amino acids failed to compete with PB. Interestingly, PrbA has an arginine residue instead of the aspartate residue, which is present in all OppA-like proteins involved in peptide binding and which forms a salt bridge with the N-terminal α-amino group of the bound peptide (18).
Disruption of the prbB gene significantly reduces PB transport in cells grown at low or high osmolarity. In stationary-phase cells grown at low osmolarity, Prb contributes to PB uptake and could thus enhance PB acquisition when the main exogenous carbon and nitrogen sources become limited for the cells and when their capacity for amino acid biosynthesis becomes diminished. In addition, under high-salinity conditions, Prb uptake activity has a major role in osmoprotection, since a strain inactivated in both Prb and BetS systems is no longer able to use exogenously provided PB as an osmoprotectant. Moreover, the demonstration that Prb is also involved in the uptake of choline and, to a lesser extent, of glycine betaine highlights the importance of Prb in the adaptation to salinity stress.
In terms of the uptake of osmoprotective compounds, S. meliloti shows important differences compared to other bacterial models. In most bacteria, osmoprotectants are metabolically inert molecules and are not actively transported at a low osmolarity. The uptake of the few compounds that play a double role in energy supply and osmoprotection is mediated through separate transport systems. For example, the acquisition of proline as a carbon and nitrogen source is mediated in E. coli by the PutP permease for which expression is proline induced (39, 40) and is mediated in B. subtilis by an uncharacterized system, while the utilization of proline for osmoprotective purposes requires the osmotically regulated ProP and ProU transporters in E. coli and the OpuE system in B. subtilis (15, 62). Similarly, in S. meliloti, the uptake of PB at low and high osmolarities involves distinct systems, Hut for energy supply and BetS for immediate adaptation to an osmotic upshock. Interestingly, Prb is, up to now, the first uptake system active under both conditions. It has been shown previously that the uptake of ectoine, a nonaccumulated osmoprotectant, is also fulfilled by a unique system (Ehu) in S. meliloti cells grown either at a low or high osmolarity. However, the expression of ehu genes is induced only by ectoine (28), whose uptake is not increased in cells grown at high osmolarity (59).
A prominent finding of this work concerns the dual regulation of Prb. Data regarding the expression of a prbB::Tn5-luxAB fusion have led to the proposal that this regulation occurs, at least partly, at the transcriptional level and is responsive to both increases in salinity and the presence of PB in the growth medium. A modest increase (twofold) in prb gene expression was observed in cells maintained in the presence of 0.3 M NaCl, whereas a strong synergistic enhancement was obtained in the presence of PB (Fig. 2), causing massive prb transcription when PB was available. Such PB and NaCl transcriptional control, combined with the osmotic posttranslational activation of BetS (7), allows the rapid and long-term uptake of PB in salt-stressed cells with a very beneficial effect on the adaptation to salinity stress. Salt stimulation by the activation of preexisting Prb proteins could additionally occur, as observed for other osmotically induced osmoprotectant transporters (31).
The capacity to accumulate PB at high osmolarity is necessarily controlled to prevent an excessive accumulation of the osmoprotectant and to maintain the cell turgor within certain limits. In many bacteria, the presence of osmoprotectants in the growth medium frequently reduces the transcription of osmotically induced genes encoding components for the uptake of these solutes (50). An opposite situation is observed in S. meliloti, and that original prb regulation could come from the capacity of the cells to catabolize PB, restraining the accumulated betaine to a certain threshold. Whereas the induction of stc2 gene expression by PB is strongly reduced in cells subjected to increased osmolarities, substantial PB degradation is still likely to occur at a lower salt concentration, such as 0.3 M NaCl (G. Alloing, unpublished results).
The affinity and specificity of ABC transporters usually depend on at least one periplasmic binding protein. In S. meliloti, periplasmic binding activities for glycine betaine and choline have already been observed in cells grown at high osmolarity (35), and our present results demonstrate the existence of a PB-binding activity. The measured binding activity is rather low and might indicate an unstable PB-protein complex, which is nevertheless associated with effective uptake. Similarly, in L. lactis (17), poor binding activity of short peptides to OppA has previously been observed, whereas these compounds are efficiently transported by the Opp system. In that case, the dissociation rate constant is an important determinant of the transport rate, and even poorly bound peptides, associated with high dissociation rate constant values, are relatively well transported (17, 33). However, the poor PB-binding activity might also be the consequence of the preloading of periplasmic proteins with PB present in the growth medium. Obviously, in S. meliloti, additional analyses of PrbA-binding activity are needed, and its potential role in PB, glycine betaine, and choline binding and uptake remains to be clarified.
Collectively, the remaining PB uptake activity observed in a prb mutant, as well as our inability to detect any difference between the wild-type, prb, and hut mutant strains in the kinetics of growth in minimal low-osmolarity medium containing PB as the only carbon or nitrogen source, indicates the existence of as-yet-uncharacterized PB transport systems. The existence of several systems dedicated to PB transport is particularly helpful for the adaptation of S. meliloti to various environments, in the soil as a heterotrophic bacterium and in planta in an endosymbiotic form. In both situations, PB synthesized by alfalfa, a host plant of S. meliloti, is present as a root exudate in the rhizosphere and in the nodule, cytosol, and bacteroids (45, 60). Finally, how prb expression is regulated in planta and whether Prb activity is crucial for the endosymbiotic form of S. meliloti when the host plant is subjected or not subjected to salt stress are interesting questions that require additional investigation.
Acknowledgments
We gratefully thank Donald Phillips, who generously provided the Tn5-luxAB mutants, and the other colleagues cited in Table 1 for the gifts of strains. We thank David Tepfer for the supply of [14C]PB.
This work was supported by the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Abouhamad, W. N., and M. D. Manson. 1994. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 14:1077-1092. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, L., T. Kühler, and M. Nilsson. 1981. Preparation of 3-carboxy-N-N-N-trimethylpropanaminium chloride (γ-butyrobetaine hydrochloride). Synthesis 6:468-469. [Google Scholar]
- 4.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, T., J. A. Pocard, B. Perroud, and D. Le Rudulier. 1986. Variations in the response of salt-stressed Rhizobium strains to betaines. Arch. Microbiol. 143:359-364. [Google Scholar]
- 6.Boncompagni, E., L. Dupont, T. Mignot, M. Osteras, A. Lambert, M. C. Poggi, and D. Le Rudulier. 2000. Characterization of a Sinorhizobium meliloti ATP-binding cassette histidine transporter also involved in betaine and proline uptake. J. Bacteriol. 182:3717-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boscari, A., K. Mandon, L. Dupont, M. C. Poggi, and D. Le Rudulier. 2002. BetS is a major glycine betaine/proline betaine transporter required for early osmotic adjustment in Sinorhizobium meliloti. J. Bacteriol. 184:2654-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvier, J., P. Bordes, Y. Romeo, A. Fourcans, I. Bouvier, and C. Gutierrez. 2000. Characterization of OpuA, a glycine-betaine uptake system of Lactococcus lactis. J. Mol. Microbiol. Biotechnol. 2:199-205. [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Bremer, E., and R. Krämer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p. 79-97. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 11.Burnet, M. W., A. Goldmann, B. Message, R. Drong, A. El Amrani, O. Loreau, J. Slightom, and D. Tepfer. 2000. The stachydrine catabolism region in Sinorhizobium meliloti encodes a multi-enzyme complex similar to the xenobiotic degrading systems in other bacteria. Gene 244:151-161. [DOI] [PubMed] [Google Scholar]
- 12.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter, R. A., K. H. Yeoman, A. Klein, A. H. Hosie, G. Sawers, P. S. Poole, and A. W. Johnston. 2002. dpp genes of Rhizobium leguminosarum specify uptake of delta-aminolevulinic acid. Mol. Plant-Microbe Interact. 15:69-74. [DOI] [PubMed] [Google Scholar]
- 14.Choquet, G., N. Jehan, C. Pissavin, C. Blanco, and M. Jebbar. 2005. OusB, a broad-specificity ABC-type transporter from Erwinia chrysanthemi, mediates uptake of glycine betaine and choline with a high affinity. Appl. Environ. Microbiol. 71:3389-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 16.Detmers, F. J., F. C. Lanfermeijer, and B. Poolman. 2001. Peptides and ATP binding cassette peptide transporters. Res. Microbiol. 152:245-258. [DOI] [PubMed] [Google Scholar]
- 17.Doeven, M. K., R. Abele, R. Tampe, and B. Poolman. 2004. The binding specificity of OppA determines the selectivity of the oligopeptide ATP-binding cassette transporter. J. Biol. Chem. 279:32301-32307. [DOI] [PubMed] [Google Scholar]
- 18.Doeven, M. K., J. Kok, and B. Poolman. 2005. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Mol. Microbiol. 57:640-649. [DOI] [PubMed] [Google Scholar]
- 19.Dupont, L., I. Garcia, M.-C. Poggi, G. Alloing, K. Mandon, and D. Le Rudulier. 2004. The Sinorhizobium meliloti ABC transporter Cho is highly specific for choline and expressed in bacteroids from Medicago sativa nodules. J. Bacteriol. 186:5988-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finan, T. M., E. Hartweig, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gage, D. J., and S. R. Long. 1998. α-Galactoside uptake in Rhizobium meliloti: isolation and characterization of agpA, a gene encoding a periplasmic binding protein required for melibiose and raffinose utilization. J. Bacteriol. 180:5739-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gloux, K., and D. Le Rudulier. 1989. Transport and catabolism of proline betaine in salt-stressed Rhizobium meliloti. Arch. Microbiol. 151:143-148. [Google Scholar]
- 25.Goldmann, A., L. Lecoeur, B. Message, M. Delarue, E. Schoonejans, and D. Tepfer. 1994. Symbiotic plasmid genes essential to the catabolism of proline betaine, or stachydrine, are also required for efficient nodulation by Rhizobium meliloti. FEMS Microbiol. Lett. 115:305-312. [Google Scholar]
- 26.Hayman, G. T., S. B. von Bodman, H. Kim, P. Jiang, and S. K. Farrand. 1993. Genetic analysis of the agrocinopine catabolic region of Agrobacterium tumefaciens Ti plasmid pTiC58, which encodes genes required for opine and agrocin 84 transport. J. Bacteriol. 175:5575-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiles, I. D., L. M. Powell, and C. F. Higgins. 1987. Peptide transport in Salmonella typhimurium: molecular cloning and characterization of the oligopeptide permease genes. Mol. Gen. Genet. 206:101-109. [DOI] [PubMed] [Google Scholar]
- 28.Jebbar, M., L. Sohn-Bosser, E. Bremer, T. Bernard, and C. Blanco. 2005. Ectoine-induced proteins in Sinorhizobium meliloti include an ectoine ABC-type transporter involved in osmoprotection and ectoine catabolism. J. Bacteriol. 187:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203-216. [DOI] [PubMed] [Google Scholar]
- 30.Kempf, B., and E. Bremer. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701-16713. [DOI] [PubMed] [Google Scholar]
- 31.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolarity environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 32.Lamarque, M., P. Charbonnel, D. Aubel, J. C. Piard, D. Atlan, and V. Juillard. 2004. A multifunction ABC transporter (Opt) contributes to diversity of peptide uptake specificity within the genus Lactococcus. J. Bacteriol. 186:6492-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanfermeijer, F. C., A. Picon, W. N. Konings, and B. Poolman. 1999. Kinetics and consequences of binding of nona- and dodecapeptides to the oligopeptide binding protein (OppA) of Lactococcus lactis. Biochemistry 38:14440-14450. [DOI] [PubMed] [Google Scholar]
- 34.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Rudulier, D., K. Gloux, and N. Riou. 1991. Identification of an osmotically induced periplasmic glycine betaine-binding protein from Rhizobium meliloti. Biochim. Biophys. Acta 1061:197-205. [DOI] [PubMed] [Google Scholar]
- 36.Le Rudulier, D., K. Mandon, L. Dupont, and J.-C. Trinchant. 2002. Salinity effects on the physiology of soil microorganisms, p. 2774-2789. In G. Bitton (ed.), The encyclopedia of environmental microbiology. John Wiley & Sons, Inc., Hoboken, N.J.
- 37.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milcamps, A., D. M. Ragatz, P. Lim, K. A. Berger, and F. J. de Bruijn. 1998. Isolation of carbon- and nitrogen-deprivation-induced loci of Sinorhizobium meliloti 1021 by Tn5-luxAB mutagenesis. Microbiology 144:3205-3218. [DOI] [PubMed] [Google Scholar]
- 39.Mogi, T., H. Yamamoto, T. Nakao, I. Yamato, and Y. Anraku. 1986. Genetic and physical characterization of putP, the proline carrier gene of Escherichia coli K12. Mol. Gen. Genet. 202:35-41. [DOI] [PubMed] [Google Scholar]
- 40.Nakao, T., I. Yamato, and Y. Anraku. 1988. Mapping of the multiple regulatory sites for putP and putA expression in the putC region of Escherichia coli. Mol. Gen. Genet. 214:379-388. [DOI] [PubMed] [Google Scholar]
- 41.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 42.Neu, H. C., and L. A. Heppel. 1965. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J. Biol. Chem. 240:3685-3692. [PubMed] [Google Scholar]
- 43.Østeräs, M., B. T. Driscoll, and T. M. Finan. 1995. Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J. Bacteriol. 177:1452-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perroud, B., and D. Le Rudulier. 1985. Glycine betaine transport in Escherichia coli: osmotic modulation. J. Bacteriol. 161:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips, D. A., C. M. Joseph, and C. A. Maxwell. 1992. Trigonelline and stachydrine released from alfalfa seeds activate NodD2 protein in Rhizobium meliloti. Plant Physiol. 99:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips, D. A., E. S. Sande, J. A. C. Vriezen, F. J. de Bruijn, D. Le Rudulier, and C. M. Joseph. 1998. A new genetic locus in Sinorhizobium meliloti is involved in stachydrine utilization. Appl. Environ. Microbiol. 64:3954-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips, D. A., J. Wery, C. M. Joseph, A. D. Jones, and L. R. Teuber. 1995. Release of flavonoids and betaines from seeds of seven Medicago species. Crop Sci. 35:805-808. [Google Scholar]
- 48.Pocard, J. A., T. Bernard, L. T. Smith, and D. Le Rudulier. 1989. Characterization of three choline transport activities in Rhizobium meliloti: modulation by choline and osmotic stress. J. Bacteriol. 171:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 50.Romeo, Y., D. Obis, J. Bouvier, A. Guillot, A. Fourcans, I. Bouvier, C. Gutierrez, and M. Y. Mistou. 2003. Osmoregulation in Lactococcus lactis: BusR, a transcriptional repressor of the glycine betaine uptake system BusA. Mol. Microbiol. 47:1135-1147. [DOI] [PubMed] [Google Scholar]
- 51.Ruberg, S., Z. X. Tian, E. Krol, B. Linke, F. Meyer, Y. Wang, A. Puhler, S. Weidner, and A. Becker. 2003. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J. Biotechnol. 106:255-268. [DOI] [PubMed] [Google Scholar]
- 52.Ruvkun, G. B., and F. M. Ausubel. 1981. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85-88. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Schmees, G., A. Stein, S. Hunke, H. Landmesser, and E. Schneider. 1999. Functional consequences of mutations in the conserved ‘signature sequence’ of the ATP-binding-cassette protein MalK. Eur. J. Biochem. 266:420-430. [DOI] [PubMed] [Google Scholar]
- 55.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 56.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 57.Smith, L. T., J. A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stirling, D. A., C. S. Hulton, L. Waddell, S. F. Park, G. S. Stewart, I. R. Booth, and C. F. Higgins. 1989. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol. Microbiol. 3:1025-1038. [DOI] [PubMed] [Google Scholar]
- 59.Talibart, R., M. Jebbar, G. Gouesbet, S. Himdi-Kabbab, H. Wroblewski, C. Blanco, and T. Bernard. 1994. Osmoadaptation in rhizobia: ectoine-induced salt tolerance. J. Bacteriol. 176:5210-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trinchant, J. C., A. Boscari, G. Spennato, G. Van de Sype, and D. Le Rudulier. 2004. Proline betaine accumulation and metabolism in alfalfa plants under sodium chloride stress. Exploring its compartmentalization in nodules. Plant Physiol. 135:1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verkamp, E., V. M. Backman, J. M. Bjornsson, D. Soll, and G. Eggertsson. 1993. The periplasmic dipeptide permease system transports 5-aminolevulinic acid in Escherichia coli. J. Bacteriol. 175:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 63.Wolk, C. P., Y. Cai, and J. M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood, J. M., E. Bremer, L. N. Csonka, R. Kraemer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437-460. [DOI] [PubMed] [Google Scholar]