Abstract
Mycothiol (MSH) (acetyl-Cys-GlcN-Ins) is the major low-molecular-mass thiol in Mycobacterium tuberculosis. MSH has antioxidant activity, can detoxify a variety of toxic compounds, and helps to maintain the reducing environment of the cell. The production of MSH provides a potential novel target for tuberculosis treatment. Biosynthesis of MSH requires at least four genes. To determine which of these genes is essential in M. tuberculosis, we have been constructing targeted gene disruptions. Disruption in the mshC gene is lethal to M. tuberculosis, while disruption in the mshB gene results in MSH levels 20 to 100% of those of the wild type. For this study, we have constructed a targeted gene disruption in the mshD gene that encodes mycothiol synthase, the final enzyme in MSH biosynthesis. The mshD mutant produced ∼1% of normal MSH levels but high levels of the MshD substrate Cys-GlcN-Ins and the novel thiol N-formyl-Cys-GlcN-Ins. Although N-formyl-Cys-GlcN-Ins was maintained in a highly reduced state, Cys-GlcN-Ins was substantially oxidized. In both the wild type and the mshD mutant, cysteine was predominantly oxidized. The M. tuberculosis mshD mutant grew poorly on agar plates lacking catalase and oleic acid and in low-pH media and had heightened sensitivity to hydrogen peroxide. The inability of the mshD mutant to survive and grow in macrophages may be associated with its altered thiol-disulfide status. It appears that N-formyl-Cys-GlcN-Ins serves as a weak surrogate for MSH but is not sufficient to support normal growth of M. tuberculosis under stress conditions such as those found within the macrophage.
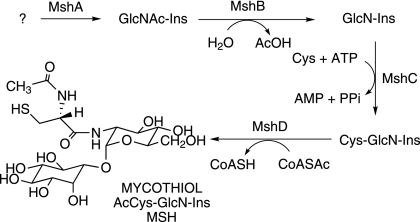
Mycothiol (MSH) (acetyl-Cys-GlcN-Ins) (Fig. 1) is the major thiol in mycobacteria (13) and contains the core amino acid cysteine similar to the predominant thiol in eukaryotes, glutathione (6). MSH is a unique product of the actinomycetes and is present at millimolar levels in Mycobacterium tuberculosis (10). Small thiols such as MSH play an important role in a number of essential functions, which include maintaining the reducing environment within the cell, protecting the cellular contents from oxidants, and detoxifying thiol-reactive compounds (4, 13, 18, 20, 25). In addition, MSH has the ability to detoxify antibiotics (4, 13, 18, 25). Failure to recover M. tuberculosis mutants in the MSH biosynthetic gene mshC has indicated that MSH is essential for the growth of M. tuberculosis (21, 23). Thus, the MSH biosynthetic pathway provides a novel antimycobacterial target, and the enzymes essential for MSH biosynthesis are potential drug targets for the treatment of tuberculosis.
FIG. 1.
Pathway for mycothiol biosynthesis. Ac, acetyl.
Four genes involved in the biosynthesis of MSH have been identified in studies of the model organism Mycobacterium smegmatis. A glycosyltransferase (MshA, encoded by Rv0486 in the M. tuberculosis genome) is involved in the production of GlcNAc-Ins, but the detailed biochemistry of this step is still being elucidated (Fig. 1). The deacetylase MshB (Rv1170) produces GlcN-Ins (11), which is ligated to Cys under catalysis by MshC (Rv2130c) (22). Mycothiol synthase (MshD, Rv0819) utilizes acetyl coenzyme A (CoA) to generate MSH (8). As each MSH biosynthetic gene has been identified in M. smegmatis, we have undertaken to generate the corresponding mutant in M. tuberculosis to ascertain its importance for growth and survival under various stresses.
The first mycothiol biosynthesis gene to be identified was mshB, and its targeted gene disruption in M. tuberculosis generated a mutant that produced ∼20% of wild-type levels of MSH during log-phase growth (4). MSH levels in the mutant increased substantially during prolonged culture, eventually reaching wild-type levels in stationary phase. In an mshB mutant of M. smegmatis, Rawat et al. (17) found that the MSH level was 5 to 10% of the wild-type level during exponential growth and did not increase significantly in early stationary phase. The ability of these mutants to synthesize significant amounts of MSH indicated the presence of a second deacetylase activity that is capable of substituting for MshB. An M. tuberculosis mshB mutant grew poorly on agar plates made with Middlebrook 7H9 medium lacking OADC (oleic acid, albumin, dextrose, and catalase) and had a heightened sensitivity to the toxic oxidant cumene hydroperoxide and to the antibiotic rifampin (4).
The second MSH biosynthesis gene identified was mshC, and its targeted mutation in M. tuberculosis resulted in no viable colonies (21), in agreement with results from transposon site hybridization studies (23). In contrast, disruption of the mshC gene in M. smegmatis produced viable mutants that were highly deficient or devoid of both Cys-GlcN-Ins and MSH (18). Thus, while M. tuberculosis apparently requires MSH for growth, M. smegmatis does not, even though the MSH-deficient mutants are highly sensitized to oxidants and certain antibiotics. Apparently, the larger genome of M. smegmatis includes genes absent in M. tuberculosis that facilitate its growth in the absence of MSH.
The third MSH biosynthesis gene identified was mshD (8). Shortly thereafter, the crystal structure of the M. tuberculosis acetyl-CoA-MshD complex was reported (27), and the recent structural analysis of a ternary complex produced from Cys-GlcN-Ins and CoA provides insights into the conformational changes involved in catalysis (28). Transposon site hybridization studies identified this gene (Rv0819) as nonessential for the growth of M. tuberculosis (23). However, mutants in the mshD gene were found to be among the four most sensitive genes among 2,154 genes characterized for survival in unactivated and activated macrophages (19). Thus, MshD seems to be a promising target for a drug against intracellular M. tuberculosis, and it is therefore important to understand the impact of the inactivation of the mshD gene upon M. tuberculosis.
The mshD mutant in M. smegmatis was recently characterized and was reported to grow normally despite having a substantially altered redox status (14). Levels of Cys-GlcN-Ins, the substrate for MshD, were elevated by orders of magnitude in the mutant, as expected from the mutation. However, the observed production of the novel thiols N-formyl-Cys-GlcN-Ins (fCys-GlcN-Ins) and N-succinyl-Cys-GlcN-Ins (succ-Cys-GlcN-Ins) in the mutant was unexpected. Given the importance of this gene for the survival of M. tuberculosis in the macrophage (19), an examination of the thiols and thiol redox status of the mshD mutant in M. tuberculosis was undertaken. We report here the construction of a directed knockout of the mshD gene in M. tuberculosis, a comparison of its thiol and disulfide composition with those of the wild-type strain and M. smegmatis, and the characterization of its requirements for growth and survival under various conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Mycobacterium tuberculosis strain Erdman (ATCC 35801) was cultured unshaken in Middlebrook 7H9 (BBL) medium supplemented with either OADC (BBL) or albumin, dextrose, and saline (ADS) and with 0.05% Tween 80 (9) or in Sauton medium containing 1.5% glycerol and 0.05% Tween 80 (1). Solid medium was made with Middlebrook 7H9 medium plus 1.4% Bacto agar (BD) and supplemented with 10% OADC enrichment medium (BBL) unless otherwise indicated.
Construction of the mshD mutant.
A targeted gene disruption in the mshD gene (Rv0819) of M. tuberculosis was produced via allelic exchange using the conditionally replicating mycobacteriophage phAE87 as previously described (21). The mutagenic construct lacked bases 43 to 938 of mshD and contained the gene for hygromycin resistance. Bacteria were infected with a phage containing the mshD mutant construct and then plated onto 7H9 plates supplemented with OADC and containing hygromycin (50 μg/ml). Plates were incubated at 37°C until hygromycin-resistant colonies appeared. DNA was prepared from hygromycin-resistant bacteria and was used for screening by Southern hybridization to identify bacteria with a disruption in mshD. The probe used for hybridization contained the region 248 bases upstream of mshD and the first 256 bases of the gene.
Because we were uncertain whether a mutation in the mshD gene would be lethal in M. tuberculosis, a strain with two copies of mshD was constructed by using a method previously described (21) and was also mutated. To construct the strain with two copies of mshD, the mshD open reading frame plus the ribosomal binding site of M. tuberculosis were amplified by PCR and incorporated into the chromosome of M. tuberculosis at the att site under the control of the aph promoter using the integrative vector pCV125 (3). The M. tuberculosis strain containing two copies of mshD (wild type + pCV125::mshD) was infected with the mshD mutagenic phage, and clones with a disruption in the native copy of mshD were identified by Southern hybridization.
Thiol and disulfide analyses.
Samples for thiol and disulfide analyses were prepared from bacteria grown in Middlebrook 7H9 medium supplemented with ADS and Tween 80. The optical density of each culture was determined at 600 nm, and an appropriate amount of bacteria was pelleted. For thiol analysis, pellets were resuspended in 50% acetonitrile-20 mM HEPES (pH 8.0) containing 2 mM monobromobimane (Molecular Probes). For disulfide analysis, the pellets were resuspended in 50% acetonitrile-20 mM HEPES (pH 8.0) containing 5 mM N-ethylmaleimide (Sigma). Bacteria were inactivated, and the cell contents were extracted by placing the samples in a 60°C water bath for 15 min and then chilling the samples. Cell debris was then pelleted, and the supernatants were stored at −70°C until analysis. Thiol analysis using high-performance liquid chromatography was performed according to protocols previously described for mycothiol determination (4), Cys-GlcN-Ins determination (2), and CoA analysis by method 2 (5). Disulfide analysis was performed according to a previously described protocol (14).
Sensitivity to hydrogen peroxide.
Sensitivity to killing by hydrogen peroxide was tested using early- to mid-log-phase bacteria grown in Sauton medium. Bacteria were adjusted to an optical density at 600 nm (OD600) of 0.08 in Sauton medium, and hydrogen peroxide was added to the test bacteria to give a final concentration of 5.0 mM. Control and test bacteria were incubated at 37°C until sampling at 4 h and 6 h. For sampling, bacteria were diluted in phosphate-buffered saline and plated onto Middlebrook 7H9 plates supplemented with OADC. Tests for each sample were performed in triplicate, and data are expressed as means plus standard errors of the means.
Growth at low pH.
The pH of Middlebrook 7H9 medium plus ADS was buffered with 100 mM 4-morpholinepropanesulfonic acid adjusted to pH 7 and filter sterilized or buffered with 100 mM 4-morpholineethanesulfonic acid adjusted to pH 5.5, 6.0, or 6.5 with NaOH and filter sterilized. Mutant and wild-type bacteria were adjusted to an OD600 of 0.08 in each of the media, and growth was monitored over time by changes in the OD600.
RESULTS
Generation of an M. tuberculosis mshD mutant.
The MshD protein of M. tuberculosis belongs to the GCN5-related family of N-acetyltransferases (GNAT) (8). The GNAT domain functions as the binding site for the substrate acetyl-CoA. MshD contains two GNAT domains at amino acids 1 to 140 and at amino acids 151 to 315 (27, 28). The C-terminal GNAT domain contains the catalytic site, while the N-terminal domain contributes to the structural stability of the protein (28). For our targeted disruption of mshD, both GNAT domains were removed by deleting amino acids 14 through 312 of the protein (bases 43 to 938) and replacing them with the gene for hygromycin resistance. This mutagenic construct replaced the wild-type mshD gene by allelic exchange using a temperature-sensitive phage, and potential mutants were selected by hygromycin resistance (4). A mutant containing an mshD gene disruption (strain ΔmshD) was identified by Southern hybridization. In the strain wild type + pCV125::mshD, carrying two copies of mshD, a clone with a gene disruption in the wild-type copy of mshD was identified. Two samples of the ΔmshD mutant strains were grown up, and their thiol levels were compared with those of wild-type M. tuberculosis and with those of bacteria containing two copies of mshD. We looked for thiols that had previously been observed in the mshD mutant of M. smegmatis, including fCys-GlcN-Ins and Cys-GlcN-Ins (14). In our initial analysis, the ΔmshD mutant produced <0.1% to 2% of the wild-type levels of MSH and significant amounts of fCys-GlcN-Ins and Cys-GlcN-Ins (Table 1). Bacteria with a mutation in the wild-type gene but containing a functional complementing copy of mshD produced wild-type levels of MSH and insignificant amounts of fCys-GlcN-Ins and Cys-GlcN-Ins. Thiol levels in the ΔmshD mutant were higher in the denser culture, suggesting a relationship with growth phase. This question was addressed in further experiments.
TABLE 1.
Major thiols in M. tuberculosis wild-type and mshD mutant strains and in their pCV125::mshD complement strains
| Strain | OD600 | Major thiol content (nmol/109 cells)
|
||
|---|---|---|---|---|
| MSH | Cys-GlcN-Ins | fCys-GlcN-Ins | ||
| Wild type | 0.28 | 27 | <0.02 | <0.02 |
| Wild type | 0.47 | 27 | <0.02 | <0.02 |
| ΔmshD mutant | 0.22 | <0.03 | 0.71 | 1.36 |
| ΔmshD mutant | 1.07 | 0.59 | 4.8 | 4.4 |
| Wild type + pCV125::mshD | 0.36 | 31 | <0.02 | <0.02 |
| Wild type + pCV125::mshD | 0.92 | 35 | <0.02 | <0.02 |
| ΔmshD mutant + pCV125::mshD | 0.42 | 39 | <0.02 | <0.02 |
| ΔmshD mutant + pCV125::mshD | 1.01 | 46 | <0.02 | <0.02 |
Thiol analysis of wild-type and ΔmshD mutant strains.
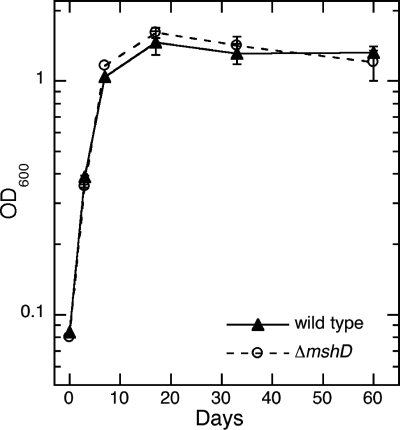
Thiol determinations with monobromobimane were used to assay the thiol composition of the wild-type and ΔmshD strains cultured over a 60-day period, enabling us to sample during all phases of growth, from early exponential phase to long-term stationary phase. The ΔmshD mutant grew in liquid medium at a rate identical to that of wild-type M. tuberculosis under these conditions (Fig. 2). In addition to the thiol data shown (Table 2), hydrogen sulfide was detected, presumably derived from iron-sulfur proteins, but was not quantitated.
FIG. 2.
Growth (OD600) of wild-type (▴) and ΔmshD (○) M. tuberculosis cells in Middlebrook 7H9 medium plus ADS versus time.
TABLE 2.
Thiol levels in wild-type and ΔmshD strains of M. tuberculosis and M. smegmatis
| Thiol | Strain | Thiol level (nmol per 109 cells)
|
|||||
|---|---|---|---|---|---|---|---|
|
M. tuberculosisa (mean ± SEM)
|
M. smegmatisb (range) | ||||||
| Day 3 | Day 7 | Day 17 | Day 33 | Day 60 | |||
| MSH | Wild type | 18 ± 1 | 22 ± 2 | 64 ± 5 | 75 ± 5 | 65 ± 11 | 20-28 |
| ΔmshD | 0.25 ± 0.23 | 0.21 ± 0.13 | 0.65 ± 0.10 | 1.7 ± 0.3 | 1.9 ± 0.3 | 0.07-0.32 | |
| Cys-GlcN-Ins | Wild type | ≤0.02 | ≤0.01 | 0.038 ± 0.003 | 0.06 ± 0.03 | 0.09 ± 0.03 | <0.02 |
| ΔmshD | 2.7 ± 0.2 | 3.4 ± 0.14 | 11 ± 3 | 11 ± 3 | 5.8 ± 0.4 | 1.0-3.3 | |
| fCys-GlcN-Ins | Wild type | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.02 |
| ΔmshD | 4.9 ± 0.3 | 2.7 ± 0.1 | 8.9 ± 0.8 | 6.6 ± 0.5 | 17 ± 2 | 3.8-7.2 | |
| succCys-GlcN-Ins | Wild type | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.02 |
| ΔmshD | <0.02 | 0.03 ± 0.03 | 0.14 ± 0.01 | 0.38 ± 0.06 | <0.02 | 0.06-0.36 | |
| Cys | Wild type | ≤0.06 | ≤0.04 | ≤0.03 | ≤0.03 | ≤0.03 | 0.14-0.46 |
| ΔmshD | ≤0.06 | 0.17 ± 0.01 | 0.21 ± 0.02 | 0.21 ± 0.05 | 0.12 ± 0.01 | 0.04-0.16 | |
| CoA | Wild type | ≤0.8 | ≤0.8 | 2.1 ± 0.3 | 4.1 ± 0.2 | 5.1 ± 0.6 | 0.7-5 |
| ΔmshD | 1.3 ± 0.2 | ≤0.8 | 2.2 ± 0.1 | 2.4 ± 1.7 | 4.9 ± 0.5 | 1.0-4 | |
Means and standard errors from triplicate determinations.
Range of values calculated from data in reference 16, based upon the formula 1 μmol per g residual dry weight = 2 nmol per 109 cells.
Mycothiol is the dominant thiol in wild-type M. tuberculosis, and the MSH level increases more than threefold in stationary phase (Table 2), consistent with previous reports (4, 10). The second most prevalent thiol was CoA. Cys-GlcN-Ins is an intermediate in MSH biosynthesis and is the immediate substrate for MshD. Small amounts of Cys-GlcN-Ins were detected in wild-type bacteria, but these amounts were generally 2 orders of magnitude lower than those in the ΔmshD mutant. In the mutant, the dominant thiols were fCys-GlcN-Ins and Cys-GlcN-Ins, followed by CoA, MSH, and Cys, in that order of prevalence (Table 2). The MSH level in the mutant increased from ∼1% of the wild-type level during exponential growth to ∼3% of the wild-type level in late stationary phase, a nearly eightfold increase in MSH content. The levels of Cys-GlcN-Ins, fCys-GlcN-Ins, Cys, and CoA all increased at least severalfold during the course of growth of the ΔmshD mutant. The other novel thiol, succ-Cys-GlcN-Ins, was also detected, but the levels were significantly above background only during the central stages of the growth cycle.
The ranges of thiol values determined previously for wild-type and ΔmshD mutant strains of M. smegmatis (14) converted to the current units are included in Table 2 for comparison. The MSH and CoA values for the wild-type strains are remarkably similar for M. tuberculosis and M. smegmatis, although the MSH content of M. smegmatis does not increase so dramatically in stationary phase. The ΔmshD mutant of M. tuberculosis produces the same spectrum of thiols as the ΔmshD mutant of M. smegmatis, despite the faster growth rate and larger genome of the latter species. The levels of fCys-GlcN-Ins, CoA, and succ-Cys-GlcN-Ins were largely the same in the two species, but the levels of MSH and Cys-GlcN-Ins were roughly threefold higher in the M. tuberculosis ΔmshD strain than in the M. smegmatis mutant (Table 2).
Previous reports had failed to detect Cys for exponential-phase cells in various M. tuberculosis strains (10, 29), and the present results also show Cys to be undetectable in the Erdman strain (Table 2). However, significant levels of Cys were found in the M. tuberculosis ΔmshD mutant. A similar range of Cys values was found previously (14) for the M. smegmatis mutant, but wild-type M. smegmatis also had measurable levels of Cys (Table 2).
Disulfide content and thiol redox status.
The thiol redox status for M. tuberculosis has not previously been reported. The disulfide or oxidized forms of thiols are produced in the cell by autoxidation as well as by peroxide reductions. In the present study, the individual disulfide contents (RSS) were determined by the reduction of the disulfides with dithiothreitol and high-performance liquid chromatography analysis of the resulting thiols (RSH) as their monobromobimane derivatives (14). Information on the specific composition of the disulfide, the identity of the R and R′ groups in RSSR′, is lost using this protocol, and the data are reported as half-disulfide content. The results for the wild-type and ΔmshD strains are reported in Table 3. No data are provided for CoA because the treatment of acyl-CoAs with dithiothreitol releases CoA through transacylation reactions, and the present protocol would therefore give misleading data for disulfide levels. The results of Tables 2 and 3 were utilized to calculate the thiol-to-disulfide redox ratios (RSH-RSS) presented in Table 4.
TABLE 3.
Disulfide (RSS) levels in wild-type and ΔmshD strains of M. tuberculosis and M. smegmatis
| Thiol | Strain | RSS level (nmol per 109 cells)
|
|||||
|---|---|---|---|---|---|---|---|
|
M. tuberculosisa (mean ± SEM)
|
M. smegmatisb (range) | ||||||
| Day 3 | Day 7 | Day 17 | Day 33 | Day 60 | |||
| MSH | Wild type | 0.10 ± 0.03 | 0.14 ± 0.04 | 0.48 ± 0.07 | 0.78 ± 0.12 | 1.3 ± 0.3 | 0.016-0.16 |
| ΔmshD | 0.008 ± 0.005 | 0.002 ± 0.001 | 0.002 ± 0.001 | 0.005 ± 0.001 | 0.04 ± 0.02 | 0.0016-0.0031 | |
| Cys-GlcN-Ins | Wild type | 0.011 ± 0.002 | 0.011 ± 0.002 | 0.044 ± 0.003 | 0.011 ± 0.005 | 0.016 ± 0.001 | NDc |
| ΔmshD | 0.5 ± 0.06 | 0.89 ± 0.15 | 1.5 ± 0.2 | 0.87 ± 0.12 | 3.0 ± 0.1 | 0.1-2.0 | |
| fCys-GlcN-Ins | Wild type | <0.005 | <0.005 | <0.005 | <0.005 | <0.02 | ND |
| ΔmshD | 0.11 ± 0.01 | ≤0.05 | ≤0.07 | 0.16 ± 0.02 | 0.30 ± 0.03 | 0.02-0.10 | |
| succCys-GlcN-Ins | Wild type | <0.04 | <0.04 | <0.04 | <0.04 | <0.04 | ND |
| ΔmshD | <0.04 | <0.04 | <0.04 | <0.04 | <0.04 | 0.0006-0.008 | |
| Cys | Wild type | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.15 ± 0.11 | 0.22 ± 0.04 | 0.14 ± 0.01 | 0.002-0.020 |
| ΔmshD | 0.02 ± 0.01 | 0.08 ± 0.01 | 0.78 ± 0.11 | 1.1 ± 0.1 | 1.0 ± 0.2 | 0.004-0.018 | |
Means and standard errors from triplicate determinations.
Range of values calculated from data in reference 16 based upon the formula 1 μmol per g residual dry weight = 2 nmol per 109 cells.
ND, not determined.
TABLE 4.
Ratio of thiol content to disulfide content in wild-type and ΔmshD strains of M. tuberculosis and M. smegmatis
| Thiol | Strain | RSH-RSS ratio
|
|||||
|---|---|---|---|---|---|---|---|
|
M. tuberculosisa (mean ± SEM)
|
M. smegmatisb (range) | ||||||
| Day 3 | Day 7 | Day 17 | Day 33 | Day 60 | |||
| MSH | Wild type | 180 ± 50 | 160 ± 50 | 133 ± 22 | 96 ± 16 | 50 ± 14 | 200-1,300 |
| Cys-GlcN-Ins | ΔmshD | 5.4 ± 0.8 | 3.8 ± 0.7 | 7.3 ± 2.2 | 12.6 ± 3.9 | 1.9 ± 0.2 | 1.7-15 |
| fCys-GlcN-Ins | ΔmshD | 45 ± 5 | >54 ± 2 | >130 ± 10 | 41 ± 6 | 57 ± 9 | 39-180 |
| Cys | Wild type | ≤2 ± 1 | ≤2 ± 1 | ≤0.2 ± 0.15 | ≤0.14 ± 0.03 | ≤0.21 ± 0.02 | 30-150 |
| ΔmshD | ≤3 ± 2 | 2.1 ± 0.3 | 0.27 ± 0.05 | 0.19 ± 0.05 | 0.12 ± 0.03 | 10-44 | |
The mycothiol disulfide content of wild-type M. tuberculosis increased markedly, some 13-fold, from early exponential phase to late stationary phase (Table 3), resulting in a substantial fall in the thiol redox ratio (Table 4). A similar pattern was observed for M. smegmatis, but its redox ratios were four- to sevenfold higher. Although Cys in the thiol form was not detectable in M. tuberculosis Erdman (Table 2), the CySS level was measurable (Table 3), allowing an upper limit to be determined for the thiol redox ratio of Cys (Table 4). Although the errors are large, it is evident that a substantial fraction of the Cys is in the disulfide form during exponential growth and that most of the Cys is in the disulfide form during stationary phase. A major difference between M. tuberculosis and M. smegmatis appears to be in the ability of the latter to maintain its Cys content in a substantially reduced state.
The total disulfide content in the M. tuberculosis ΔmshD mutant was substantially higher than that found in the wild-type strain (Table 3). fCys-GlcN-Ins was maintained in a substantially reduced state in the mutant, with the thiol-to-disulfide ratio never falling below 40 (Table 4). However, for the other prominent thiol, Cys-GlcN-Ins, the redox ratio was substantially lower (Table 4), owing to markedly higher disulfide levels (Table 3). As in the wild-type strain, Cys disulfide in the mutant is a significant fraction of the total Cys content in the exponential phase and is the dominant form in the stationary phase (Table 4). The levels of Cys disulfide and Cys-GlcN-Ins disulfide are both high in the mutant during stationary phase, and it seems likely that much of the Cys is in the form of a mixed disulfide with Cys-GlcN-Ins.
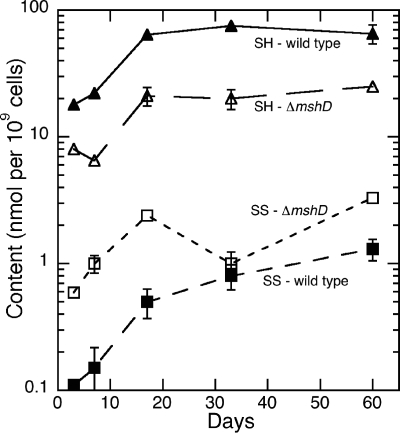
A comprehensive view of the intracellular redox potential of M. tuberculosis wild-type and ΔmshD strains can be made by summing each bacteria's thiol amount and disulfide amount and then determining the ratio. Using this calculation, the impact of a mshD knockout on the overall redox status of M. tuberculosis is apparent (Fig. 3). Production of the novel thiol fCys-GlcN-Ins and elevated levels of Cys-GlcN-Ins in the ΔmshD mutant are not sufficient to compensate for the lack of MSH synthesis. The mutant's thiol content was consistently low, ranging from 27% to 48% of wild-type levels over the 60-day period. At the same time, cellular disulfide levels were significantly elevated in the ΔmshD mutant, with levels about fourfold higher than wild-type values. The combination of these two factors resulted in a combined thiol redox ratio for the mutant of about 13 during exponential phase and falling to about 6 in late stationary phase. The combined redox ratio in wild-type M. tuberculosis was ∼130 in log-phase cells, with a decrease to about 45 at 60 days (Table 4).
FIG. 3.
Total thiol (▴ and ▵) and disulfide (▪ and □) content of wild-type (▴ and ▪) and ΔmshD M. tuberculosis cells grown in Middlebrook 7H9 medium as a function of time. CoA data were not included.
Lack of growth on plates lacking OADC.
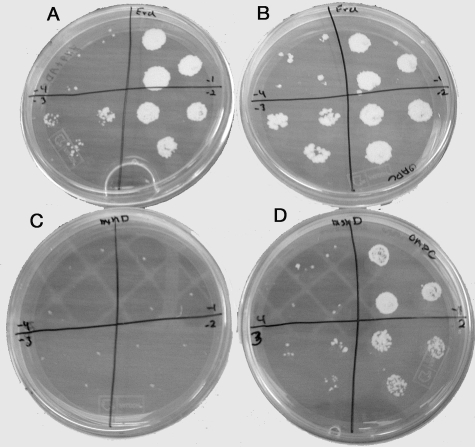
In a previous study of the M. tuberculosis mshB mutant, we had observed that a lack of normal MSH synthesis resulted in poor growth on Middlebrook 7H9 agar plates that lacked OADC (4). The number of mshB mutant colonies that developed on a 7H9 plate supplemented with ADS was 100-fold less than that on plates supplemented with OADC. We therefore wanted to assess the growth of the MshD mutant on Middlebrook 7H9 plates lacking OADC. Wild-type and ΔmshD mutant bacteria were grown to mid-log phase and adjusted to an OD600 of 0.08. Tenfold serial dilutions of each bacterium were spotted onto 7H9 plates supplemented with either OADC or ADS. The plates were then incubated for 3 weeks. On the ADS plate, the ΔmshD mutant failed to grow at even the lowest dilution but grew like the wild type on the OADC plate (Fig. 4). The calculated plating efficiencies for wild-type bacteria were 2.2 × 107 CFU per ml on OADC plates and 2.6 × 107 CFU per ml on ADS plates. The calculated plating efficiencies for the ΔmshD mutant were 8.7 × 106 colonies per ml on OADC plates and <1.2 × 104 colonies per ml on ADS plates. The addition of either oleic acid or catalase to the ADS plates at the same level as that present in the commercial OADC medium produced a moderate increase in growth of the mutant, suggesting that both contribute to the mutant's growth (data not shown).
FIG. 4.
Colony appearance of wild-type (A and B) and ΔmshD M. tuberculosis (C and D) cells grown on Middlebrook 7H9 plates supplemented with ADS (A and C) or OADC (B and D). Cells were plated at dilutions of 10−1 (upper right quadrant), 10−2 (lower right quadrant), 10−3 (lower left quadrant), and 10−4 (upper left quadrant). Plates were photographed after 4 weeks of incubation.
Increased sensitivity to hydrogen peroxide.
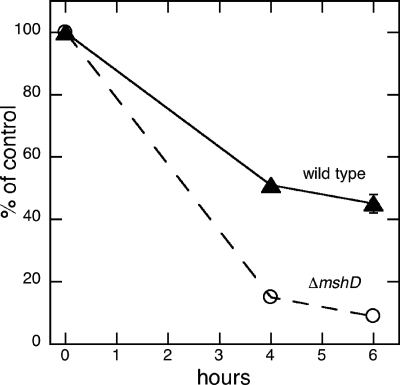
The susceptibility of the ΔmshD mutant to killing by hydrogen peroxide was compared to that of wild-type M. tuberculosis in order to determine whether changes in the thiol makeup of the ΔmshD mutant would alter its response to a toxic oxidant. Bacteria grown in Sauton medium were exposed to 5 mM hydrogen peroxide for 4 h or 6 h and then plated onto 7H9 Middlebrook plates plus OADC to quantitate number of surviving bacteria. The MshD mutant was moderately more sensitive to killing by hydrogen peroxide after both 4 and 6 h of incubation (Fig. 5). When surviving numbers of bacteria were compared with the numbers at time zero, 58% of the wild-type cells survived at 6 h, while 13% of the ΔmshD mutant cells survived.
FIG. 5.
Survival (percentage of control) for wild-type and ΔmshD M. tuberculosis cells incubated at 37°C in 5 mM H2O2 as a function of time.
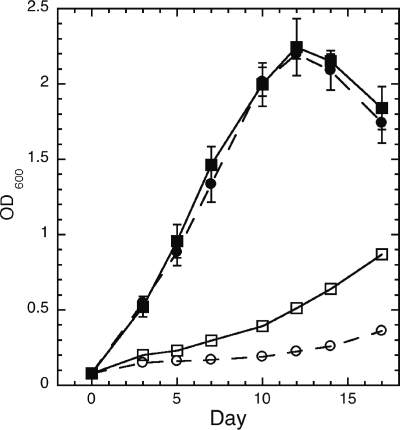
Restricted growth in acidic medium.
A moderately acidic environment in the macrophage is one of the stresses encountered by M. tuberculosis. For that reason, we compared the ability of wild-type M. tuberculosis and the ΔmshD mutant to grow in acidic media. Over a 17-day period, growth of the ΔmshD mutant was more restricted in the pH 5.5 medium compared to growth of the wild type (Fig. 6). Growth was unaltered at pH 7.0 (Fig. 6), 6.5, and 6.0 (data not shown). One possible reason for the reduced growth of the mutant at pH 5.5 is that the formation of necessary thiols was reduced at pH 5.5. Therefore, we analyzed the thiol content of bacteria grown at pH 7.0 and 5.5. Surprisingly, levels of most thiols in both bacteria were elevated in the pH 5.5 medium (Table 5). In wild-type bacteria, mycothiol levels increased 1.8-fold, and in the MshD mutant, fCys-GlcN-Ins increased 3.3-fold and Cys-GlcN-Ins increased 1.9-fold in the pH 5.5 medium. The exception was MSH in the ΔmshD mutant, which fell about twofold.
FIG. 6.
Growth of wild-type (▪ and □) and ΔmshD (• and ○) M. tuberculosis cells at pH 7.0 (▪ and •) and pH 5.5 (□ and ○) in Middlebrook 7H9 medium. Error bars show standard errors of the means for triplicate determinations (not shown where smaller than symbol).
TABLE 5.
Effect of pH upon thiol content of wild-type and ΔmshD M. tuberculosis strains grown in Middlebrook 7H9 medium
| pH | Wild type
|
ΔmshD
|
||||||
|---|---|---|---|---|---|---|---|---|
| OD600 | nmol per 109 cells
|
OD600 | nmol per 109 cells
|
|||||
| MSH | CoA | MSH | Cys-GlcN-Ins | fCys-GlcN-Ins | CoA | |||
| 7.0 | 0.43 | 14 ± 1 | 0.19 ± 0.03 | 0.41 | 0.25 ± 0.16 | 0.83 ± 0.02 | 1.65 ± 0.04 | 0.15 ± 0.05 |
| 5.5 | 0.63 | 26 ± 3 | 0.26 ± 0.02 | 0.36 | 0.13 ± 0.02 | 1.6 ± 0.2 | 5.5 ± 0.3 | 0.33 ± 0.03 |
DISCUSSION
In order to understand the role of MSH in M. tuberculosis, we have constructed gene knockouts in the MSH biosynthesis genes. Inactivation of the mshB gene reduces, but does not eliminate, MSH production, and the mutant grew poorly on solid medium lacking catalase and oleic acid (4). Viable mutants with the native mshC gene inactivated could be obtained only when a second copy of mshC was present (21). The present studies show that an mshD mutant of M. tuberculosis can be grown in rich liquid culture but that it grows poorly under conditions of oxidative stress. The mutant accumulates substantial levels of Cys-GlcN-Ins, which serves as a precursor for the production of a low level of MSH via a chemical transacetylation reaction with acetyl-CoA and of a substantial level of fCys-GlcN-Ins as reported previously for M. smegmatis (14). The production of these thiols apparently permits the growth of the ΔmshD mutant in rich medium, whereas an mshC mutant cannot produce any of these thiols and is unable to grow.
There are substantial reasons to believe that fCys-GlcN-Ins can serve as a modest replacement for MSH in at least some MSH-dependent enzymatic processes and that Cys-GlcN-Ins is a significantly poorer substitute. As a substrate for M. tuberculosis mycothiol S-conjugate amidase, the bimane derivative of fCys-GlcN-Ins (fCySmB-GlcN-Ins) had 5% of the activity of the bimane derivative of mycothiol (MSmB), whereas the derivative of Cys-GlcN-Ins (CySmB-GlcN-Ins) had only 0.1% of the activity (25). Another enzyme of M. tuberculosis mycothiol metabolism, mycothiol disulfide reductase (16), reduces the symmetrical disulfide of fCys-GlcN-Ins at 13% of the rate with which it reduces MSSM, but the rate for reduction of the symmetrical disulfide of Cys-GlcN-Ins was not measurable, ≤4% (14). Thus, it appears likely that fCys-GlcN-Ins, in which the acyl group attached to Cys is HCO, can partially substitute for MSH (acyl group = CH3CO) with various enzymes of mycothiol metabolism, whereas Cys-GlcN-Ins, with the positively charged NH3+ group of the Cys residue, is a significantly poorer surrogate.
The present studies represent the first examination of the thiol redox status in M. tuberculosis, and therefore, the results with the wild-type strain are important. The thiol redox status is dominated by mycothiol, and MSH increases substantially in stationary phase (Table 2). However, the disulfide level undergoes an even greater increase (Table 3), with the result that the MSH-MSS ratio falls in stationary phase (Table 4). A striking difference between M. smegmatis and M. tuberculosis was the failure to detect reduced cysteine in M. tuberculosis (Table 2), a result in accord with measurements for other strains of M. tuberculosis (10, 29). Surprisingly, Cys could be measured in the disulfide form (Table 3). As a consequence, the Cys RSH-RSS ratios could be given upper limits only, but these fall from ∼2 in exponential phase to ∼0.2 in stationary phase. This ratio reflects the balance between the rates of oxidative processes producing CySS and the rates of reductive processes that regenerate Cys from CySS forms together with new Cys synthesis. Autoxidation of Cys is a rather rapid process catalyzed by heavy metals for which the thiol and amino groups of Cys serve as good ligands. Auto-oxidation of MSH, in which the amino group is blocked, is much slower (12). Oxidation of Cys and other thiols can also occur through chemical reactions with peroxides and other reactive oxygen species.
How CySS forms are reduced in mycobacteria is unclear. Since the removal of either the inosityl (15) or acetyl (14) residues from mycothiol disulfide markedly decreases the activity with mycothiol disulfide reductase, it seems unlikely that CySS forms are efficiently reduced by this enzyme. In systems where glutathione is the dominant thiol, glutaredoxins play an important role in reducing mixed disulfides of Cys and glutathione with glutathione as a cofactor (7). It is possible that analogous enzymes (mycoredoxins) utilizing MSH to reduce CySSM, the form expected to dominate in wild-type cells, occur in mycobacteria, but they remain to be identified.
The thiol redox status is substantially perturbed in the ΔmshD mutant of M. tuberculosis, compared with the wild-type strain, as a consequence of decreased thiol levels and increased disulfide levels (Fig. 3). In the mutant, fCys-GlcN-Ins and Cys-GlcN-Ins are detected at similar levels, and together, they account for at least ∼80% of the total thiol content. However, the disulfide content of the ΔmshD mutant derives primarily from Cys-GlcN-Ins (Table 3). The thiol redox status of fCys-GlcN-Ins is maintained in a reduced state (Table 4) because fCys-GlcN-Ins disulfide is a reasonable substrate for mycothiol disulfide reductase (14). By contrast, the disulfide of Cys-GlcN-Ins is a poorer substrate for the reductase, and, like Cys, it undergoes rapid autoxidation (14). As a consequence, its thiol redox ratio is substantially lower. The high CySS content in the mutant is likely to be significantly associated with a mixed disulfide of Cys and Cys-GlcN-Ins or with cysteine, both expected to be poor substrates for mycothiol disulfide reductase.
The ΔmshD mutant of M. tuberculosis is more sensitive to various stresses than the wild-type strain. Plating factors for aerobic growth of the ΔmshD mutant on plates containing catalase and oleic acid (Middlebrook 7H9 agar supplemented with OADC) were ∼40% of those for the wild-type strain, but on agar lacking catalase and oleic acid (Middlebrook 7H9 agar supplemented with ADS), growth was severely limited (plating efficiency was 2,000-fold lower than that for the wild type). The mutant was more sensitive to hydrogen peroxide than the wild-type strain (Fig. 5). The altered thiol-disulfide status of the ΔmshD mutant is not sufficiently severe to prevent its growth in rich liquid medium but does limit its ability to grow and survive in environments where oxidative stress is more severe. Growth of wild-type and ΔmshD strains was severely slowed at pH 5.5, with the effect on the mutant being more dramatic (Fig. 6). The mechanism underlying the poorer growth of the mutant at low pH is not apparent, but the phenomenon may contribute to its limited growth in the macrophage.
An important limiting condition during in vivo growth of M. tuberculosis is the environment of the macrophage. The mshD gene was recently reported to be necessary for growth in macrophages during a genome-wide screen of nonessential M. tuberculosis genes (19). In that study, the mshD mutant failed to grow in cultures of primary murine macrophages that modeled all stages of the host immune response. These included inactivated macrophages, gamma interferon-preactivated macrophages, and gamma interferon-postactivated macrophages. However, in a previous study aimed at identifying genes required for growth in vivo using a murine model of infection after an intravenous route of infection, an mshD mutant was not identified as being defective for growth in the spleen (24). This suggests that the mshD gene is critical for survival under some in vivo conditions but not others. Growth within alveolar macrophages in the lungs is critical in tuberculosis infections. The environment of the lung differs considerably from that of the spleen in mice and would likely place more severe demands upon the MSH-dependent protective systems of M. tuberculosis.
The thiol compositions of M. tuberculosis and M. smegmatis and those of their ΔmshD mutants are remarkably similar (Table 2). However, there are some notable differences in the thiol redox ratios. The mycothiol redox ratios for M. smegmatis are greater than those for M. tuberculosis and Mycobacterium bovis BCG (26), and the cysteine redox ratios are dramatically higher (Table 4). Ung and Av-Gay (26) previously showed that the mycothiol redox status of M. smegmatis is markedly more resistant to challenge by hydrogen peroxide and diamide than is that of M. bovis BCG. It therefore appears that M. smegmatis, which has a larger genome size than members of the M. tuberculosis complex, has a greater capacity to resist thiol oxidation, but the basis for this resistance has not been identified.
In conclusion, the present study has shown that the dominant thiols produced by an ΔmshD mutant of M. tuberculosis are Cys-GlcN-Ins and fCys-GlcN-Ins, while MSH is present at a very low level. fCys-GlcN-Ins serves as a modest surrogate for mycothiol in some MSH-dependent reactions and is maintained in a substantially reduced state, whereas Cys-GlcN-Ins is a much poorer surrogate and is found in a less-reduced state. Surprisingly, Cys was found to be substantially oxidized in both wild-type and ΔmshD strains. The altered thiol-disulfide status is adequate for the ΔmshD mutant to grow normally in liquid culture but restricts its ability to grow on plates and at low pH and to resist challenge by hydrogen peroxide. Finally, the thiol compositions of the mshD mutants of M. tuberculosis and M. smegmatis are similar, further demonstrating that M. smegmatis serves as a useful model for the thiol biochemistry of M. tuberculosis.
Acknowledgments
This research was supported by Public Health Service grant AI49174 from the National Institute of Allergy and Infectious Diseases.
We thank Philong Ta for technical assistance in performing the thiol and disulfide analyses.
REFERENCES
- 1.Allen, B. W. 1998. Mycobacteria. General culture methodology and safety considerations. Methods Mol. Biol. 101:15-30. [DOI] [PubMed] [Google Scholar]
- 2.Anderberg, S. J., G. L. Newton, and R. C. Fahey. 1998. Mycothiol biosynthesis and metabolism. Cellular levels of potential intermediates in the biosynthesis and degradation of mycothiol. J. Biol. Chem. 273:30391-30397. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier, N., A. Blanc-Potard, S. Ehrt, D. Piddington, L. Riley, and E. A. Groisman. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35:1375-1382. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., G. L. Newton, T. Koledin, and R. C. Fahey. 2003. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 47:1723-1732. [DOI] [PubMed] [Google Scholar]
- 5.Fahey, R. C., and G. L. Newton. 1987. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 143:85-96. [DOI] [PubMed] [Google Scholar]
- 6.Hand, C. E., and J. F. Honek. 2005. Biological chemistry of naturally occurring thiols of microbial and marine origin. J. Nat. Prod. 68:293-308. [DOI] [PubMed] [Google Scholar]
- 7.Johansson, C., C. H. Lillig, and A. Holmgren. 2004. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J. Biol. Chem. 279:7537-7543. [DOI] [PubMed] [Google Scholar]
- 8.Koledin, T., G. L. Newton, and R. C. Fahey. 2002. Identification of the mycothiol synthase gene (mshD) encoding the acetyltransferase producing mycothiol in actinomycetes. Arch. Microbiol. 178:331-337. [DOI] [PubMed] [Google Scholar]
- 9.Larsen, M. H. 2000. Some common methods in mycobacterial genetics, p. 313-320. In G. F. Hatfull and W. R. Jacobs (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 10.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. delCardayré, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. N-Acetyl-1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside deacetylase (MshB) is a key enzyme in mycothiol biosynthesis. J. Bacteriol. 182:6958-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton, G. L., C. A. Bewley, T. J. Dwyer, R. Horn, Y. Aharonowitz, G. Cohen, J. Davies, D. J. Faulkner, and R. C. Fahey. 1995. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur. J. Biochem. 230:821-825. [DOI] [PubMed] [Google Scholar]
- 13.Newton, G. L., and R. C. Fahey. 2002. Mycothiol biochemistry. Arch. Microbiol. 178:388-394. [DOI] [PubMed] [Google Scholar]
- 14.Newton, G. L., P. Ta, and R. C. Fahey. 2005. A mycothiol synthase mutant of Mycobacterium smegmatis produces novel thiols and has an altered thiol redox status. J. Bacteriol. 187:7309-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel, M. P., and J. S. Blanchard. 1999. Expression, purification, and characterization of Mycobacterium tuberculosis mycothione reductase. Biochemistry 38:11827-11833. [DOI] [PubMed] [Google Scholar]
- 16.Patel, M. P., and J. S. Blanchard. 2001. Mycobacterium tuberculosis mycothione reductase: pH dependence of the kinetic parameters and kinetic isotope effects. Biochemistry 40:3119-3126. [DOI] [PubMed] [Google Scholar]
- 17.Rawat, M., S. Kovacevic, H. Billman-Jacobe, and Y. Av-Gay. 2003. Inactivation of mshB, a key gene in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Microbiology 149:1341-1349. [DOI] [PubMed] [Google Scholar]
- 18.Rawat, M., G. L. Newton, M. Ko, G. J. Martinez, R. C. Fahey, and Y. Av-Gay. 2002. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 102:8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55:21-48. [DOI] [PubMed] [Google Scholar]
- 21.Sareen, D., G. L. Newton, R. C. Fahey, and N. A. Buchmeier. 2003. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 185:6736-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sareen, D., M. Steffek, G. L. Newton, and R. C. Fahey. 2002. ATP-dependent L-cysteine:1D-myo-inosityl 2-amino-2-deoxy-α-D-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry 41:6885-6890. [DOI] [PubMed] [Google Scholar]
- 23.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 24.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffek, M., G. L. Newton, Y. Av-Gay, and R. C. Fahey. 2003. Characterization of Mycobacterium tuberculosis mycothiol S-conjugate amidase. Biochemistry 42:12067-12076. [DOI] [PubMed] [Google Scholar]
- 26.Ung, K. S., and Y. Av-Gay. 2006. Mycothiol-dependent mycobacterial response to oxidative stress. FEBS Lett. 580:2712-2716. [DOI] [PubMed] [Google Scholar]
- 27.Vetting, M. W., S. L. Roderick, M. Yu, and J. S. Blanchard. 2003. Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases. Protein Sci. 12:1954-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vetting, M. W., M. Yu, P. M. Rendle, and J. S. Blanchard. 2006. The substrate-induced conformational change of Mycobacterium tuberculosis mycothiol synthase. J. Biol. Chem. 281:2795-2802. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler, P. R., N. G. Coldham, L. Keating, S. V. Gordon, E. E. Wooff, T. Parish, and R. G. Hewinson. 2005. Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic mycobacteria. J. Biol. Chem. 280:8069-8078. [DOI] [PubMed] [Google Scholar]