Abstract
The tad locus of Actinobacillus actinomycetemcomitans encodes a molecular transport system required for tenacious, nonspecific adherence to surfaces and formation of extremely strong biofilms. This locus is dedicated to the biogenesis of Flp pili, which are required for colonization and virulence. We have previously shown that 11 of the 14 tad locus genes are required for adherence and Flp pilus production. Here, we present genetic and phylogenetic analyses of flp-2, tadV, and rcpB genes in biofilm formation. We show that tadV, predicted to encode prepilin peptidase, is required for adherence. In contrast, targeted insertional inactivation of flp-2, a gene closely related to the prepillin gene flp-1, did not abrogate biofilm formation. Expression studies did not detect Flp2-T7 protein under standard laboratory conditions. We present phylogenetic data showing that there is no significant evidence for natural selection in the available flp-2 sequences from A. actinomycetemcomitans, suggesting that flp-2 does not play a significant role in the biology of this organism. Mutants with insertions at the 3′ end of rcpB formed biofilms equivalent to wild-type A. actinomycetemcomitans. Surprisingly, 5′ end chromosomal insertion mutants in rcpB were obtained only when a wild-type copy of the rcpB gene was provided in trans or when the Tad secretion system was inactivated. Together, our results strongly suggest that A. actinomycetemcomitans rcpB is essential in the context of a functional tad locus. These data show three different phenotypes for the three genes.
Actinobacillus actinomycetemcomitans, a facultative anaerobic, gram-negative coccobacillus that belongs to the Pasteurellaceae family, is strongly associated with human disease, such as localized aggressive periodontitis and abscess formation and is also implicated in gram-negative infective endocarditis (4, 9, 15, 26, 41). Clinical isolates of A. actinomycetemcomitans display a rough colony morphology phenotype with star-like centers and adhere tenaciously to surfaces to form extremely strong biofilms (8, 19, 39). The tenacious biofilm of A. actinomycetemcomitans is critical for colonization and persistence in the oral cavity (7, 8) and has been shown to be required for pathogenesis in a rat model (33). The ability to form such tenacious biofilms is encoded by the tight adherence (tad) locus (18, 27, 28), which encodes the constituents of a secretion system (28) for the production of long, bundled Flp pili, also referred to as fibrils (16, 17, 19). The secretion and assembly of Flp pili are also required for cellular autoaggregation and rough colony morphology (20).
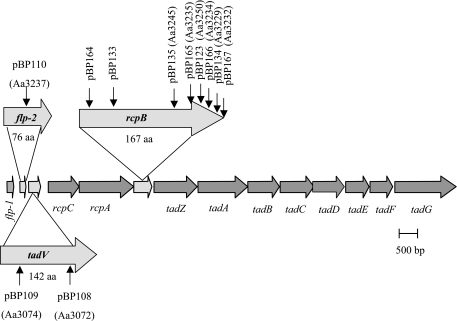
The tad locus is arranged as an operon that extends from flp-1 to tadG (Fig. 1) and is expressed as a single transcriptional unit from flp-1 through at least tadD (11). The product of the flp-1 prepilin gene is the major component of purified pili (17, 19). Flp1 is a type IVb prepillin protein (20) that is posttranslationally modified, most likely by glycosylation, and processed into a mature form prior to pilus assembly (16). The tadA gene product is homologous to secretion NTPases of type IV secretion, such as VirB11, and has been shown to have ATPase activity (1, 27). Two outer membrane components encoded by the tad locus, RcpA and RcpB, were identified in rough strains of A. actinomycetemcomitans (12). RcpA is a member of the GspD/OutD/PulD secretin family of proteins (12, 19) and is predicted to form the outer membrane channel for the secretion of pili (19). Many of the proteins encoded by the tad locus have no assigned functions and have no obvious homology to other known members of prokaryotic secretion systems.
FIG. 1.
Arrangement of the tad genes in A. actinomycetemcomitans. The location of the EZ::TN<KAN-2> insertions in specific plasmids and mutant strains (in parentheses) is shown by vertical arrows.
Comparative phylogenetic analysis has strongly suggested that the tad locus is carried on a mobile genomic island, referred to as the widespread colonization island (WCI) (28). The WCI is found in diverse bacterial and archaeal species. Bacteria that harbor the WCI include animal and plant pathogens and environmental bacteria (19, 28). Microcolony formation and adhesion by Haemophilus ducreyi, the etiological agent of chancroid, is dependent on an intact tad locus (24), and infection by a tadA mutant of H. ducreyi did not result in pustule formation in a human chancroid model (37). Similarly, signature tag mutagenesis studies revealed that a tadD mutant and a predicted flp-1 promoter mutant of Pasturella multocida, the causative agent of fowl cholera, were attenuated for virulence in mouse and chicken infection models (10, 14). A recent study in Yersinia ruckeri, which causes redmouth disease in fish, has shown that tadD expression is upregulated in vivo (6). In the environmental organism Caulobacter crescentus, the WCI, known as the cpa locus, has been shown to be required for the assembly of a polar pilus that likely mediates adhesion of the motile swarmer cell to surfaces (35, 36). Many other human and plant pathogens, including Pseudomonas aeruginosa and Mycobacterium tuberculosis, as well as the Yersinia, Burkholderia, and Bordetella species, harbor homologous WCI regions, raising the possibility that the tad genes may be important for these organisms to colonize their respective environments.
In previous studies, random mutagenesis and genetic complementation were utilized to demonstrate the requirement of 11 tad locus genes (flp-1, rcpA, rcpC, and tadZABCDEFG) for adherence (18, 20, 28). Mutations in any of these 11 tad genes resulted in the loss of all adherence-related phenotypes of biofilm formation, autoaggregation, rough colony morphology, and Flp pili production. We were unable to identify transposon insertions in flp-2, tadV, and rcpB. A recent study by Wang and Chen (40), in which insertional mutations were targeted to every gene in the tad locus of A. actinomycetemcomitans (40), also did not show a role for flp-2, tadV, and rcpB in tenacious biofilm formation or Flp pilus production. The authors did not genetically complement the mutations in flp-2, tadV, and rcpB; therefore, the involvement of all three genes remains unclear.
Sequence similarities suggested potential roles for flp-2 and tadV. The predicted product of flp-2 is similar to type IV pilins and demonstrates 50% amino acid identity to Flp1, the major pilin subunit (17, 20). The predicted product of tadV is similar to prepillin peptidases found in type II secretion systems (28), and its homolog in C. crescentus, cpaA, has been shown to be required for pilus formation (35). Here we show that tadV is also essential for adherence and Flp fibril production. Our original mutagenesis scheme failed to detect it (18, 28). The rcpB gene is unique to the tad loci and found only in close relatives of A. actinomycetemcomitans. The rcpB gene has no clear predicted function.
Here we use a targeted mutagenesis strategy (M. Bhattacharjee and D. H. Figurski, unpublished data), coupled with genetic complementation analysis, to test whether flp-2, tadV, and rcpB are required for biofilm formation. We show that tadV is required for adherence, whereas mutations in flp-2 do not disrupt tenacious biofilm formation, autoaggregation, rough colony morphology, or Flp pilus formation. We also present expression and phylogenetic studies that strongly suggest that flp-2 is no longer expressed in A. actinomycetemcomitans. We were unable to disrupt rcpB in wild-type adherent A. actinomycetemcomitans, but we were able to mutagenize rcpB in a tad mutant strain of A. actinomycetemcomitans that is spontaneously nonadherent, makes no Flp fibrils, does not autoaggregate, and does not make rough colonies. These findings suggest that rcpB is essential to A. actinomycetemcomitans in the context of a functional Tad secretion system. The phenotypes displayed by both flp-2 and rcpB mutants explain why random transposon insertions were not found in flp-2 and rcpB in our early mutagenesis studies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are described in Table 1. A. actinomycetemcomitans strains were stored and cultured in A. actinomycetemcomitans growth medium (AAGM), as previously described (39). DF2200N (serotype a) (22) is a nalidixic acid-resistant variant of the rough clinical isolate DF2200. Aa3138 is a spontaneous, nonadherent variant of DF2200N. When appropriate, the following concentrations of antibiotics were used: chloramphenicol (CHL), 2 μg/ml; kanamycin (KAN), 40 μg/ml; spectinomycin (SPT), 20 μg/ml; and nalidixic acid, 20 μg/ml. Because IncQ plasmid copy numbers are lower than normal for A. actinomycetemcomitans strains Aa3118 and Aa3119, we decreased the concentration of antibiotics in the growth medium. We used 10 μg/ml SPT and 2 μg/ml CHL.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| A. actinomycetemcomitans strains | ||
| Aa0886 | CU1000N tadZ0005::IS903 kan; Km | 28 |
| Aa3072 | DF2200N tadV::KAN-2 (codon position 127) | This study |
| Aa3074 | DF2200N tadV::KAN-2 (codon position 31) | This study |
| Aa3076 | Aa3074 containing pJAK17 | This study |
| Aa3078 | Aa3072 containing pJAK17 | This study |
| Aa3079 | Aa3074 containing pBP114 | This study |
| Aa3081 | Aa3072 containing pBP114 | This study |
| Aa3118 | DF2200N transformation strain containing pJAK12 | This study |
| Aa3119 | DF2200N transformation strain containing pBP152 | This study |
| Aa3138 | Spontaneous smooth, nonadherent variant of DF2200N | This study |
| Aa3229 | DF2200N rcpB:: KAN-2 (codon position 159) | This study |
| Aa3232 | DF2200N rcpB KAN-2 (codon position 167) | This study |
| Aa3234 | DF2200N rcpB:: KAN-2 (codon position 137) | This study |
| Aa3235 | DF2200N rcpB:: KAN-2 (codon position 132) | This study |
| Aa3237 | DF2200N flp-2:: KAN-2 (codon position 42) | This study |
| Aa3245 | DF2200N rcpB:: KAN-2 (codon position 118) | This study |
| Aa3250 | DF2200N rcpB:: KAN-2 (codon position 134) | This study |
| CU1000N | Spontaneous Nalr variant of CU1000 (serotype f) | 7, 18 |
| DF2200 | Wild-type clinical isolate (serotype a) | 22 |
| DF2200N | Spontaneous Nalr variant of DF2200 | 22 |
| E.coli strains | ||
| BP038 | Top10 containing pBP026 | This study |
| BP039 | Top10 containing pBP027 | This study |
| SK108 | Top10 containing pJAK16 | This study |
| SK363 | Top10 containing pSK163 | This study |
| Top10 | F−mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔM15ΔlacX74 deoR nupG recA1 araD139Δ(ara-leu)7697 galU galK rpsL(StrR) endA1-λ− | Invitrogen |
| Plasmids | ||
| pBP022 | pCR2.1 containing flp-2SD -T7 | This study |
| pBP024 | pCR2.1 containing flp-2-T7 | This study |
| pBP026 | pJAK16 containing flp-2SD-T7 | This study |
| pBP027 | pJAK16 containing flp-2-T7 | This study |
| pBP098 | pMB78 containing flp-1- rcpC′ | This study |
| pBP108 | pMB78 containing flp-1- rcpC′ tadV::KAN-2 (codon position 127) | This study |
| pBP109 | pMB78 containing flp-1- rcpC′ tadV::KAN-2 (codon position 31) | This study |
| pBP110 | pMB78 containing flp-1- rcpC′ flp-2::KAN-2 (codon position 42) | This study |
| pBP113 | pCR2.1 containing rcpA-tadZ′ | This study |
| pBP114 | pJAK17 containing tadV | This study |
| pBP120 | pMB78 containing rcpA-tadZ′ | This study |
| pBP123 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 134) | This study |
| pBP131 | pCR2.1 containing rcpB | This study |
| pBP133 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 59) | This study |
| pBP134 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 159) | This study |
| pBP135 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 118) | This study |
| pBP152 | pJAK12 containing rcpB | This study |
| pBP164 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 11) | This study |
| pBP165 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 132) | This study |
| pBP166 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 137) | This study |
| pBP167 | pMB78 containing rcpA-tadZ′ rcpB::KAN-2 (codon position 167) | This study |
| pCR2.1TOPO | E. coli cloning vector; Kmr Apr | Invitrogen |
| pJAK12 | IncQ broad-host-range expression vector; tacp Spr | J. A. Kornacki |
| pJAK16 | IncQ broad-host-range expression vector; tacp Cmr | J. A. Kornacki |
| pJAK17 | IncQ broad-host-range expression vector; tacp Cmr | J. A. Kornacki |
| pMB78 | Vector for allelic exchange and A. actinomycetemcomitans transformations | M. K. Bhattacharjee |
| pSK163 | pJAK16 containing flp-1- T7 | 20 |
| pSK273 | pCR2.1 containing flp-1-rcpC′ | This study |
| pSK288 | pCR2.1 containing tadV | This study |
Nalr, nalidixic acid resistance; Kmr, kanamycin resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Spr, spectinomycin resistance. Codon position refers to the first codon disrupted by the transposon.
Escherichia coli strain Top10 (Invitrogen) was used for DNA subcloning procedures and as a conjugative donor for the mobilization of broad-host-range IncQ expression vectors into A. actinomycetemcomitans. As described previously (34), an RK2 oriT-defective mutant plasmid, pRK21761, was used for mobilization of IncQ vectors. E. coli transformants were cultured in Luria-Bertini (LB) medium and grown at 37°C. When appropriate the following concentrations of antibiotics were used: CHL, 50 μg/ml; KAN, 50 μg/ml; SPT, 50 μg/ml.
DNA procedures.
All plasmid preparations were done using Miniprep Spin Kits (QIAGEN), and DNA manipulations with restriction enzymes and T4 DNA ligase were done according to the manufacturer's recommendations (New England Biolabs). PCR amplification was done with either high-fidelity Triplemaster Taq polymerase or Taq polymerase (Eppendorf) for 30 cycles. Agarose gel electrophoresis was used for purification of PCR amplification products and plasmid digests. Subsequently, SpinX tubes (Corning) or a QIAexII kit (QIAGEN) was used for agarose gel extractions according to the manufacture's instructions. E. coli transformations were done as described previously (3). All cloned PCR products and transposon insertions were confirmed by nucleotide sequencing at the Columbia University DNA sequencing facility using an Applied Biosystem 3100 capillary sequencer.
flp-2 and tadV mutagenesis.
Vectors for allelic exchange were constructed as follows. First, the segment including flp-1-flp-2-tadV-rcpC′ was PCR amplified from genomic DNA extracted from CU1000N using the primers 5′-AACAACAATAGGAGCATTAAGACA-3′ (flpcompUP) and 5′-GGTCATTGTCGACTAGTGCAC-3′ (AaOCL1). This 1,344-bp PCR fragment, which includes 21 bp upstream of flp-1 through 269 bp into rcpC, was cloned into pCR2.1 TOPO (Invitrogen) to generate pSK273. This plasmid was then digested with EcoRI and ligated into pMB78 to generate pBP098, which contains the A. actinomycetemcomitans uptake signal sequence for transformation (39) (Bhattacharjee and Figurski, unpublished data).
The plasmid pBP098 was mutagenized using an EZ::TN <KAN-2> insertion kit (Epicenter), which randomly inserts a 1,221-bp Tn5-based Kmr transposon into the plasmid. Insertions in flp-2 and tadV were screened by PCR and confirmed by nucleotide sequencing. Plasmids containing insertions, in which the transposon's Kmr gene was transcribed in the same direction as the flp-rcp-tad genes, were selected for further studies. A. actinomycetemcomitans strain DF2200N was transformed with linearized pBP108, pBP109, and pBP110 (Table 1) and selected for Kmr transformants. The transformation protocol was provided by M. Bhattacharjee and D. H. Figurski (unpublished data). Transformations resulted in strains that have chromosomal insertions in flp-2 and tadV (designated Aa3237, Aa3072, and Aa3074) (Table 1). To confirm the mutations, genomic DNA was purified from the transformants using the DNAeasy genomic prep kits (QIAGEN) and screened by PCR using the following primers: 5′-TTTTTCAGTACTATAATGCCAC-3′ (flp-pro-UP3), RP-1 (provided with the EZ::TN <KAN-2> insertion kit; Epicenter), 5′-GGGCTATCTACTGCTATATTTTG-3′ (Flp2up), 5′-CATTAATAACCCAGTTCATTTTTTATC-3′ (Flp2dwn), 5′-CTATAGTTAGCAAAAGTAGCTAAAAAATGATAAAAAATG-3′ (AaUOBR1), and 5′-CTGCTCATCAAGACAATATTAG-3′ (AaUOCL1).
Genetic complementation of the tadV mutation.
The tadV gene was amplified from genomic DNA extracted from CU1000N by PCR using the AaUOBR1 and AaUOCL1 primers (see above) and cloned into pCR2.1 TOPO to generate pSK288. This plasmid was digested with BamHI and XhoI, and the tadV fragment was ligated into pJAK17 (J. A. Kornacki, unpublished results) (39), a derivative of pMMB67. The ligated plasmid with insert, pBP114, was mobilized by conjugation into the tadV mutant strains, Aa3072 and Aa3074.
Construction of flp-2-T7 expression vectors.
The flp-2-T7 and flp-2-T7SD (where SD indicates that the construct carries the flp-1 Shine-Dalgarno sequence) open reading frame (ORFs) were PCR amplified from CU1000N genomic DNA using the primer pair 5′-GGGCTATCTACTACTGCTATATTTTG-3′ (Flp2-up) and 5′-TTAACCCATTTGCTGTCCACCAGTCATGCTAGCCATGCTACTTTTGCTAACTATAGCGCC-3′ (Flp2-T7-down) and the pair 5′-AACAACAATAGGAGCATTAAGACAATGATGGATTTACTGGATTACTTTTAC-3′ (Flp1 + Flp2-up) and Flp2-T7-down, respectively. The primer Flp2-T7-down added a DNA coding sequence for the T7 epitope tag to the 3′ end of the flp-2 gene. The PCR fragments were cloned into pCR2.1 TOPO (Invitrogen) to generate pBP024 (flp-2-T7) and pBP022 (flp-2SD-T7). The plasmids were digested with EcoRI, and the flp-2-T7 and flp-2SD-T7 fragments were ligated into pJAK16 (J. A. Kornacki, unpublished results) to generate pBP027 and pBP026, respectively. Plasmids pBP27 and pBP026 were transformed into E. coli Top10 resulting in strains BP039 and BP038.
flp-2 expression and detection.
Expression experiments and detection of Flp2-T7, Flp2SD-T7, and Flp1-T7 proteins were done as described previously (20) with some modifications. All E. coli strains harboring expression vectors were grown overnight in 5 ml of LB broth supplemented with CHL. These overnight cultures were diluted 1:100 into fresh LB broth supplemented with CHL and grown for 3 h. Cultures were supplemented with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to induce flp-2-T7, flp-2SD-T7, and flp-1-T7 expression. Bacterial cells were harvested and lysed by brief sonication. The cell pellet was resuspended and boiled for 5 min in Laemmli buffer. The volumes of bacterial cultures harvested were determined by taking absorbance readings of the optical density at 600 nm (OD600). Equivalent amounts of total protein were loaded on gels for immunoblot analysis.
Immunoblot analysis.
Protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by hybridizing with an α-T7 epitope-tagged antibody (dilution of 1:10,000; Novagen). An α-rabbit immunoglobulin G antibody (Sigma), conjugated to horseradish peroxidase (HRP), was used as a secondary antibody at a dilution of 1:50,000. Femto Western chemiluminescence reagent (Pierce) was used as a substrate for HRP.
Mutagenesis of rcpB.
The rcpA′-rcpB-tadZ′ DNA coding region was PCR amplified from genomic DNA extracted from CU1000N using the primers 5′-ATGGTAAGGTTCTGGCCGAAC-3′ (AaRAR2) and 5′-CGCCCTTACAACTTAATACAGC-3′ (AaXL1). The 1,556-bp PCR fragment includes the last 607 bp of rcpA through 453 bp into tadZ. This PCR generated fragment was cloned into pCR2.1 TOPO to generate pBP113, which was then digested with KpnI and XbaI. The resulting DNA fragment was ligated into pMB78 (digested with KpnI and SpeI) to generate pBP120. The plasmid pBP120 was mutagenized using an EZ::TN <KAN-2> insertion kit. As described above, insertions in the correct orientation were selected for further studies.
Adherent strain DF2200N and the isogenic, nonadherent strain Aa3138 were transformed (as previously described) with linearized and mutagenized pBP164, pBP133, pBP135, pBP165, pBP123, pBP166, pBP134, and pBP167, and we selected for Kmr transformants. To confirm the mutations, purified genomic DNA was screened by PCR using the following primers: 5′-CCCATCACGACCTAAGTTAA-3′ (AaTadXL2), FP-1 (provided with the EZ::TN <KAN-2> insertion kit, Epicenter), 5′-TTCTTAAAAAATAGTGGGTTCATACAATG-3′ (RcpBup), and 5′-GATCTAGTAATAACATAAAAAATACCTTC-3′ (RcpBdwn).
Cloning of rcpB.
The rcpB gene was PCR amplified from genomic DNA extracted from CU1000N using the following primers: 5′-AGATTCTTCCATACGACTCC-3′ (AaRAR3) and 5′-GTCGACTTAATACTTCAATTGAACACGCTGATT-3′ (RcpBSalIdwn). The primer RcpBSalIdwn added an SalI restriction site at the 3′ end of the rcpB gene. The rcpB PCR-generated fragment was cloned into pCR2.1 to generate pBP131, which was digested with BamHI and SalI. The rcpB-containing fragment was ligated into pJAK12 (J. A. Kornacki, unpublished results). This ligated plasmid, pBP152, and pJAK12 were mobilized by conjugation into A. actinomycetemcomitans strain DF2200N to generate Aa3119 and Aa3118, respectively.
Mutagenesis of rcpB with wild-type rcpB expressed in trans.
A. actinomycetemcomitans strains Aa3119 and Aa3118 were transformed as described above with linearized pBP164, pBP133, pBP135, pBP165, and pBP166 and selected for Kmr transformants. To allow the loss of plasmid pBP152 in the transformants, we inoculated each transformant in AAGM without selection, and the transformants were allowed to grow overnight. The transformants were then streaked onto AAGM plates without selection and passaged once or twice on AAGM plates. Ten individual colonies from each transformation strain were spotted on AAGM plates that contained CHL, SPT, KAN, or no antibiotic.
Transmission electron microscopy (TEM).
A. actinomycetemcomitans strains were grown on AAGM plates without antibiotics or with appropriate antibiotics for 3 days at 37°C in 10% CO2, and cells were examined for pili, as described previously (18, 20).
Crystal violet adherence assays.
To assay adherence, we used a previously described 96-well polystyrene crystal violet assay with some modifications (21, 25). A. actinomycetemcomitans strains were inoculated into 5 to 10 ml of AAGM broth. These cultures were allowed to grow for 24 h at 37°C. If necessary, the cultures were scraped to remove adherent bacteria. These bacterial suspensions (20 μl) were added to 96-well polystyrene plates that contained 100 μl of fresh AAGM and allowed to grow for 16 h. The medium was removed, and the cells were washed 15 times with double-distilled H2O to remove loosely adherent or nonadherent cells. The cells were stained with 25 μl of 1% crystal violet for 15 min. The wells were washed another 15 times with double-distilled H2O to remove excess crystal violet, and 200 μl of dimethyl sulfoxide was added to each well. The wells were incubated with dimethyl sulfoxide for 1.5 h to solubilize the crystal violet. The amount of released crystal violet was measured at the optical density at 590 nm in a plate reader (Molecular Devices SpectraMAX 340pc). Samples were tested in triplicate wells, and error bars indicate standard error.
Assay for autoaggregation.
To assay for autoaggregation, A. actinomycetemcomitans strains were physically scraped with sterile wooden sticks to remove adherent bacteria from the walls of the culture tube and vortexed vigorously for several seconds. Autoaggregation was inspected visually for the presence of clumping and by light microscopy using phase-contrast optics.
Database sequence collection and phylogenetic analysis.
We used the application ORFCurator (31) to search all available γ-proteobacteria with the full set of genes from the WCI in A. actinomycetemcomitans HK1651 and C. crescentus. We accepted all loci with two or more ORFs with BLAST hits (e-value less than 0.01) to any WCI gene. A maximum spacing of 5,000 base pairs between ORFs was allowed. We extracted flp gene sequences from these loci and examined each for the existence of the Flp pilin motif (GXXXXEY). We then excluded any flp genes that were not closely linked to those tadA sequences belonging to the γ-proteobacterial clade arising from the horizontal transfer event from the α-proteobacteria (28). This led to the exclusion of all pseudomonad flp sequences and to several flp sequences from members of the Vibrionaceae [Vibrio vulnificus YJ016 (VV2665:NP_935458; VV2664:NP_935457) and Vibrio parahaemolyticus (VP2423:NP_798802)]. No tadA gene was linked to the flp-like ORF extracted from the Shigella flexneri 2a genome, but this gene was included in our analysis based on its similarity to other flp sequences from Enterobacteriaceae and the close relationship of the closely linked tadV-like ORF to other tadV ORFs from this group; flp sequences from close relatives of the γ-proteobacterial clade (as determined by a tadA phylogenetic analysis) from Burkholderia pseudomallei and Magnetospirillum magnetotacticum were included as outgroups. All included sequences with their database sources and accession numbers, if available, are shown in Table 2.
TABLE 2.
The sources of sequences data and accession numbers
| Organism | Strain | Gene name | Genome identifier | Accession no. | Sequencing sourcea |
|---|---|---|---|---|---|
| Actinobacillus actinomycetemcomitans | HK1651 | flp-1 | UO | ||
| flp-2 | |||||
| CU1000 | flp-1 | AAL93266 | |||
| flp-2 | AAN75205 | ||||
| DS7 | flp-1 | AAP43981 | |||
| flp-2 | AAP43982 | ||||
| D9 | flp-1 | AAS00698 | |||
| flp-2 | AAS00699 | ||||
| ATCC29523 | flp-1 | AAS00710 | |||
| flp-2 | AAS00711 | ||||
| ATCC29522 | flp-1 | AAS00708 | |||
| flp-2 | AAS00709 | ||||
| ATCC29524 | flp-1 | AAS00712 | |||
| flp-2 | AAS00713 | ||||
| 310a | flp-1 | BAA21831 | |||
| flp-2 | BAA21931 | ||||
| DF2200 | flp-1 | AAL93276.1 | |||
| flp-2 | AF487926 | ||||
| 29R | flp-1 | AAS00702 | |||
| flp-2 | AAS00703 | ||||
| IDH781 | flp-1 | AAL93271 | |||
| flp-2 | AF487921 | ||||
| Actinobacillus pleuropneumoniae | Serotype 1 | flp-1 | Not annotated | Not annotated | |
| flp-2 | Aple02001968 | ZP_00135453 | |||
| Serotype 5 | flp-1 | UO | |||
| flp-2 | |||||
| Burkholderia pseudomallei | K96243 | flp-1 | BPSL1821 | YP_108420 | |
| Citrobacter rodentium | flp-1 | WTSI | |||
| Erwinia carotovora subsps. atroseptica | flp-1 | BX950851.1 | |||
| Haemophilus aphrophilus | flp-1 | AMNH | |||
| flp-2 | |||||
| Haemophilus ducreyi | flp-1 | HD1312 | NP_873744 | ||
| flp-2 | HD1311 | AAP96132 | |||
| flp-3 | HD1310 | AAP96131 | |||
| Magnetospirillum magnetotacticum | flp-1 | Magn0301062 | ZP_00055934 | ||
| flp-2 | Magn03010625 | ZP_00055935 | |||
| Mannheimia succiniciproducens | MBEL55E | flp-1 | MS1782 | YP_088974 | |
| Pasteurella multocida | Pm 70 | flp-1 | PM0855 | NP_245792 | |
| flp-2 | Not annotated | Not annotated | |||
| Photobacterium profundum | SS9 | flp-1 | Not annotated | Not annotated | |
| Shigella flexneri | 2a 301 | flp-1 | SF3008 | NP_708782 | |
| Vibrio parahaemolyticus | RIMD 2210633 | flp-1 B | VPA0719 | NP_800229 | |
| Vibrio vulnificus | CMCP6 | flp-1 A | Not annotated | Not annotated | |
| flp-1 B | VV20084 | AAO07059 | |||
| YJ016 | flp-1 A | VV2013 | NP_934806 | ||
| flp-1 B | VVA0590 | NP_936646 | |||
| Yersinia enterocolitica | flp-1 | WTSI | |||
| Yersinia pseudotuberculosis | IP32953 | flp-1 | YPTB3369 | YP_071858 |
UO, University of Oklahoma; WTSI, Wellcome Trust Sanger Institute; AMNH, American Museum of Natural History.
We aligned all amino acid sequences using default ClustalW settings in the application BioEdit 7.0.4.1 (13) and then converted the sequences back to the original nucleic acids. Nucleic acids and amino acid alignments were concatenated into a single alignment for further analysis. We used the program PAUP, version 4.0b10 (38), for phylogenetic analysis using parsimony as an optimality criterion. Heuristic searches for the optimal tree topology consisted of 1,000 replicates of random addition followed by the tree branch reconnection technique. All characters and state transformations were given equal weight. To calculate confidence in the resulting trees, we generated Bremer decay indices using the program Autodecay (5) and 1,000 bootstrap replicates in PAUP, with 10 iterations of random addition followed by tree branch reconnection. We used default settings in single-likelihood ancestor counting (SLAC) in the Datamonkey package (23, 29, 30) to produce selection plots of dN-dS (where dN is the value for nonsynonymous mutations and dS is the value for synonymous mutations) scores. Flp amino acid sequences and their alignments are included in Fig. S1 in the supplemental material.
Nucleotide sequence accession numbers.
The DNA sequences of flp-2, tadV, and rcpB from A. actinomycetemcomitans strains DF2200N and IDH781N have been deposited in the GenBank database under the following accession numbers: DQ650320, DQ650321, DQ650322, DQ650323, DQ650324, and DQ650325.
RESULTS
flp-2 and tadV mutagenesis.
To determine if flp-2 and tadV are required for the adherent phenotype in A. actinomycetemcomitans, we made targeted insertional transposon mutations in flp-2 and tadV in the naturally transformable, nalidixic acid-resistant clinical isolate of A. actinomycetemcomitans strain DF2200. We transformed wild-type adherent strain DF2200N with plasmid pBP110, which contains a transposon insertion at codon 42 in flp-2, and with plasmids pBP109 and pBP108, which have transposon insertions at codons 31 and 127 in tadV, respectively (Fig. 1). We obtained many potential recombinants for both flp-2 and tadV. Using PCR amplification, we confirmed that the mutant flp-2 and tadV alleles had recombined in the expected location in the chromosome of each mutant. Primers were designed to the 5′ end of the transposon and to a region upstream of flp-1 in the chromosome that is not found on the suicide vectors, pBP108, pBP109, or pBP110. We also confirmed the presence of each insertional mutation using PCR with primer pairs designed within the coding region of either flp-2 or tadV. In each case, we observed the appropriate shift in molecular weight corresponding to the size of the transposon (1,221 bp) (data not shown).
Phenotypic analysis of tadV and flp-2 mutants.
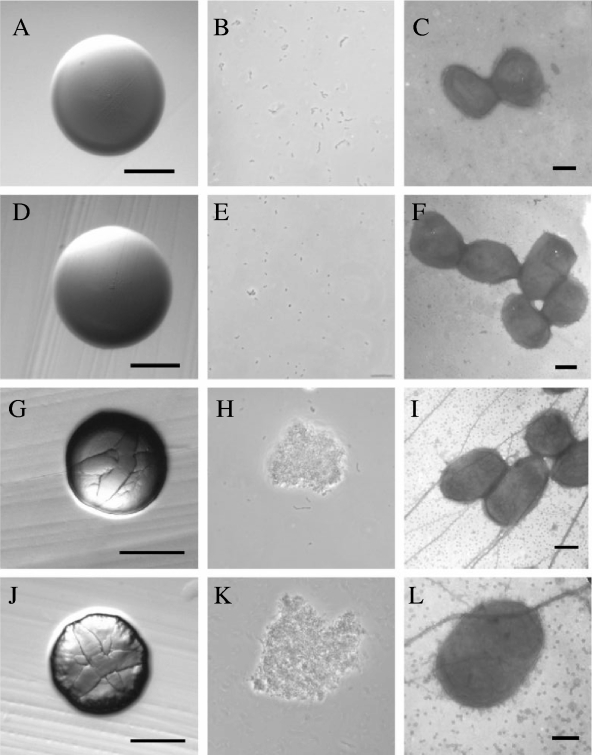
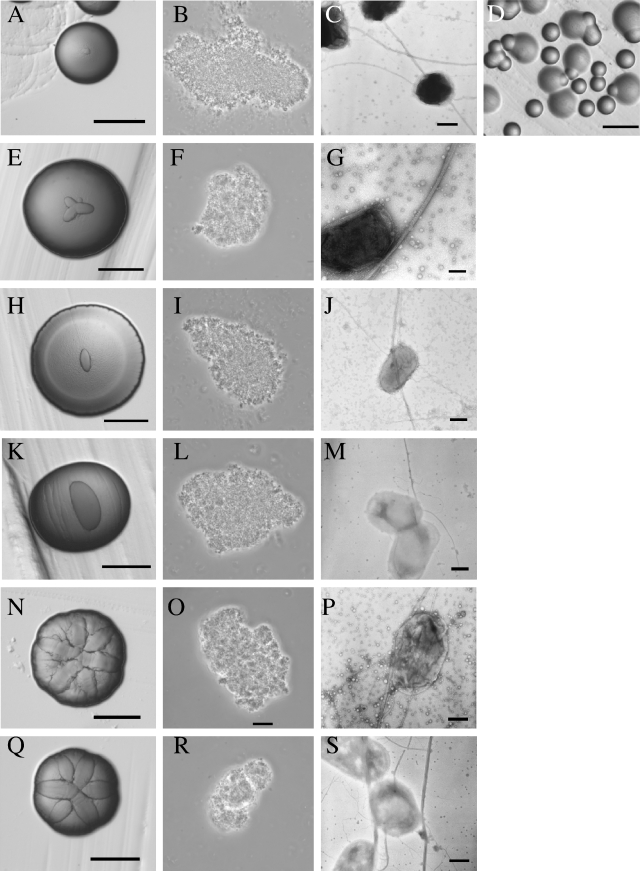
Like other previously examined mutants in the tad gene locus, all tadV mutants (e.g., Aa3074 and Aa3072) have smooth colony morphology and grow planktonically in broth (Fig. 2A and D). The ability of tadV mutants to form biofilms was severely reduced, as measured by a crystal violet biofilm assay (see Fig. 4), and individual bacteria do not clump, as observed under a light microscope (Fig. 2B and E). TEM confirmed that no fibrils are produced by these strains (Fig. 2C and F).
FIG. 2.
Phenotypes and genetic complementation of tadV mutants. tadV mutant strains Aa3074 (A to C) and Aa3072 (D to F), containing the vector pJAK17, and tadV mutant strains Aa3074 (G to J) and Aa3072 (J to L), containing pJAK17 with cloned tadV ORF from strain CU1000N, are shown (bar, 1.0 mm). Images from stereomicroscopy of 3-day-old colonies are shown in panels A, D, G, and J. Images from light microscopy of bacterial cells at magnification of ×100 are shown in panels B, E, H, and K. Images from TEM of negatively stained cells are shown in panels C, F, I, and L (bar, 0.2 μm).
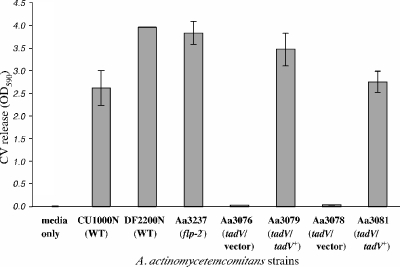
FIG. 4.
Adherence by flp-2 and tadV mutants. Shown are the adherent properties of wild-type A. actinomycetemcomitans strains CU1000N and DF2200N, the flp-2 mutant strain, the tadV mutant strains, and the complemented tadV mutant strains. Adherence was measured in the wells of 96-well microtiter plates. The amount of crystal violet (CV) released is proportional to biofilm formation (25).
In contrast to the tadV mutants, the flp-2 mutant (Aa3237) exhibited no obvious phenotypic differences compared to wild-type strains. Aa3237 displayed rough colony morphology (Fig. 3D) and grew tightly adherent to the culture vessel in broth. In the crystal violet biofilm assay, the flp-2 mutant adhered as well as the wild-type A. actinomycetemcomitans strains CU1000N and DF2000N (Fig. 4). flp-2 mutant cells showed bundled fibrils, and the fibrils appeared morphologically similar to, and as abundant as, those of the wild-type strain DF2200N (Fig. 3F). We noted no differences in autoaggregation of flp-2 mutant (Fig. 3E).
FIG. 3.
Phenotypes of flp2 mutants. Wild-type rough strain DF2200N (A to C) and flp-2 mutant strain Aa3237 (D to F) as described in the legend of Fig. 2. In panels C and F, the bars are 0.2 μm and 0.1 μm, respectively.
Genetic complementation of tadV mutations.
To confirm that tadV is required for biofilm formation by A. actinomycetemcomitans and to ensure that our targeted insertions had no polar effects on the expression of downstream genes in the tad locus, we genetically complemented the tadV mutations. We cloned tadV from the wild-type, adherent strain CU1000N onto a broad-host-range plasmid (pJAK17) that has an IPTG-inducible tacp promoter. The resulting plasmid (pBP114) was transferred to tadV mutants (Aa3074 and Aa3072) using DNA conjugation from an E. coli donor. Genetic complementation of the tadV mutants showed that the transposon insertions in the tadV mutants are not polar. Complemented tadV mutants (Aa3081 and Aa3079) displayed rough colony morphology with star-like centers (Fig. 2G and J). Low levels of tadV expression from the tacp promoter in the absence of IPTG was sufficient to fully restore autoaggregation (Fig. 2H and K), adherence (Fig. 4), and fibril secretion (Fig. 2I and L) in tadV mutants. tadV mutant strains with empty vector (Aa3078 and Aa3076) retained their smooth colony morphology and did not complement for any of the other adherence-related phenotypes (Fig. 2A to F).
flp-2 expression.
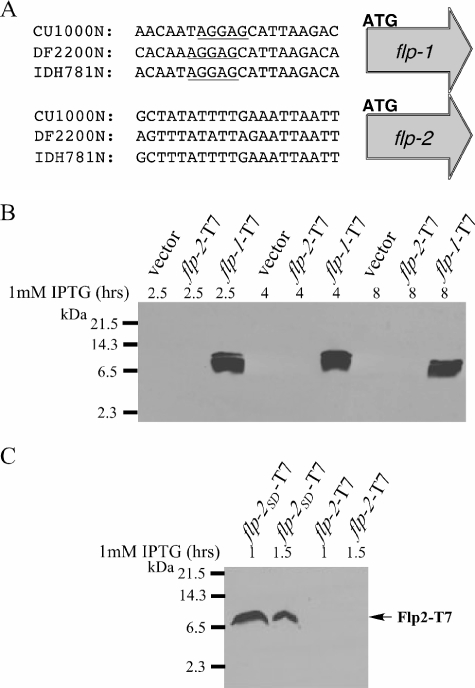
Examination of the upstream region of flp-2 revealed the lack of an obvious Shine-Dalgarno sequence, suggesting that the gene may be expressed poorly or not expressed (Fig. 5A). In contrast, there is a strong Shine-Dalgarno sequence 10 bp upstream of the flp-1 start codon (Fig. 5A). To test whether or not the flp-2 gene product can be detected, we cloned flp-2 with a C-terminal T7 epitope tag into a broad-host IPTG-inducible vector. We induced expression of Flp2-T7 and analyzed cell lysates by Western blotting with anti-T7-tag antibodies. We were unable to detect expression of Flp2-T7 in either E. coli or A. actinomycetemcomitans (Fig. 5B). To test if the lack of or lower expression of Flp2 was due to the absence of a Shine-Dalgarno sequence, we fused the sequence upstream of flp-1 to the flp-2-T7 coding sequence, and induced expression of the resulting construct, designated Flp2SD-T7 from the tacp promoter using IPTG. We were able to detect Flp2SD-T7 protein expression in E. coli within 1 h after the addition of IPTG (Fig. 5C), but expression was not detectable after 2 h postinduction, suggesting that the protein is unstable in this host. We were unable to detect expression of Flp2SD-T7 in A. actinomycetemcomitans (data not shown), indicating that the Flp2 protein is unstable and does not behave similarly to Flp1 (20).
FIG. 5.
Sequence comparison of the upstream regions of flp-1 and flp-2 from A. actinomycetemcomitans strains CU1000N, DF2200N, and IDH781N. (A) The underlined sequence represents the ribosome binding site or Shine-Dalgarno of A. actinomycetemcomitans (http://www.oralgen.lanl.gov/oralgen/bacteria/aact/). (B) Analysis of flp-2-T7 and flp-1-T7 expression. Whole-cell extracts of E. coli strains SK108, SK363, BP039 were prepared and analyzed by α-T7 immunoblotting. Cultures were induced with 1 mM IPTG, as discussed in Materials and Methods, for the indicated amount of time. (C) Analysis of flp-2SD-T7 and flp-2-T7 expression. Whole-cell extracts of E. coli strains BP039 and BP038 were immunoblotted using the α-T7 antibody. Cultures were induced with 1 mM IPTG for the indicated amount of time. The black arrow indicates the position of Flp2-T7. The predicted molecular sizes for immature flp-1 and flp-2 are ∼8 kDa and ∼8.3 kDa, respectively.
flp phylogeny and studies on selection.
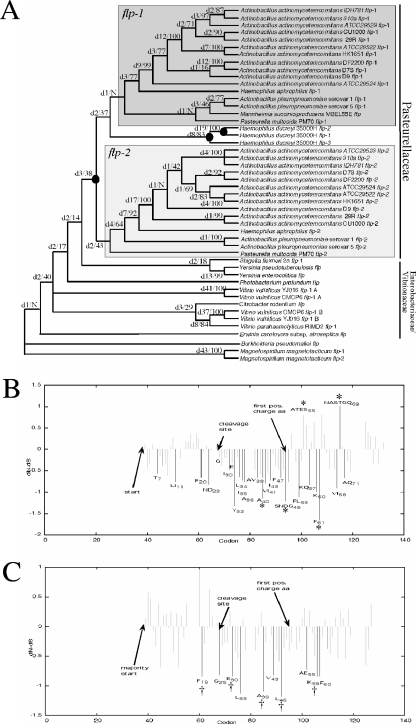
To test if there is evidence that the flp-2 sequence may be acted on by natural selection and, hence, that the gene may have some unrecognized function in the biology of A. actinomycetemcomitans, we did a battery of phylogenetic tests. We first used parsimony analysis to infer the phylogeny of all flp genes recovered from γ-proteobacterial genomes. The phylogeny (Fig. 6A) confirmed an earlier analysis, which suggested that, with the exception of flp alleles found in H. ducreyi, all flp gene alleles in the second position in the gene cluster (i.e., flp-2) formed a monophyletic group, and all flp alleles in the first position in the gene cluster (i.e., flp-1) are also monophyletic. These results indicate that all flp-1 alleles are related to all flp-2 alleles by a single duplication event; i.e., every flp-1 is a paralog of every other flp-2, and vice versa. Our findings also suggested that flp-2 has been conserved and maintained as an independent ORF over a long period of time.
FIG. 6.
Evolution of flp-1 and flp-2 in the γ-proteobacteria. (A) Shown is the strict consensus of the most parsimonious trees found using a concatenated alignment of the amino acid and nucleotide sequences of flp genes. Support for branches is shown on each branch as Bremer decay index/bootstrap percentages. Bremer support values are preceded with the letter “d.” Nodes that did not exist in the bootstrap tree are labeled N. Probable duplication events are indicated with black ovals. Note the duplication that separates flp-1 and flp-2 clades. (B and C) Shown are plots of the normalized value of dN-dS (y axis) along the nucleotide alignment/sequence length (x axis) as computed by the SLAC program (29, 30) for the flp-1 clade and the flp-2 clade, respectively. In this analysis, natural selection was calculated as a difference (dN=dS). The flp-1 analysis (B) also includes the three flp gene sequences from H. ducreyi (24). Positive deflections indicate selection pressure to change, and negative deflections represent selection pressure not to change. Gray lines indicate values with nonsignificant P values (>0.1), and black lines are values with significant P values (<0.1). Codons with significant P values and those in the Flp prepilin motif are designated with their single-letter amino acid codes. The numbers in both panels refer to the position in flp-1 and flp-2, respectively, of A. actinomycetemcomitans HK1651. Asterisks indicate residues with statistically significant dN-dS values when the analysis was restricted to either flp-1 or flp-2 genes from A. actinomycetemcomitans only. Note the lack of asterisks in panel C as no selection was demonstrated for flp-2 genes from A. actinomycetemcomitans. Daggers in panel C indicate residues in flp-2 genes from the Pasteurellaceae shown to have statistically significant selection values even with the exclusion of all A. actinomycetemcomitans flp-2 genes except for flp-2 from HK1651.
Using the clades in the phylogeny, we then divided flp-2 and flp-1 sequences for further analysis. Evidence for natural selection can be inferred from the ratio or difference of rates in hypothesized mutations that lead to changes in amino acid identity (dN) to those that conserve amino acid identity (dS). A ratio of 1 or a difference of 0 suggests that the sequences are neutral to selection. A ratio of >1 or a positive difference is evidence for positive selection, i.e., selection that favors change. A ratio of <1 or a negative difference is evidence of negative selection, i.e., selection that favors conservation.
We used a likelihood-based technique (SLAC) (29, 30) that uses inferences of nucleotide change reconstructed on a given phylogeny using a model of sequence evolution to calculate dN and dS. This test can also assign a P value to the measured ratio to gauge the possibility that the result may have been observed by chance alone. Using the phylogeny as a guide, we tested the following five sequence groupings for evidence of selection: flp-1 from all Pasteurellaceae, flp-1 from A. actinomycetemcomitans strains alone, flp-2 from all Pasteurellaceae, flp-2 from A. actinomycetemcomitans strains alone, and flp-2 from all Pasteurellaceae but with only one A. actinomycetemcomitans strain (HK1651).
For flp-1, each test showed statistically significant evidence for both positive and negative selection both across all Pasteurellaceae and within A. actinomycetemcomitans (Fig. 6B). The negatively selected residues are concentrated in the hydrophobic domain of flp-1. There is more evidence of positive selection in the variable C terminus (Fig. 6B).
Across all Pasteurellaceae, flp-2 also showed evidence for negative (conservative) selection (Fig. 6C), which is consistent with the maintenance of the flp-2 duplicate over a long period of time. However, there was no statistically significant evidence that flp-2 is positively or negatively selected in strains of A. actinomycetemcomitans, suggesting that in these strains flp-2 may be evolving neutrally and may not have a function that is strongly acted on by selection.
rcpB mutagenesis and phenotypic characterization.
To determine if rcpB is required for biofilm formation, we transformed A. actinomycetemcomitans strain DF2200N with eight different plasmids containing insertions throughout the length of the rcpB gene (Fig. 1 and Table 1). The presence of mutant rcpB alleles in the chromosome was determined by PCR amplification using a primer designed to the 3′ end of the transposon and a second primer that anneals at the 3′ end of the coding region of tadZ, a sequence not present on the transformation plasmids. These results were confirmed by observing a 1,221-bp shift in the size of the amplified rcpB gene, corresponding to the size of the transposon (data not shown).
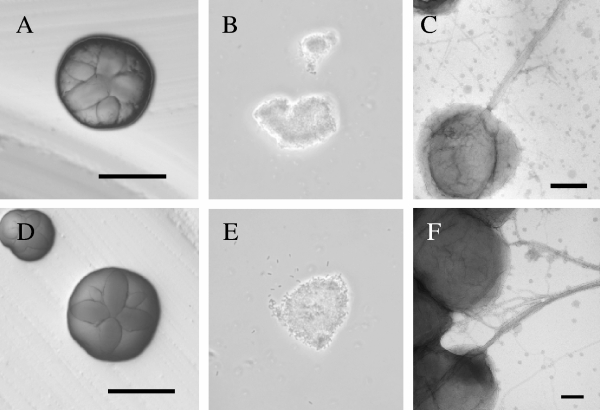
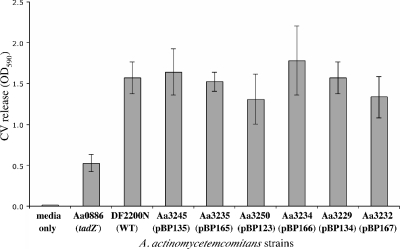
We frequently obtained recombinants with mutant plasmids harboring insertions at the 3′ end or in the last third of the rcpB gene (pBP135, pBP165, pBP123, pBP166, pBP134, and pBP167). Mutant strains with chromosomal insertions in these locations (Aa3245, Aa3235, Aa3250, Aa3234, Aa3229, and Aa3232) have rough colony morphologies and are adherent (Fig. 7 and 8). We rarely obtained recombinants using plasmids in which the transposon insertions were located at the 5′ end of rcpB (pBP164 and pBP133) (Table 3). These recombinants displayed smooth colony morphologies and were nonadherent (data not shown). However, we observed no restoration of any adherence-associated phenotypes in these mutants when rcpB was expressed from a plasmid containing a tacp IPTG-inducible promoter. These results suggested that these mutations either had polar effects on the expression of downstream genes or that each of the 5′ end mutants had undergone an additional mutation event resulting in the loss of the adherent phenotype.
FIG. 7.
Phenotypes of 3′ end rcpB mutants. Shown are images from mutant strain Aa3245 (A to D), mutant strain Aa3235 (E to G), mutant strain Aa3250 (H to J), mutant strain Aa3234 (K to M), mutant strain Aa3229 (N to P), and mutant strain Aa3232 (Q to S). Note the emergence of spontaneous, smooth mutants (D). These figures were done as described in the legend of Fig. 2. Images from TEM of negatively stained cells have bars of 0.1 μm (C and G) and 0.2 μm (J, M, P and S).
FIG. 8.
Adherence by 3′-end rcpB mutants. Adherent properties of wild-type A. actinomycetemcomitans strains DF2200N, the tadZ mutant strain, and the rcpB mutant strains. Adherence was measured in the wells of 96-well microtiter plates, as described in the legend of Fig. 4.
TABLE 3.
Transformation of A. actinomycetemcomitans with rcpB mutant plasmids
| Construct
|
Transformant obtained with DF2200N strains
|
|||||
|---|---|---|---|---|---|---|
| rcpB mutant plasmid | Site of insertion (bp)a | Location | Codon positionb | Adherent wild typec | Nonadherent tad mutantd | Merodiploid (rcpB+ in trans)e |
| pBP164 | 34 | 5′ end | 11 | +/− | + | + |
| pBP133 | 174 | 5′ end | 59 | +/− | + | + |
| pBP135 | 353 | 3′ end | 118 | + | + | + |
| pBP165 | 394 | 3′ end | 132 | + | + | + |
| pBP123 | 401 | 3′ end | 134 | + | NDf | ND |
| pBP166 | 409 | 3′ end | 137 | + | + | + |
| pBP134 | 475 | 3′ end | 159 | + | ND | ND |
| pBP167 | 500 | 3′ end | 167 | + | ND | ND |
Site of insertion refers to the position of EZ::TN<KAN-2> within rcpB (504 bp).
Codon position refers to the codon disrupted by the transposon insertion in RcpB (167 amino acids).
Transformation of wild-type A. actinomycetemcomitans strain DF2200N. +/−, rarely obtained transformants that were nonadherent and unable to be complemented.
Transformation of an isogenic DF2200N spontaneous nonadherent mutant (Aa3138).
Transformation of a DF2200N strain carrying a plasmid expressing a wild-type copy of rcpB (pBP152).
ND, experiment not done.
rcpB mutagenesis in nonadherent and merodiploid A. actinomycetemcomitans.
The low frequency of recombination of the 5′ end rcpB mutants raised the possibility that inactivation of rcpB is toxic to wild-type A. actinomycetemcomitans. Transformation of an isogenic, spontaneous nonadherent, tad mutant of DF2200N (Aa3138) readily yielded recombinants with all rcpB mutant plasmids, regardless of the location of the insertion. Both the 5′ and 3′ end insertional rcpB mutant alleles transformed at comparable frequencies of approximately 4 × 105 transformants/μg of DNA (Table 3). These results suggested that rcpB is only essential in tad+ wild-type A. actinomycetemcomitans. We also transformed another naturally transformable, clinical isolate of A. actinomycetemcomitans, strain IDH781N, and an isogenic nonadherent tad mutant derivative strain. We rarely obtained recombinants when the wild-type IDH781N strain was transformed with rcpB mutant plasmids (e.g., pBP164 and pBP133) harboring 5′ end insertions in rcpB. We obtained recombinants when the nonadherent IDH781N strain was transformed with both 5′ end and 3′ end rcpB mutant plasmids.
We also used the 5′ and 3′ end insertional rcpB mutant alleles to transform a DF2200N strain carrying a plasmid expressing a wild-type copy of rcpB (pBP152; Spr). We readily obtained transformants that were both Kmr and Spr for all rcpB mutant alleles. These results show that allelic exchange can occur with both 5′ and 3′ end insertional rcpB mutant alleles in A. actinomycetemcomitans strains that have rcpB in trans. (Table 3).
We then asked if we could promote the loss of rcpB (pBP152) in these recombinant DF2200N strains. Previous experiments have shown that in the absence of selection, nonessential IncQ-based plasmids are rapidly lost in A. actinomycetemcomitans (M. Bhattacharjee and Figurski, unpublished). After several passages on nonselective medium, recombinants transformed with rcpB mutants with 3′ end insertions were Sps and Kmr or both Kms and Sps, indicating recombination in the chromosome or recombination in the rcpB plasmid pBP152, respectively (Table 3). The only mutants that appeared to require pBP152 (i.e., those that always retained Spr) were those with 5′ end mutations. This result indicates that the loss of the wild-type rcpB gene in rcpB mutants with insertions at the 5′ end may be lethal (Table 3).
Characterization of 3′ end rcpB insertion mutants.
To look for any defects in the 3′ end of rcpB insertion mutants (e.g., Aa3245, Aa3235, Aa3250, Aa3234, Aa3229, and Aa3232), we assayed for biofilm formation, the presence of fibrils, and autoaggregation. In a crystal violet biofilm assay, the rcpB mutants did not appear defective for adherence, relative to the wild-type A. actinomycetemcomitans strain DF2000N (Fig. 8). We observed the presence of bundled fibrils in mutant cells by TEM (Fig. 7C, G, J, M, P, and S), and the abundance and quality of the fibrils are comparable to wild-type A. actinomycetemcomitans. We also observed no change in autoaggregation of cells (Fig. 7B, F, I, L, O, and R). However, careful examination of colony morphology yielded some reproducible differences between strains. Mutants that harbor insertions closer to the 3′ end of rcpB (Aa3229 and Aa3232) display a rough star-like colony morphology, as observed in wild-type DF2200N (Fig. 7N and Q). Colony morphology appears to have less structure with smoother surfaces in mutants that harbor insertions that are closer to the 5′ end of rcpB (e.g., Aa3245, Aa3235, Aa3250, and Aa3234) (Fig. 7A, E, H, and K). We observed the emergence of spontaneous smooth, nonadherent colonies when these “smoother” rcpB mutants (e.g., Aa3245, Aa3235, and Aa3250) were grown on solid medium. We did not observe the emergence of spontaneous-smooth colonies in the mutants with insertions at the very 3′ end of rcpB (e.g., Aa3234, Aa3229, and Aa3232) (Fig. 7D).
DISCUSSION
Transposon mutagenesis studies demonstrated that 11 of 14 genes in the tad locus are required for biofilm formation in A. actinomycetemcomitans (18, 28). In previous work, we did not obtain mutations in flp-2, tadV, or rcpB using a promoterless random transposon. The lack of transposon insertions in flp-2, tadV, and rcpB may be due to the possibility that these genes are not required for adherence, are not expressed, or are essential for viability. A recent mutational analysis of the tad locus in A. actinomycetemcomitans, which involved targeting insertional mutations to each gene, failed to show the requirement of flp-2, tadV, and rcpB for biofilm formation (40). Therefore, the roles of these genes in biofilm formation remained unclear.
To investigate if these genes are required for adherence in A. actinomycetemcomitans, we directed insertional mutations to flp-2, tadV, and rcpB in conjunction with genetic complementation analysis. Our studies tested for all known adherence-related phenotypes: biofilm formation, presence of fibrils, rough colony morphology, and autoaggregation. We determined that tadV is required for biofilm formation. In contrast, flp-2 is not required for any adherence-related phenotypes in A. actinomycetemcomitans and may not be expressed. Mutations in rcpB are lethal to wild-type, adherent A. actinomycetemcomitans.
Similarly to the previously described tad mutants (18-20, 28), tadV mutants are smooth and nonadherent, do not autoaggregate, and do not produce pili. Mutants of tadV were recently described by Wang and Chen as having smooth-colony morphology (40). However, the authors were unable to further characterize these mutants due to an apparent polar effect on the expression of downstream genes. To confirm that tadV is required for adherence and to ensure that our mutants are not polar, we genetically complemented our tadV mutant strains with the wild-type tadV gene in trans. We observed complete complementation of all adherence-related phenotypes. Therefore, tadV is required for biofilm formation in A. actinomycetemcomitans. Recent studies (M. Tomich and D. H. Figurski, submitted for publication) indicate that tadV is the prepillin peptidase required for processing of the Flp1 major pilin subunit (17, 20).
Unlike the phenotypes displayed by previously described tad mutant strains, flp-2 mutants are phenotypically indistinguishable from wild-type A. actinomycetemcomitans. The flp-2 gene is the second flp allele in the tad locus, and its gene product is very similar to Flp1 (17, 20). The loss of adherence in the flp-1 mutant suggested that flp-2 may be a minor pilin subunit that is not sufficient for the assembly of fibrils. Here we have demonstrated that targeted insertional mutations in flp-2 do not affect biofilm formation, autoaggregation, colony morphology, or pilus biogenesis in A. actinomycetemcomitans. Findings by Wang and Chen in the A. actinomycetemcomitans strain D7S suggested that flp-2 mutants are defective for autoaggregation (40); however, we could not detect any differences in autoaggregation between our flp-2 mutant and wild-type A. actinomycetemcomitans. Despite the very close evolutionary relationship between DF2200N and D7S (serotype a) and their flp-2 alleles (Fig. 6A), it is formally possible that these results are due to differences in the strains used in these studies. To confirm our results, we constructed an flp-2 mutation in another wild-type clinical isolate of A. actinomycetemcomitans, IDH781 (serotype d) and observed no change in autoaggregation or any of the adherence-related phenotypes. We speculate that the flp-2 mutation constructed in the Wang and Chen study may be at least partially polar on downstream genes in the locus, explaining the observed partial phenotype (40).
Our results demonstrate clearly that flp-2 is not required for adherence. However, the striking similarity between Flp2 and Flp1, the major pilin subunit (17, 20), suggests that flp-2 may encode a pilin. We hypothesized that flp-2 may be required for adherence under different environmental conditions or that Flp2 might function as a minor pilin. To address these questions, we performed a series of flp-2 expression experiments. We found that inducing expression of flp-2 in trans cannot suppress a flp-1 mutation in A. actinomycetemcomitans and that flp-2 is not expressed. We determined that flp-2 lacks a convincing Shine-Dalgarno sequence, though limited expression of Flp2 is detected from our flp-1 Shine-Dalgarno-flp-2 fusion. Our expression studies suggest that Flp2 does not accumulate in A. actinomycetemcomitans, which is consistent with the flp-2 mutant phenotypes.
Previous work on the flp gene phylogeny suggested that all flp-1 alleles were related to all flp-2 alleles by a single, ancient duplication event that occurred either before the emergence of the Pasteurellaceae lineage or early in the family's history. The two flp alleles suggested that there is, or was, a functional benefit in maintaining duplicate genes. The predicted protein products of both flp-1 and flp-2 have identical or similar residues at equivalent positions, suggesting conservative maintenance of key functional amino acids by natural selection.
From an evolutionary perspective, it could be argued that flp-2 has been conserved as a necessary part of the tad gene secretion system for production of Flp pili. However, our molecular studies strongly suggest that flp-2 has no function in A. actinomycetemcomitans. We offer two possible explanations to reconcile these results: flp-2 might be required for Flp pilus production in A. actinomycetemcomitans only under circumstances absent from our laboratory assays, or flp-2 might be required in other Pasteurellaceae family members but not in A. actinomycetemcomitans. Both possibilities can be addressed by examining the evolution of flp genes in the Pasteurellaceae family and in A. actinomycetemcomitans strains. For instance, natural selection acting on flp-2 in A. actinomycetemcomitans could signal that the gene product is required in circumstances not accounted for in our laboratory assays. We used a test that examines sequence data for evidence of a preponderance of nonneutral evolutionary changes signaled by codon changes that either change (nonsynonymous) or do not change (synonymous) the amino acid. If either change predominates, then natural selection may be acting at that sequence position. An advantage of this type of analysis is that both conservative (negative) selection and selection that favors change (positive) can be detected.
We examined flp-1 and flp-2 alleles for all available Pasteurellaceae family members and then restricted each analysis to available sequence from A. actinomycetemcomitans only. We found significant evidence for selection of flp-1 both in the Pasteurellaceae and when the analysis was limited to A. actinomycetemcomitans. This analysis also showed that the hydrophobic portion of the Flp1 molecule was negatively selected (i.e., selected not to change), while the C terminus was positively selected (i.e., selected to change). This is consistent with the idea that the hydrophobic amino terminal domain is buried in the center of the pilus, where it is constrained by the structural contacts it must make to form the pilus, while the carboxyl terminus might be exposed to the outside and subject to selective pressures in the host or environment.
The flp-2 clade in the Pasteurellaceae also showed evidence for negative selection, albeit less than flp-1. This suggested that the flp-2 gene is or was expressed and acted on by selection at some point in the history of Pasteurellaceae. However, when the analysis was restricted to include only flp-2 from A. actinomycetemcomitans strains, there was no statistically significant evidence of nonneutral selection. In contrast, when the analysis excluded A. actinomycetemcomitans flp-2 genes, evidence for selection was present, suggesting that flp-2 may be a functioning gene in other Pasteurellaceae species. This is consistent with our molecular data that show no apparent role for flp-2 in A. actinomycetemcomitans and suggests that the conservation of flp-2 may be simply vestigial in this species.
The pattern of selection of flp-2 and flp-1 may explain the seemingly paradoxical observation made in some strains of A. actinomycetemcomitans (40) that certain flp-2 ORFs are more similar to one another than their neighboring flp-1 genes are to each other. If flp-2 ORFs are evolving randomly without significant selection while portions of flp-1 genes (e.g., the C terminus) are being positively selected to change, then the active and functional flp-1 may have changed more over a given amount of time than the defunct flp-2. This result illustrates that similarity values are not always the best indicators of sequence conservation through natural selection.
It should be noted, however, that the test employed above can only detect selection at the amino acid level; if the active product of flp-2 is RNA, its selection would not be detected. It is also possible that flp-2 is only selected under extremely rare circumstances and/or that changes in flp-2 only create very subtle defects in fitness.
We also show that rcpB appears to be essential in adherent (tad+) A. actinomycetemcomitans. In this study, we readily obtained recombinants that harbor insertions at the 3′ third of the rcpB gene. These transformants are rough and as adherent as wild-type A. actinomycetemcomitans. This finding suggests that the RcpB protein produced in these mutants is either still functional or not required. Based on the location of the insertions in these mutants, we speculate that the last 48 C terminal residues of RcpB, which comprise 167 amino acids (Fig. 1), may not be required for adherence in A. actinomycetemcomitans.
We only infrequently obtained recombinants using mutant plasmids that have transposon insertions at the 5′ end of the rcpB gene. These recombinants are nonadherent and exhibit smooth-colony morphology, but we did not observe complementation of adherence or adherence-related phenotypes with wild-type rcpB gene provided in trans. These mutants may be polar or have additional mutations that result in the loss of adherence. Wang and Chen (40) described mutations of rcpB to be nonadherent and to have smooth-colony morphologies. They also noted that recombinants in rcpB were rare (40). However, the authors did not complement their rcpB mutations with wild-type rcpB in trans. The 5′-end mutant plasmids harbor insertions that are predicted to eliminate the function of rcpB. The low rate of transformation of these rcpB mutant alleles suggested that loss of rcpB is toxic to wild-type A. actinomycetemcomitans. We readily obtained recombinants when we transformed an isogenic, spontaneous-smooth, nonadherent derivative of DF2200N. We conclude that an intact, wild-type rcpB gene is essential in the context of a functional tad locus.
Although all the chromosomal rcpB mutants (all 3′ end insertion mutations) constructed in this study appeared to be adherent, we did observe slight changes in colony morphology. Additionally, the emergence of spontaneous smooth and nonadherent colonies was only observed in strains that harbored insertions toward the 5′ end of rcpB. These results indicate that mutants that harbor insertions closer to the 5′ end of rcpB are unstable, and these mutations are slightly toxic to A. actinomycetemcomitans. It is possible that smooth, nonadherent variants may have acquired suppressor mutations to relieve the toxicity associated with insertional inactivation of rcpB.
The role of rcpB in biofilm formation by A. actinomycetemcomitans remains unknown. However, the requirement for the RcpB protein in the context of a functional Tad system raises interesting questions about the possible roles of rcpB in adherence. This situation is not unprecedented in the biology of secretion systems. For example, DotL/IcmO, a member the Icm/Dot type IVb system in Legionella pneumophila, has been demonstrated to be essential for viability (2). dotL/icmO mutations and lethality can be suppressed by mutations that inactivate the Icm/Dot secretion system (2). DotL/IcmO has no sequence similarity to RcpB, but it has been proposed to be involved the regulation of type IV secretion.
Because RcpB is located in the outer membrane (12), it is tempting to speculate that the function of RcpB is linked to the RcpA secretin protein. The function of RcpB could be similar to that of protein S (PulS/OutS) of Klebsiella oxytoca and Erwinia chrysanthemi, which is a secretin-stabilizing protein found in the Pul and Out type II secretion systems (32). Although RcpB does not exhibit similarity to protein S, it is possible that RcpB represents a new, unrelated class of secretin-stabilizing proteins.
Supplementary Material
Acknowledgments
We thank Mrinal Bhattacharjee for assistance with transformation experiments and J. Dworkin, A. Mitchell, H. A. Shuman, H. Young, and the members of D. H. Figurski's and D. H. Fine's laboratories for helpful discussions and comments. We also thank S. Silverstein for support and equipment.
This work was supported by grants from the National Institutes of Health to D. H. Figurski (5R01DE014713-04). B.A.P. is supported by the NIH Kirschstein-NRSA Individual Fellowship Program.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buscher, B. A., G. M. Conover, J. L. Miller, S. A. Vogel, S. N. Meyers, R. R. Isberg, and J. P. Vogel. 2005. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J. Bacteriol. 187:2927-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das, M., A. D. Badley, F. R. Cockerill, J. M. Steckelberg, and W. R. Wilson. 1997. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 48:25-33. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson, T. 1997. Autodecay, 2.9.9 ed. Hypercard stack distributed by the author. Botaniska Institutionen, Stockholm University, Stockholm, Sweden.
- 6.Fernandez, L., I. Marquez, and J. A. Guijarro. 2004. Identification of specific in vivo-induced (ivi) genes in Yersinia ruckeri and analysis of ruckerbactin, a catecholate siderophore iron acquisition system. Appl. Environ. Microbiol. 70:5199-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063-1076. [DOI] [PubMed] [Google Scholar]
- 8.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 9.Fives-Taylor, P. M., D. H. Meyer, K. P. Mintz, and C. Brissette. 1999. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol. 2000 20:136-167. [DOI] [PubMed] [Google Scholar]
- 10.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 11.Haase, E. M., J. O. Stream, and F. A. Scannapieco. 2003. Transcriptional analysis of the 5′ terminus of the flp fimbrial gene cluster from Actinobacillus actinomycetemcomitans. Microbiology 149:205-215. [DOI] [PubMed] [Google Scholar]
- 12.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 67:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall, T. A. 1999. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Harper, M., J. D. Boyce, I. W. Wilkie, and B. Adler. 2003. Signature-tagged mutagenesis of Pasteurella multocida identifies mutants displaying differential virulence characteristics in mice and chickens. Infect. Immun. 71:5440-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson, B., M. Wilson, L. Sharp, and J. M. Ward. 2002. Actinobacillus actinomycetemcomitans. J. Med. Microbiol. 51:1013-1020. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, T., H. Ohta, I. Tanimoto, R. Shingaki, and K. Fukui. 2000. Heterogeneous post-translational modification of Actinobacillus actinomycetemcomitans fimbrillin. Microbiol. Immunol. 44:715-718. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, T., I. Tanimoto, H. Ohta, K. Kato, Y. Murayama, and K. Fukui. 1998. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol. 42:253-258. [DOI] [PubMed] [Google Scholar]
- 18.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9:429-437. [DOI] [PubMed] [Google Scholar]
- 20.Kachlany, S. C., P. J. Planet, R. Desalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan, J. B., H. C. Schreiner, D. Furgang, and D. H. Fine. 2002. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J. Clin. Microbiol. 40:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosakovsky Pond, S. L., and S. D. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 24.Nika, J. R., J. L. Latimer, C. K. Ward, R. J. Blick, N. J. Wagner, L. D. Cope, G. G. Mahairas, R. S. Munson, Jr., and E. J. Hansen. 2002. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect. Immun. 70:2965-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 26.Paturel, L., J. P. Casalta, G. Habib, M. Nezri, and D. Raoult. 2004. Actinobacillus actinomycetemcomitans endocarditis. Clin. Microbiol. Infect. 10:98-118. [DOI] [PubMed] [Google Scholar]
- 27.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Planet, P. J., S. C. Kachlany, D. H. Fine, R. DeSalle, and D. H. Figurski. 2003. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34:193-198. [DOI] [PubMed] [Google Scholar]
- 29.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 30.Pond, S. L., S. D. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 31.Rosenfeld, J. A., I. N. Sarkar, P. J. Planet, D. H. Figurski, and R. DeSalle. 2004. ORFcurator: molecular curation of genes and gene clusters in prokaryotic organisms. Bioinformatics 20:3462-3465. [DOI] [PubMed] [Google Scholar]
- 32.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 33.Schreiner, H. C., K. Sinatra, J. B. Kaplan, D. Furgang, S. C. Kachlany, P. J. Planet, B. A. Perez, D. H. Figurski, and D. H. Fine. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA 100:7295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sia, E. A., D. M. Kuehner, and D. H. Figurski. 1996. Mechanism of retrotransfer in conjugation: prior transfer of the conjugative plasmid is required. J. Bacteriol. 178:1457-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommer, J. M., and A. Newton. 1988. Sequential regulation of developmental events during polar morphogenesis in Caulobacter crescentus: assembly of pili on swarmer cells requires cell separation. J. Bacteriol. 170:409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 71:7178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0 beta ed. Sinauer, Sunderland, Mass.
- 39.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Y., and C. Chen. 2005. Mutation analysis of the flp operon in Actinobacillus actinomycetemcomitans. Gene 351:61-71. [DOI] [PubMed] [Google Scholar]
- 41.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.