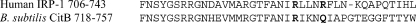Abstract
Bacillus subtilis aconitase, encoded by the citB gene, is homologous to the bifunctional eukaryotic protein IRP-1 (iron regulatory protein 1). Like IRP-1, B. subtilis aconitase is both an enzyme and an RNA binding protein. In an attempt to separate the two activities of aconitase, the C-terminal region of the B. subtilis citB gene product was mutagenized. The resulting strain had high catalytic activity but was defective in sporulation. The defect was at a late stage of sporulation, specifically affecting expression of σK-dependent genes, many of which are important for spore coat assembly and require transcriptional activation by GerE. Accumulation of gerE mRNA and GerE protein was delayed in the aconitase mutant strain. Pure B. subtilis aconitase bound to the 3′ untranslated region of gerE mRNA in in vitro gel mobility shift assays, strongly suggesting that aconitase RNA binding activity may stabilize gerE mRNA in order to allow efficient GerE synthesis and proper timing of spore coat assembly.
Aconitase catalyzes the reversible isomerization of citrate and isocitrate in the second step of the tricarboxylic acid branch of the Krebs citric acid cycle. In Bacillus subtilis, the Krebs cycle is induced during late exponential phase in nutrient-exhausted medium and is responsible for the production of biosynthetic intermediates and ATP and for reducing power during stationary phase and sporulation. When induced, aconitase becomes one of the most abundant proteins in the cell (8). Induction of the aconitase gene, citB, is dependent on inactivation of two DNA binding proteins, CcpC (22) and CodY (25). Repression by CcpC is antagonized by citrate (22), whereas CodY loses repressing activity when the intracellular pools of GTP (32) and branched-chain amino acids (38) decrease. A citB null mutant is severely defective in spore formation and has a relative sporulation efficiency of 1.4 × 10−4% compared to its wild-type parent (6). Previous work by Craig et al. (6) showed that the defect in sporulation of a citB null mutant is caused, in part, by the accumulation of citrate, a chelator of Mn2+ and Fe2+. Chelation of divalent cations disrupts phosphorylation (activation) of Spo0A, the major transcription factor for early sporulation-specific genes (19). However, addition of excess Mn2+ and Fe2+ only partially overcomes the sporulation defect of an aconitase null mutant (6).
When a constitutively active allele of spo0A is introduced into the citB null mutant, cells proceed past the stage 0 block in sporulation but are still blocked at a later stage (6). A similar result is seen when an aconitase null mutant is genetically modified to prevent synthesis of citrate (6); such cells proceed past stage 0 but do not complete sporulation efficiently. Taken together, these results suggest that citrate accumulation may be responsible for the stage 0 block, but aconitase may have an additional function unrelated to citrate metabolism that is important during the later stages of sporulation. The only other known activity of B. subtilis aconitase and related proteins is the ability to bind to RNA (2, 42-44).
B. subtilis aconitase is structurally related to eukaryotic IRP-1 (iron regulatory protein 1), also known as cytosolic aconitase (3). IRP-1 is an RNA binding protein that regulates, posttranscriptionally, the expression of iron receptor, utilization, and storage proteins (3, 5, 15, 16, 24). IRP-1 exists in two forms and acts as a sensor of iron concentration and oxidative stress. Under iron-replete, reducing conditions, the catalytic 4Fe-4S cluster of IRP-1 is intact and the protein has aconitase enzyme activity. During conditions of iron limitation or oxidative stress, however, the 4Fe-4S cluster is oxidized or depleted and IRP-1 becomes an RNA binding protein (5, 16-18, 30, 31). IRP-1 binds iron-responsive elements (IREs), stem-loop structures located in the 5′ or 3′ untranslated regions (UTRs) of mRNAs, and directly affects transcript stability or translation (15, 34, 35). B. subtilis aconitase also binds to RNA, including putative IREs encoded in some B. subtilis mRNAs, as well as the rabbit ferritin IRE (2).
Escherichia coli aconitases A and B are also RNA binding proteins, are autoregulatory at the level of protein production, and are involved in the oxidative stress response (42, 44). Salmonella enterica serovar Typhimurium AcnB is also an RNA binding protein that binds to the transcript for a protease important for flagellar control (43). In addition, aconitase activity in other prokaryotes, such as Staphylococcus aureus, Pseudomonas aeruginosa, and Xanthomonas campestris pv. campestris, has been correlated with toxin production and pathogenicity (39, 40, 45). Whether the role of aconitase in the latter cases is enzymatic or nonenzymatic remains to be determined.
In an accompanying paper (37), we show that expression in a B. subtilis citB null mutant of an aconitase incapable of RNA binding (the yeast mitochondrial aconitase Aco1) restores Krebs cycle function but only partially restores sporulation. Sporulation in the Aco1-expressing strain was delayed and defective at late stages, in particular at the time of σG- and σK-dependent gene expression. These results, in combination with earlier evidence, suggest that the citB null mutant not only lacks aconitase catalytic activity but also is missing a second activity of aconitase, possibly RNA binding, that is required for efficient sporulation (6, 37). Therefore, we used site-directed mutagenesis to try to create a form of B. subtilis aconitase that retained catalytic activity but lost RNA binding activity. In this paper we present evidence that the nonenzymatic role of aconitase is critical for late mother-cell-specific gene expression during sporulation and that aconitase, in this case, acts as an RNA binding regulatory protein.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and assays of sporulation and β-galactosidase.
The strains of B. subtilis used in this work are listed in Table 1. All strains were grown at 37°C with aeration, unless otherwise specified. The nutrient-broth-based medium DSM was supplemented with 0.01 mM MnCl2, 0.001 mM FeSO4, and 1 mM Ca(NO3)2 (13). TSS minimal medium has been described previously and contained glucose (0.05%) as the carbon source and NH4Cl (0.02%) as the nitrogen source (13). When required, TSS was also supplemented with tryptophan (0.004%, wt/vol), phenylalanine (0.004%, wt/vol), and glutamate (0.2%, wt/vol). Chloramphenicol was added, when required, to a final concentration of 2.5 μg/ml.
TABLE 1.
B. subtilis strains
| Strain | Genotype | Source or reference |
|---|---|---|
| JH642 | trpC2 pheA1 | J. Hoch |
| MAB160 | trpC2 pheA1 ΩcitB::spc | 6 |
| EUDC9901 | trpC2 pheA1 gerE::kan | C. Moran (7) |
| JHBS5 | trpC2 pheA ΔspoIVCB | J. Haraldsen |
| JCB36 | trpC2 pheA1 ΩcitB::spc ΦspoIIA-lacZ cat | 6 |
| JCB63 | trpC2 pheA1 ΩcitB::spc SPβc2 Δ2::pSK10Δ6::Φ(cotA′-lacZ) tet | 6 |
| SJB227 | trpC2 pheA1 SPβc2 Δ2::pSK10Δ6::Φ(sspB′-lacZ) cat | S. Jin |
| BB1783 | trpC2 pheA1 citB-His6neo | B. Belitsky |
| AWS96 | trpC2 pheA1 | MAB160 × DNA JH642 |
| AWS133 | trpC2 pheA1 citB5 (R740E Q744E F661L I809T V852A) | AWS96 × pAWS42 |
| AWS137 | trpC2 pheA1 citB5 (R740E Q744E F661L I809T V852A) SPβc2 Δ2::pSK10Δ6::Φ(cotA′-lacZ) tet | AWS133 × DNA JCB63 |
| AWS144 | trpC2 pheA1 citB-His6neo | AWS96 × DNA BB1783 |
| AWS145 | trpC2 pheA1 citB-His6neo SPβc2 Δ2::pSK10Δ6::Φ(cotA′-lacZ) tet | AWS144 × DNA JCB63 |
| AWS149 | trpC2 pheA1 ΩcitB::spc ΦspoIIA-lacZ tet | JCB36 × pP528 |
| AWS153 | trpC2 pheA1 citB5 (R740E Q744E F661L I809T V852A) SPβc2 Δ2::pSK10Δ6::Φ(spoIIAA′-lacZ) tet | AWS133 × DNA AWS149 |
| AWS154 | trpC2 pheA1 citB-His6neo SPβc2 Δ2::pSK10Δ6::Φ(spoIIAA′-lacZ) tet | AWS144 × DNA AWS149 |
| AWS198 | trpC2 pheA1 His10-citB cat | JH642 × pAWS58 |
Cells were grown in DSM and assayed for heat-resistant spore formation at various times after entry into stationary phase. Samples were heated at 80°C for 10 min and then plated on DSM agar to determine the heat-resistant spore titer, while untreated samples were plated and represent the viable cell titer. Colonies were counted after overnight incubation at 37°C unless otherwise stated. β-Galactosidase assays were performed as described previously (32).
citB C-terminal mutant construction.
E. coli strain DH5α was used as a host for cloning. The C-terminal 1.2 kb of citB was amplified and mutagenized by three-step site-directed PCR mutagenesis. Arginine residue 740 and glutamine residue 744 were both changed to glutamate. Primers citMF1 (5′ GCGTCTAGAACCGTAACTTTGAAGGACGTATTCAC 3′; XbaI restriction site is underlined) and citMR3 (5′ GGTGCGATTTCGTTTTTGATTTCAATGTTGG 3′; mutated residues are in bold) were used to PCR amplify 600 bp of the upstream C-terminal sequence. Primers citMF2 (5′ GAAATCAAAAACGAAATCGCACCGGGTACAGAAGG 3′; mutated residues are in bold) and citMR4 (5′ GCGGAGCTCCCTATTGATTCATCAGTGATGATGGTGATGGTGGGACTGCTTCAT 3′; SacI restriction site is underlined) were used to PCR amplify 545 bp of downstream sequence and to add six histidines at the C terminus of the protein. The products of these reactions were mixed, and the entire 1.2-kb fragment was amplified with primers citMF1 and citMR4. The PCR product was cloned by XbaI/SacI restriction digestion and ligation to the B. subtilis integrative vector pJPM1 (29), creating pAWS42. pAWS42 was used to transform B. subtilis, leading to homologous recombination at the citB locus.
Construction of in vitro transcription templates.
The pBluescript II SK(−) and KS(−) vectors (Stratagene) were used for cloning of DNA sequences that would serve as templates for in vitro transcription. The multiple cloning sites in these vectors are bracketed by promoters recognized by phage T3 and phage T7 RNA polymerases. The gerE gene was PCR amplified with 250 bp flanking each end of the coding sequence with primers gerEF5 (5′ GGACTCTAGATCTCGGAGAGAACCTCGAAGG 3′) and gerER6 (5′ GGACGGTACCTTGTCCATCCCTCACTCAAGG 3′) (with appended restriction sites XbaI and KpnI underlined), resulting in a product of approximately 700 bp. The digested product was ligated to XbaI/KpnI-digested pSK and pKS, creating plasmids pAWS60 and pAWS80, respectively. The 3′ end of gerE, with 250 bases of the 3′ UTR, was also PCR amplified with primers gerER6 and gerEIN (5′ GGACTCTAGAGGAGATTGCAAGCGAGCTATTTAT 3′), with the same appended restriction sites (underlined), resulting in a product of approximately 330 bp. This product was cloned in pKS, creating pAWS100.
The 3′ end of fliT, with 250 bases of the 3′ UTR, was PCR amplified with primers fliTR8 (5′ GGACGGTACCTCAGCACACCGCGAATCCATAATG 3′) and fliTIN (5′ GGACTCTAGAACATGGGATCAGCTGATTGTGAAGG 3′), with the same appended restriction sites (underlined), resulting in a product of approximately 350 bp. This product was cloned in pKS, creating pAWS106.
N-terminal His tag of citB.
A 10-histidine N-terminal tag was introduced immediately after the start codon of citB by a three-step PCR procedure. Two separate reactions were used to amplify approximately 400 bp upstream of the start codon, with primers citBF6 (5′ GGGCATGCGAGAACCTCCTTAAAAGAGTTCGGTGTTATT 3′) and citBR8 (5′ TGCTGCAGTTTTTTGCTCGTTTGCGTGATGATGGTGGTGATGATGGTGATGGTGCATTCTCCAAAATCCCCCTTCAGA 3′), and approximately 550 bp downstream of the start codon, with primers citBF7 (5′ TCTGAAGGGGGATTTTGGAGAATGCACCATCACCATCATCACCACCATCATCACGCAAACGAGCAAAAAACTGCAGCA 3′) and citBR9 (5′ GGCCCGGGCAATACCTGTTGCAGGCGGTACTGCCTGATAATT 3′), with both regions containing 10-histidine codons (shown in bold) as appendages. An annealing PCR was then performed with the two products plus the farthest upstream and downstream oligonucleotides primers (citBF6 and citBR9) to generate a 950-bp product.
The 950-bp PCR product was purified, digested with restriction enzymes XmaI/SphI (sites embedded in primers citBF6 and citBR9 and underlined), and ligated to the B. subtilis integrative plasmid pJPM1 (29), creating pAWS50. Integration at the citB locus by homologous recombination was selected for by chloramphenicol resistance. Five of six transformants tested expressed N-terminal His10-tagged aconitase as determined by immunoblotting; one such transformant was AWS198. This strain was a glutamate prototroph and sporulated like the wild type. AWS198 was used for aconitase purification.
Purification of aconitase by cobalt affinity chromatography.
AWS198 (His10-citB) was grown in DSM and harvested upon entry into stationary phase, at an optical density at 600 nm (OD600) of approximately 1.0. Cells were pelleted at 4°C for 15 min at 5,000 rpm. Following centrifugation, the pellet was washed twice with ice-cold 20 mM Tris-citrate, pH 7.35. The cell pellet was stored at −80°C.
Cells were thawed at room temperature and resuspended in 10 ml of buffer containing 200 mM KCl, 50 mM Tris-HCl (pH 7.5), 10% glycerol, 10% Nonidet P-40, 0.2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.5 mM dithiothreitol (32). The resuspended cells were subjected to two rounds of breakage in a French pressure cell (15,000 lb/in2). The lysate was sonicated briefly to fragment chromosomal DNA and then centrifuged at 4°C at 13,000 rpm for 20 min.
The supernatant fluid was dialyzed against the ice-cold resuspension buffer (described above) without EDTA or dithiothreitol for 1 h and then incubated with preequilibrated cobalt resin (Talon) by tumbling at 4°C. The resin was gently pelleted by centrifugation and washed twice with the same buffer containing imidazole at 10 mM. The resin was then loaded onto a column, and the wash step was repeated. Pure aconitase was eluted in a buffer containing 300 mM imidazole. Aconitase was dialyzed against buffer containing 50% glycerol, 50 mM KCl, and 20 mM Tris-HCl (pH 7.5) and stored at −20°C. The protein concentration was determined with the Bio-Rad protein assay reagent.
Partial purification of aconitase by DEAE-Sephacel chromatography.
AWS144 and AWS133 cell pellets, prepared as stated above, were thawed in ice-cold buffer containing 20 mM Tris-citrate (pH 7.35), 1 mM PMSF, and 7 mM β-mercaptoethanol. The resuspended cells were subjected to two rounds of breakage in a French pressure cell (15,000 lb/in2). The cell lysate was then centrifuged at 4°C at 13,000 rpm for 20 min.
Ammonium sulfate precipitation was performed, first at 40% saturation and then at 85%, in order to precipitate aconitase. This pellet, after centrifugation, was dialyzed against buffer containing 20 mM Tris-citrate (pH 7.35) 1 mM PMSF, and 7 mM β-mercaptoethanol for 3 h at 4°C and then again against fresh buffer overnight.
Following dialysis, the protein extract was loaded onto a DEAE-Sephacel column that had been preequilibrated with 20 mM Tris-citrate, pH 7.35. The protein was eluted with a gradient of 30 to 100 mM Tris-citrate, pH 7.35. Fractions were stored at 4°C and analyzed by aconitase enzyme assays and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The eluate fractions containing partially pure aconitase were pooled, stored at a final concentration of 50% glycerol, and concentrated with the Biomax-30K ultrafree centrifugal filter supplied by Millipore. The protein concentration was determined as stated above. The concentration of aconitase in the partially pure preparations was quantitated by immunoblotting with antibody to B. subtilis aconitase.
Crude extract preparation.
Crude extracts for determination of aconitase specific activity and for Western blotting of aconitase and σK were prepared from cells pelleted by centrifugation at 6,000 rpm at 4°C. Pellets were washed with ice-cold 50 mM Tris-HCl, pH 8.0, supplemented with 0.5 mM PMSF, frozen immediately on dry ice, and stored at −80°C. Cells were lysed by incubation for 15 min at 37°C with 50 mM Tris-HCl, pH 8.0, containing 0.4 mg of lysozyme per ml, 0.03 mg DNase I per ml, and 0.1 mM PMSF. Cell debris was eliminated by centrifugation, and the resulting supernatant fluid was applied to SDS-polyacrylamide gels.
For Western blots of GerE protein, cells were grown in DSM, harvested at the indicated times, washed with ice-cold 10 mM Tris-HCl, pH 7.6 (supplemented with 1× complete protease inhibitors [Roche]), frozen immediately, and stored at −80°C. Resuspended pellets were incubated at 37°C in 10 mM Tris-HCl, pH 7.6, containing 4 mg lysozyme per ml, 50 mM glucose, and 1× complete protease inhibitors. After 15 min, the extracts were mixed with SDS-PAGE sample buffer, boiled for 5 min, and centrifuged to remove debris. Samples of the supernatant fluids were applied to SDS-polyacrylamide gels.
For immunoblot analysis, samples subjected to SDS-PAGE were transferred to an Immobilon polyvinylidene difluoride membrane (Millipore Corp.). Immunoblotting was performed with rabbit polyclonal primary antibody to σK (a gift from D. Rudner, Harvard Medical School, Boston, MA), aconitase (36), or GerE (27; the generous gift of H. Takamatsu, Setsunan University, Japan). Secondary goat anti-rabbit immunoglobulin G antibody, conjugated with alkaline phosphatase, was supplied by Bio-Rad, as were reagents for enzymatic detection of alkaline phosphatase activity. For immunoblots of GerE protein, the secondary antibody was goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Upstate Biotechnology, Inc.) and detected with the ECL Plus reagent kit (GE-Amersham).
Aconitase activity.
Samples of cell extract were incubated with substrate, 20 mM dl-isocitrate, in 90 mM Tris-HCl, pH 8.0. The accumulation of cis-aconitase was measured as the change in absorbance at 240 nm. One unit of activity was defined as a change in absorbance of 0.0033 per minute (9).
RNA isolation.
Cells grown in DSM were transferred to ice-cold tubes and pelleted by centrifugation at 6,000 rpm for 3 min at 4°C. Pellets were washed with ice-cold 50 mM Tris-HCl, pH 8.0-1 mM EDTA, frozen immediately on dry ice, and stored at −80°C. Cells were resuspended in a protoplasting buffer (25% sucrose-50 mM Tris-HCl, pH 8.0-1 mM EDTA) containing 0.4 mg lysozyme per ml and incubated at 37°C for 5 min. RNA was isolated by the TRIzol method (Invitrogen) and resuspended in diethyl pyrocarbonate (DEPC)-treated deionized H2O.
For reverse transcriptase (RT) PCR, 25 μg of RNA was treated with DNase by using the Turbo DNA-free kit supplied by Ambion. DNA-free RNA was confirmed by negative PCR. For microarray analysis, the RNA was treated with a QIAGEN RNeasy purification kit for removal of any contaminating chromosomal DNA and checked for integrity by electrophoresis on a 2% agarose gel. RNA concentrations were determined spectrophotometrically.
RT-PCR.
cDNA was synthesized from 1 μg of RNA with primers rRNA16SR (5′ CCCCAGTTTCCAATGACC 3′) and gerER (5′ CTCCTTCCAAAAGAAGGAATACC 3′) for 50 min at 42°C with Superscript II RT (Sigma). Following inactivation of RT at 70°C, PCRs were performed on the cDNA with oligonucleotides rRNA16SF (5′ GGGTGATCGGCCACAC 3′) and rRNA16SR, to amplify a 300-bp product from cDNA representing the rrnA-16S transcript, while primers gerEFRT (5′ AAACCCATTCGTTTCTGATTCGC 3′) and gerER were used to amplify a 250-bp product representing the gerE transcript. PCR products were electrophoresed on 1.5% agarose gels and analyzed with the ImageQuant program.
cDNA synthesis for microarray analysis.
cDNA was synthesized from 15 μg of RNA in a 30-μl reaction volume by incubation with random hexamers (Roche) (0.5 μg/μl) and Superscript II RT (Invitrogen) for 2 h at 42°C. The reaction mix contained dATP, dCTP, dGTP, and aminoallyl-dUTP (Sigma). Following inactivation of RT at 70°C, RNA was degraded by incubation at 70°C with 0.1 M NaOH for 10 min and the reaction was neutralized with 0.1 M HCl. cDNA was purified with the QIAGEN MinElute kit and eluted with 10 μl of DEPC-treated deionized H2O.
Coupling of the N-hydroxysuccinimide-ester Cy3 and Cy5 dyes (Amersham) to the aminoallyl-dUMP residues in the cDNA was carried out in the presence of 0.05 M NaHCO3. The coupling reaction was quenched after 1 h by addition of hydroxylamine to 1.7 M and further incubation for 15 min. The Cy3- and Cy5-labeled cDNA probes for each experiment were mixed, treated with a QIAquick PCR purification kit, eluted with 60 μl of deionized H2O, vacuum dried, and resuspended in 10.5 μl of deionized H2O.
Microarray analysis.
Microarray slides were printed with the B. subtilis OligoLibrary supplied by Sigma-Genosys. The library contains 4,128 65-mer oligonucleotides which represent 4,106 B. subtilis genes, 10 controls from E. coli and Brome mosaic virus, and 12 random oligonucleotides.
Microarray slides were UV cross-linked by exposure to 600,000 μJ (Stratalinker). Slides were prehybridized for 1 h at 42°C in a buffer composed of 5× SSC (sodium chloride-sodium citrate; 1× SSC is 0.15 M NaCl-0.015 M Na3 citrate, pH 7.0), 0.1% SDS, and 1% bovine serum albumin. Prehybridized slides were rinsed in deionized H2O and dried by brief centrifugation at 1,000 rpm.
Ten microliters of the mixed probe was incubated at 100°C for 5 min in the presence of salmon testes DNA (1 μg/μl) and yeast tRNA (1 μg/μl). The probe was mixed with 12 μl of 0.1% SDS-10× SSC-50% formamide and pipetted immediately onto the prehybridized microarray slides.
Hybridization was performed overnight at 42°C. Hybridized slides were washed first with 1× SSC-0.2% SDS, next with 0.1× SSC-0.2% SDS, and then six times with 0.1× SSC. Slides were dried by brief centrifugation at 1,000 rpm and scanned with the ScanArray program. The raw data were analyzed with the Imagene (BioDiscovery Inc.) and Genespring (Silicon Genetics) programs.
In vitro transcription and gel mobility shift assays.
Linearized plasmid DNAs were used as templates for in vitro transcription reactions driven by either T3 or T7 RNA polymerase using kits provided by either Sigma or Ambion. Nascent RNA was labeled by carrying out synthesis in the presence of [α-32P]UTP (NEN). Radioactive RNA was purified by phenol-chloroform extraction and precipitated with isopropanol and 0.3 M sodium acetate.
Radioactive RNA samples were heated to 85°C, allowed to cool slowly to room temperature, and then incubated for 15 min at room temperature with various concentrations of aconitase in a binding buffer composed of 10 mM Tris-HCl (pH 7.5), 35 mM KCl, 10% glycerol, 5 mM β-mercaptoethanol, and 0.5 μg/μl yeast tRNA (a nonspecific competitor). Samples were subjected to electrophoresis on an 8% nondenaturing polyacrylamide gel. The gel was dried and then analyzed by using a phosphorimager (Applied Biosystems).
RESULTS
Construction of a citB C-terminal mutant aconitase.
To create a B. subtilis strain expressing an enzymatically active but RNA binding-deficient aconitase, we based our approach on the mutagenesis of IRP-1 by Kaldy et al. (23). Those authors concluded that arginines 728 and 732 in IRP-1 are critical for interaction of IRP-1 with RNA targets. The corresponding residues in B. subtilis aconitase are R740 and Q744 (Fig. 1). A PCR-based site-directed mutagenesis strategy was used to convert both of these residues to glutamate in B. subtilis aconitase. A C-terminal hexahistidine tag was also incorporated into this PCR product. The cloned 1.2-kb PCR product was sequenced, and, while the intended R740E and Q744E mutations were confirmed, three additional amino acid substitutions were obtained (F661L, I809T, and V852A). Since our intention was not to define the roles of R740 and Q744 but rather to create a form of aconitase with reduced RNA binding, no attempt was made to separate the effects of the five point mutations on aconitase function. The allele of the citB gene carrying the five point mutations was called citB5.
FIG. 1.
C-terminal alignment of human IRP-1 and B. subtilis aconitase. Human IRP-1 residues in bold have been defined as important for IRP-1-RNA interaction (23). B. subtilis citB residues in bold are the correlating residues and were targeted for site-directed mutagenesis.
The mutagenized C-terminal portion of citB was cloned in the B. subtilis integrative vector pJPM1 (29), creating plasmid pAWS42. AWS96 (wild type) was transformed with this construct, and integration by homologous recombination at the citB locus was selected for by chloramphenicol resistance. The resulting strain, AWS133, appeared to be sporulation defective on plates; i.e., colonies were translucent rather than having the characteristic opaque morphology and brown pigmentation of wild-type colonies. However, AWS133 grew on minimal medium plates without the addition of glutamate, indicating that aconitase catalytic activity was intact.
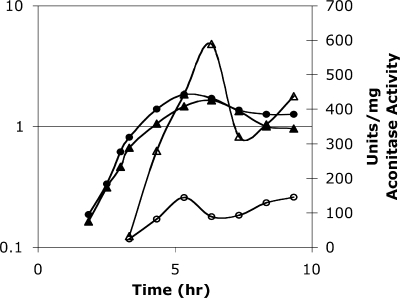
The kinetics of appearance of aconitase activity in strain AWS133 was analyzed in nutrient-rich medium during exponential growth and stationary phase. Cell extracts from AWS133 (citB5-His6) and AWS144 (citB+-His6) exhibited similar profiles of activity (Fig. 2). Interestingly, the specific activity of the mutant aconitase was approximately sixfold higher than that of the wild-type aconitase.
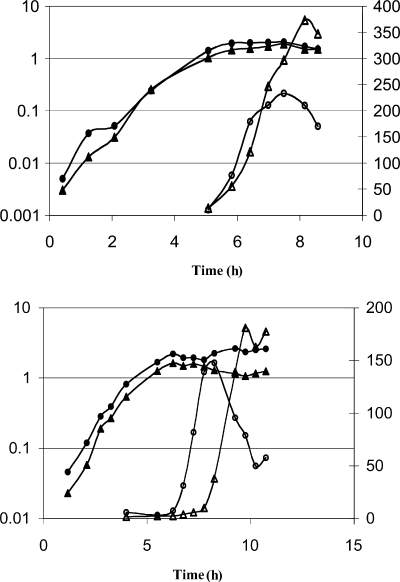
FIG. 2.
Specific activity of aconitase in cell extracts. AWS144 (citB+; closed circles) and AWS133 (citB5; closed triangles) were grown in DSM, and cell extracts were prepared at the time points indicated and analyzed for aconitase enzyme activity (open symbols).
The higher aconitase specific activity in extracts of AWS133 might be due to an increase in the amount of aconitase protein. However, immunoblots with antibody specific to aconitase, as well as with antibody specific to the hexahistidine tag present on both the wild-type and mutant aconitases, showed that the amounts of aconitase protein were similar in the AWS133 and wild-type strains throughout growth and stationary phase (data not shown). Therefore, we concluded that the mutant protein has a higher intrinsic specific activity. One possible explanation for this phenomenon is that the C-terminal mutations stabilize the 4Fe-4S catalytic cluster, reducing the ability of the protein to switch from its enzymatically active form to its RNA binding form. Alternatively, decreased binding of RNA might reduce hypothetical RNA-dependent inhibition of enzyme activity. Whatever the basis for the increased enzyme activity of the mutant aconitase, it is clear that any sporulation defect in this strain must be due to some factor other than a deficiency in catalytic activity.
Sporulation in the citB5 mutant is delayed at a late stage.
As described in the accompanying paper, we expressed in B. subtilis the Saccharomyces cerevisiae mitochondrial aconitase, a protein that does not bind RNA, and saw that sporulation was delayed compared to wild-type cells (37). Therefore, we tested the kinetics of heat-resistant spore formation in strain AWS133. While wild-type cells had a sporulation efficiency of nearly 60% at approximately 7 h after entry into stationary phase, AWS133 had a sporulation efficiency at that time of only 0.72%. However, at approximately 20 h after entry into stationary phase, wild-type cultures had a sporulation efficiency of 82% and AWS133 had a sporulation efficiency of 76%. We conclude that the C-terminal mutations in aconitase cause a delay in spore formation.
Microarray analysis of sporulation gene expression.
A time course microarray analysis was utilized to identify genes whose expression during sporulation is affected in strain AWS133. AWS133 and AWS144 were grown in DSM, and RNA was prepared from cells isolated during the late exponential growth phase and at roughly 5, 6, and 7 h after entry into stationary phase. Each experiment was done in triplicate, yielding a total of 24 RNA samples. The 24 individual RNA samples were pooled in equal concentrations and utilized as a reference RNA from which cDNA was synthesized. Then, each of the 24 individual RNA samples was used for cDNA synthesis, and subsequent hybridization was compared to that of the reference RNA.
Transcripts of sporulation-specific genes that are under σE- or σG-dependent control were similarly abundant in the wild type and in AWS133 compared to the reference RNA at 5, 6, and 7 h after entry into stationary phase. However, many σK-dependent transcripts were much less abundant in AWS133 at 7 h after entry into stationary phase than they were in the wild type, while σG-dependent transcripts were similarly abundant (Table 2). Most of the genes that were differentially expressed code for components of the spore coat. Interestingly, many of these genes require the DNA binding protein GerE for transcriptional activation (21). Note that alterations in the concentration of a given RNA detected by microarray analysis may reflect a change in the rate of synthesis of the RNA or a change in its stability.
TABLE 2.
Microarray analysis
| Gene | Fold changea
|
σ factor dependence | |
|---|---|---|---|
| Wild type | citB5 | ||
| cotE | 2.3 | 2.4 | σE |
| spoIVCB | 1.4 | 3.3 | σE and σK |
| gerBB | 8.8* | 2.9 | σG |
| gerBC | 7.8 | 3.0 | σG |
| spoVAA | 3.5 | 2.8 | σG |
| spoVT | 2.5 | 2.7 | σG |
| sspA | 2.7 | 2.6 | σG |
| sspB | 4.1 | 3.8 | σG |
| sspC | 7.4 | 4.5 | σG |
| sspD | 3.1 | 3.3 | σG |
| spoIVB | 8.4* | 3.8 | σG |
| cgeA | 7.1 | 1.0 | σK (GerE) |
| cgeB | 11.7 | 1.6 | σK (GerE) |
| cgeC | 8.5 | 1.5 | σK (GerE) |
| cgeD | 8.8 | 1.2 | σK (GerE) |
| cotA | 11.3 | 2.2 | σK |
| cotB | 6.4 | 2.7 | σK (GerE) |
| cotC | 5.3 | 1.9 | σK (GerE) |
| cotD | 7.7 | 1.1 | σK (GerE) |
| cotG | 7.5 | 2.1 | σK (GerE) |
| cotS | 8.5 | 1.1 | σK (GerE) |
| cotSA | 10.7 | 2.9 | σK (GerE) |
| cotT | 5.0 | 2.8 | σK |
| cotW | 4.7 | 1.8 | σK (GerE) |
| cotX | 6.6* | 1.9 | σK (GerE) |
| cotY | 4.4 | 1.8 | σK (GerE) |
| gerE | 12.1** | 1.9 | σK |
Ratios comparing wild-type and citB5 cells approximately 7 h after entry into stationary phase. Fold change represents the average change in three independent experiments, except where indicated: *, based on data from two experiments; **, based on data from one experiment.
gerE transcript is more abundant in the wild type than in the citB5 mutant.
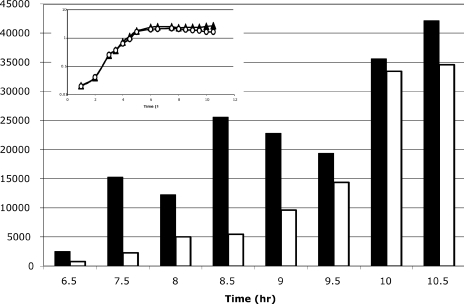
Since GerE affects the expression of almost all of the σK-dependent genes that were reduced in expression in the citB5 mutant strain, the gerE transcript would be a logical candidate target of aconitase regulation. We analyzed the levels of gerE transcript throughout stationary phase in wild-type and citB5 cells by RT-PCR. The rrnA-16S transcript was used as a control. Our results demonstrate that the gerE transcript was, on average, 3.5-fold more abundant in wild-type cells than in citB5 cells during mid-stationary phase and that this deficit was eventually overcome in the citB5 mutant strain at the latest time points tested (Fig. 3). These results confirm that the gerE transcript at mid- to late-sporulation time points is less abundant in AWS133 than in AWS144, as was seen in the microarray analysis. This result may indicate a decrease in gerE mRNA stability in strain AWS133.
FIG. 3.
RT-PCR analysis of gerE transcript levels. AWS144 (citB+; triangles) and AWS133 (citB5; circles) were grown in DSM at 37°C (inset). Samples were isolated at the time points indicated. RNA was prepared, and cDNA was synthesized with reverse primers to rrnA-16S and gerE. Eighteen cycles of amplification was determined to be within the linear range for PCR of rrnA-16S. The linear range for PCR of gerE was determined for each sample; 48 to 50 cycles were used for time points 6.5 to 8.5, and 35 cycles were used for time points 9 to 10.5. The rrnA-16S PCRs were done in duplicate for each sample. After product quantitation, the pixel numbers were averaged and normalized by determining the ratio of each sample to the wild-type rrnA-16S (h 6.5 sample). Then, all gerE transcripts were normalized to the rrnA-16S product from the same time point by determining the ratio of gerE product to the normalized rrnA-16S product. Relative gerE transcripts for AWS144 (citB+; solid bars) and AWS133 (citB5; empty bars) are shown for each time point.
GerE activates many coat protein genes, including cotD. Transcription of a cotD-lacZ fusion is sevenfold higher in the wild type than in a gerE mutant (20). Our microarray results showed that whereas AWS144 (wild-type) RNA had cotD transcript at a 7.72-fold-higher level than the reference RNA, AWS133 (citB5 mutant) had cotD transcript at only a 1.12-fold-higher level than the reference.
GerE protein level in the citB5 mutant strain.
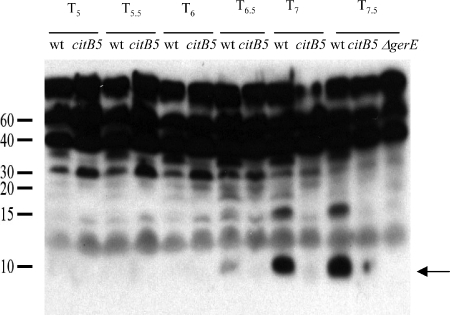
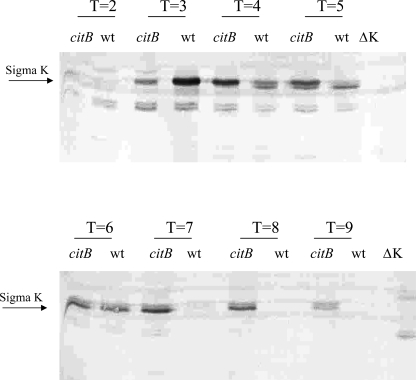
To assess the effect of the citB5 mutations on the rate of accumulation of GerE protein, extracts of stationary-phase cells of strains AWS144 (citB+), AWS133 (citB5), and EUDC9901 (gerE::kan) (7) were analyzed by immunoblotting with antibodies raised against GerE (27). As shown in Fig. 4, GerE protein (8 kDa) was first detected in the wild-type strain at 6 h and was very abundant at 7 and 7.5 h after the end of the exponential growth phase. Only a trace of GerE was detectable in strain AWS133 at these time points. In strain EUDC9901, used as a control, no band corresponding to GerE was seen.
FIG. 4.
Accumulation of GerE protein in mutant and wild-type (wt) strains. Extracts of stationary-phase cultures of strains AWS144 (citB+), AWS133 (citB5), and EUDC9901 (gerE::kan) harvested at the indicated time points (Tn = n h after the end of the exponential growth phase) were assayed for GerE protein by immunoblotting after separation by SDS-15% PAGE. GerE (8 kDa; indicated by the arrow) was the fastest-migrating band that reacted with antibody.
Further verification of microarray data.
To confirm that AWS133 is not defective in initiation of sporulation but rather is delayed at a later stage, we analyzed the expression of lacZ fused to sporulation-specific promoters. The spoIIA operon is transcribed by the σH form of RNA polymerase and is under positive control by Spo0A-phosphate. Its expression is an indicator of sporulation initiation. A spoIIA-lacZ fusion was integrated at the amyE locus of the wild-type strain and AWS133, creating strains AWS154 and AWS153, respectively. In AWS153 and AWS154, spoIIA-lacZ activity was turned on at approximately the same time in stationary phase, indicating that the citB5 mutant is not defective for sporulation initiation (Fig. 5, top).
FIG. 5.
Expression of lacZ fusions to sporulation-specific promoters. Strains AWS154 (citB+ spoIIA-lacZ; circles) and AWS153 (citB5 spoIIA-lacZ; triangles) (top) or strains AWS145 (citB+ cotA-lacZ; circles) and AWS137 (citB5 cotA-lacZ; triangles) (bottom) were grown in DSM at 37°C. Samples were removed for measurement of the OD600 (closed symbols) and for assay of β-galactosidase activity (open symbols).
A second fusion, cotA-lacZ, was integrated at the amyE locus of the wild type and AWS133, creating strains AWS145 and AWS137, respectively. cotA codes for a spore coat protein, and its transcription depends on the latest-acting σ factor, σK. Expression of cotA-lacZ was delayed approximately 2 to 3 h in AWS137 compared to the wild type. The results are consistent with the microarray data, in that the citB5 mutant showed a delay late in sporulation, particularly for σK-dependent gene activation (Fig. 5, bottom).
Immunoblotting of σK.
GerE is not only a transcriptional activator but also a repressor of some genes (20). GerE represses sigK transcription, for instance. Our microarray results showed that the sigK (spoIVCB) transcript was 3.33-fold more abundant in AWS133 (citB5 mutant) than in the reference RNA, while in AWS144 (wild type), spoIVCB was 1.44-fold more abundant than in the reference RNA. This result is reminiscent of the phenotype of a gerE mutant, which has twofold-higher sigK-lacZ expression than does the wild type (20). Since GerE-dependent repression of sigK inhibits accumulation of σK during late stages of sporulation (20), we performed immunoblotting to analyze σK levels in the wild-type strain and in AWS133.
Strains AWS133, AWS144, and JHBS5 (ΔsigK) were grown in DSM, cell extracts were prepared at time points late in exponential growth and throughout stationary phase, and the extracts were analyzed by immunoblotting with antibody that reacts with both pro-σK and σK. The results showed that σK persisted later in stationary phase in AWS133 (citB5 mutant) than in AWS144 (citB+) (Fig. 6). σK was no longer detected after approximately 7 h in strain AWS144 cells, while AWS133 retained σK.
FIG. 6.
Immunoblot with antibody to σK. AWS144 (citB+ [wt]), AWS133 (citB5), and JHBS5 (citB+ ΔsigK [ΔK]) were grown in DSM at 37°C. Samples were isolated at the time points indicated (T, indicated number of hours after entry into stationary phase). Crude cellular extracts were subjected to SDS-PAGE and analyzed by immunoblotting with antibody specific to the σK protein.
These results confirm that the citB5 mutant is unable to repress σK transcription efficiently and that the reason for the defect in activation of σK-dependent genes is not simply the lack of the σ factor. Importantly, overexpression of sigK and overproduction of sigK would occur if the mutant strain had a reduced GerE protein concentration. This might explain the persistence of the σK protein in the citB5 mutant compared to the wild type.
Aconitase binds to gerE mRNA.
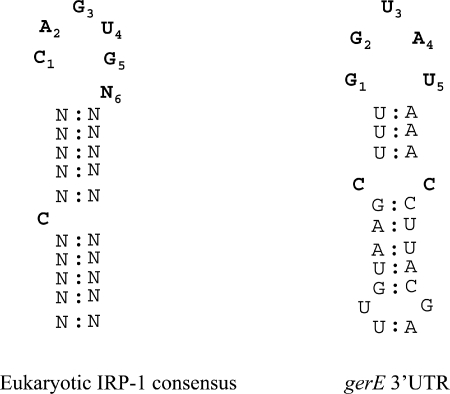
The results above suggest that aconitase functions at the level of σK-dependent gene expression, possibly by targeting specific mRNAs. Interestingly, the gerE transcript has a putative stem-loop structure that resembles, at least superficially, the eukaryotic IRE consensus sequence (Fig. 7). It is possible that aconitase binds to and stabilizes the gerE mRNA and thus allows for efficient translation and accumulation of GerE. The absence of such binding could explain the delay in sporulation of the citB5 mutant. To test whether aconitase can bind to gerE mRNA, we performed in vitro gel mobility shift assays.
FIG. 7.
Eukaryotic IRE consensus sequence and a putative stem-loop structure of the gerE mRNA 3′ UTR. The gerE putative stem-loop structure is found 54 bases after the translational stop codon.
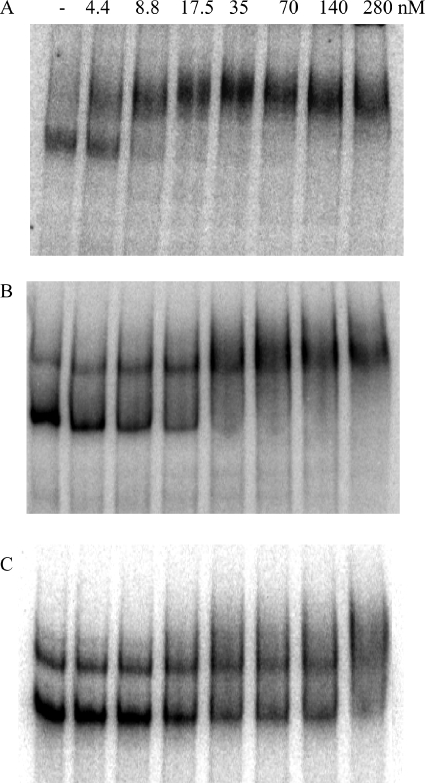
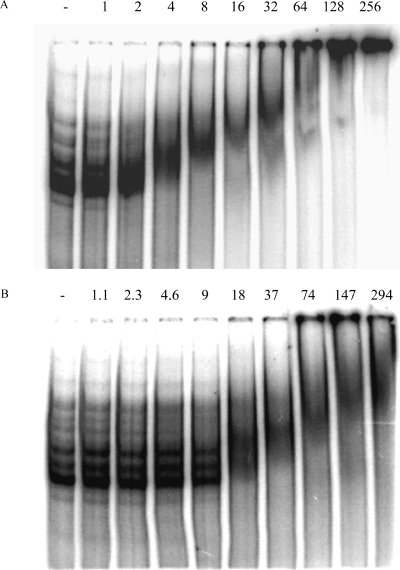
Radiolabeled RNAs of 350, 450, and 350 bases, corresponding to the 3′ end of the gerE gene and its UTR, an antisense version of this sequence, and an antisense version of the 3′ end of the fliT mRNA and its UTR, respectively, were synthesized in vitro, purified, and analyzed in a gel shift assay in the presence or absence of aconitase (see Materials and Methods). Pure aconitase shifted the gerE sense strand mRNA transcript at a concentration 4-fold lower than that needed to shift the antisense RNA and at a concentration 35-fold lower than that needed to shift the antisense fliT RNA (Fig. 8). The concentration of aconitase that shifted 50% of the sense strand RNA was 4.4 nM.
FIG. 8.
Gel mobility shift assays of aconitase binding to gerE mRNA. The gerE mRNA 3′ UTR (A), the gerE antisense RNA 3′ UTR (B), and the fliT antisense RNA 3′ UTR (C) were synthesized and radiolabeled by in vitro transcription. Radiolabeled RNA was incubated without or with increasing concentrations of pure B. subtilis His10-aconitase, as indicated, and then analyzed for complex formation by gel mobility shift assay.
We next tested whether citB5 mutant aconitase is defective in RNA binding. Since the citB5-encoded aconitase has a C-terminal His6 tag instead of the N-terminal 10-histidine tag needed for complete purification, we partially purified the equivalently tagged versions of the wild-type and mutant proteins (see Materials and Methods) for analysis in gel mobility shift assays. After purification, immunoblotting with antibody specific to aconitase was utilized to determine the relative concentrations of aconitase protein in the two preparations (data not shown). The citB5 mutant aconitase required at least a 4.5-fold-higher concentration to shift 50% of the gerE mRNA than did the wild-type aconitase (Fig. 9). These results strongly suggest that the late sporulation defect in the citB5 mutant strain AWS133 is due to defective RNA binding.
FIG. 9.
Gel mobility shift assays with partially purified aconitase. The gerE mRNA 3′ UTR was synthesized and radiolabeled by in vitro transcription. Radiolabeled RNA was incubated without or with increasing concentrations of wild-type aconitase-His6 (A) or citB5 mutant aconitase-His6 (B), as indicated, and then analyzed for complex formation by gel mobility shift assay. The amount of partially purified aconitase (aconitase units) added to each reaction was independently estimated by quantitative immunoblot analysis.
DISCUSSION
Our results demonstrate that the citB5 mutant aconitase retains catalytic activity, as shown by its ability to support growth on minimal medium without glutamate supplementation and by in vitro enzymatic assays. However, despite high-level aconitase catalytic activity, the strain is delayed late in sporulation, particularly at the level of σK-dependent gene expression, as shown by lacZ fusion assays and microarray analysis. This phenotype differs from that of a citB null mutant, which is blocked at stage 0, is unable to activate Spo0A, and is thus unable to initiate sporulation (6). These results point to the gerE transcript and other members of the σK regulon as putative targets for regulation by aconitase.
Regulation of spore coat synthesis is fine-tuned. First, synthesis of σK is regulated not only by the earlier-acting σ factor, σE, and by SpoIIID, a DNA binding protein, but also by excision of a prophage-like element, SKIN, that interrupts the coding sequence of the gene (26, 33, 41). σK is then synthesized in a pro-form, which requires cleavage for activation (28). σK is required for transcription of gerE and other genes, many of which additionally require GerE as a positive regulator (12, 20, 21). Late in sporulation, GerE represses sigK transcription and eventually, at a higher protein concentration, represses transcription of many of the genes it previously activated (20, 21).
Based on in vitro assays, we hypothesize that aconitase binds to the gerE transcript late in sporulation and stabilizes it for translation, thereby increasing the rate and level of GerE protein accumulation. The delay in sporulation of the citB5 mutant at the level of σK- and GerE-dependent gene activation may be explained in part by the absence of such stabilization. As shown in Fig. 4, the accumulation of GerE is significantly delayed in the citB5 mutant strain. In support of the microarray data, our RT-PCR results demonstrate that gerE transcript is approximately 3.5-fold reduced in the citB5 strain for several hours during sporulation compared to the wild type. In addition, our immunoblotting results demonstrate the persistence of σK in the citB5 mutant compared to the wild type, while our microarray analysis demonstrates that the sigK transcript is over twofold more abundant in the citB5 mutant than in the wild type. Both effects are presumably due to a reduced ability of GerE to repress sigK. While we have shown binding of aconitase to gerE mRNA, we cannot exclude the possibility that aconitase binds to other σK-dependent transcripts or sigK mRNA as well.
The high level of mutant aconitase catalytic activity is not likely to be a cause for the defective sporulation phenotype. In our accompanying study (37), we created a strain expressing the yeast mitochondrial aconitase Aco1, a protein that does not bind RNA (24). That strain had less catalytic activity in cell extracts than did the wild type and yet still had a late-stage sporulation delay, a defect very similar to that of the citB5 mutant strain (37). These results support the hypothesis that although catalytic activity is required for sporulation initiation, late sporulation requires a second function of aconitase.
The gel mobility shift assays suggest that aconitase binding is not only sequence specific but also structure specific. Since the sense and antisense RNAs of gerE likely form structurally analogous IRE-like structures, it was not unexpected that aconitase would bind to this RNA as well. However, the reduced binding affinity of aconitase for both the antisense gerE and antisense fliT RNAs compared to the gerE mRNA demonstrates that a specific nucleotide sequence is also necessary. This is similar to the requirements for IRP-1 binding to the eukaryotic IRE (1, 14).
At least a 4.5-fold-higher concentration of the citB5 mutant aconitase than the wild-type aconitase was required for binding to the gerE mRNA 3′ UTR. The mutations in the citB5 aconitase may interfere directly with RNA binding. Alternatively, they may stabilize the 4Fe-4S cluster such that the protein has reduced ability to switch to the RNA binding form. Since aerobic purification procedures are likely to induce some disruption of the 4Fe-4S cluster, the RNA binding function of the citB5 mutant aconitase may be even more defective in the reducing environment of the cell.
In addition, we observed a second shift when high concentrations of partially purified wild-type aconitase were incubated with the gerE mRNA 3′ UTR. No such shift was observed in the gel mobility assays with the pure wild-type aconitase or with the citB5 mutant aconitase. Since dimerization of members of the aconitase A/IRP-1 family has never been shown, we suggest that the second shift might be due to involvement of an additional protein in the interaction, acting perhaps as a chaperone. In fact, yeast aconitase utilizes Yfh1p, a homolog of frataxin, as a chaperone for maturation of the 4Fe-4S cluster, for stabilizing the interaction with citrate, and for preventing 4Fe-4S cluster oxidation (4). Perhaps B. subtilis has an analogous chaperone, and perhaps this protein is missing from the pure preparation of aconitase and is unable to interact with the citB5 mutant protein.
Iron is required for sporulation (C. Alén, R. Pagliarini, and A. Sonenshein, unpublished data), but the role that iron limitation plays in aconitase binding to gerE mRNA is unclear. In gel mobility shift assays, binding to gerE mRNA was stimulated at least twofold by the addition of an iron-specific chelator, dipyridyl (36). Aconitase is also susceptible to oxidative stress and, based on work with IRP-1, is likely to switch to the RNA binding form under such conditions (17, 18). The RNA binding form of aconitase may begin to predominate as stationary phase and sporulation proceed, because of either iron limitation or oxidative stress. In addition, Zheng and Losick (46) demonstrated that the nutritional status of early-stationary-phase cells can greatly influence late-sporulation gene expression. For instance, cotC expression was much higher when cells exhausted the nutrients themselves (DS medium) than when the cells were resuspended in a medium already lacking in nutrients (SM medium) (46). Whether this effect reflects different expression or activity of aconitase is unknown.
Assembly of the spore coat occurs in layers, as proteins build upon one another (10, 11). GerE is responsible for efficient timing of spore coat assembly through its ability to activate and then repress certain cot genes (12, 20, 21). It would not be surprising if synthesis of GerE were regulated in multiple ways to allow for timed accumulation of a protein that coordinates the assembly of the spore coat layers. The mutant aconitase of strain AWS133, with reduced ability to bind gerE mRNA, accumulates GerE protein more slowly than does the wild type, which presumably leads to aberrant expression of several σK-dependent genes and a consequent delay in spore formation.
Acknowledgments
We are very grateful to D. Rudner and H. Takamatsu for generous gifts of antibodies to σK and GerE, respectively. We also thank D. Lazinski, C. Squires, J. Coburn, B. Belitsky, D. RayChaudhuri, and D. Hava for many helpful discussions.
This work was supported by a research grant (GM036718) from the U.S. Public Health Service.
REFERENCES
- 1.Addess, K. J., J. P. Basilion, R. D. Klausner, T. A. Rouault, and A. Pardi. 1997. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 274:72-83. [DOI] [PubMed] [Google Scholar]
- 2.Alén, C., and A. L. Sonenshein. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. USA 96:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basilion, J. P., T. A. Rouault, C. M. Massinople, R. D. Klausner, and W. H. Burgess. 1994. The iron-responsive element-binding protein: localization of the RNA-binding site to the aconitase active-site cleft. Proc. Natl. Acad. Sci. USA 91:574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulteau, A. L., H. A. O'Neill, M. C. Kennedy, M. Ikeda-Saito, G. Isaya, and L. I. Szweda. 2004. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305:242-245. [DOI] [PubMed] [Google Scholar]
- 5.Constable, A., S. Quick, N. K. Gray, and M. W. Hentze. 1992. Modulation of the RNA-binding activity of a regulatory protein by iron in vitro: switching between enzymatic and genetic function? Proc. Natl. Acad. Sci. USA 89:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig, J. E., M. J. Ford, D. C. Blaydon, and A. L. Sonenshein. 1997. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J. Bacteriol. 179:7351-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crater, D. L., and C. P. Moran, Jr. 2002. Two regions of GerE required for promoter activation in Bacillus subtilis. J. Bacteriol. 184:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingman, D. W., M. S. Rosenkrantz, and A. L. Sonenshein. 1987. Relationship between aconitase gene expression and sporulation in Bacillus subtilis. J. Bacteriol. 169:3068-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingman, D. W., and A. L. Sonenshein. 1987. Purification of aconitase from Bacillus subtilis and correlation of its N-terminal amino acid sequence with the sequence of the citB gene. J. Bacteriol. 169:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driks, A. 2002. Proteins of the spore core and coat, p. 527-535. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 11.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 12.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouet, A., and A. L. Sonenshein. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray, N. K., K. Pantopoulous, T. Dandekar, B. A. Ackrell, and M. W. Hentze. 1996. Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements. Proc. Natl. Acad. Sci. USA 93:4925-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray, N. K., S. Quick, B. Goossen, A. Constable, H. Hirling, L. C. Kuhn, and M. W. Hentze. 1993. Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur. J. Biochem. 218:657-667. [DOI] [PubMed] [Google Scholar]
- 16.Haile, D. J., T. A. Rouault, J. B. Harford, M. C. Kennedy, G. A. Blondin, H. Beinert, and R. D. Klausner. 1992. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc. Natl. Acad. Sci. USA 89:11735-11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haile, D. J., T. A. Rouault, C. K. Tang, J. Chin, J. B. Harford, and R. D. Klausner. 1992. Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: role of the iron-sulfur cluster. Proc. Natl. Acad. Sci. USA 89:7536-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentze, M. W., and L. C. Kuhn. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 93:8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoch, J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441-465. [DOI] [PubMed] [Google Scholar]
- 20.Ichikawa, H., R. Halberg, and L. Kroos. 1999. Negative regulation by the Bacillus subtilis GerE protein. J. Biol. Chem. 274:8322-8327. [DOI] [PubMed] [Google Scholar]
- 21.Ichikawa, H., and L. Kroos. 2000. Combined action of two transcription factors regulates genes encoding spore coat proteins of Bacillus subtilis. J. Biol. Chem. 275:13849-13855. [DOI] [PubMed] [Google Scholar]
- 22.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865-878. [DOI] [PubMed] [Google Scholar]
- 23.Kaldy, P., E. Menotti, R. Moret, and L. C. Kuhn. 1999. Identification of RNA-binding surfaces in iron regulatory protein-1. EMBO J. 18:6073-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaptain, S., W. E. Downey, C. Tang, C. Philpott, D. Haile, D. G. Orloff, J. B. Harford, T. A. Rouault, and R. D. Klausner. 1991. A regulated RNA binding protein also possesses aconitase activity. Proc. Natl. Acad. Sci. USA 88:10109-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, H. J., S. I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. 2003. Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel, B., R. Losick, and P. Stragier. 1990. The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 4:525-535. [DOI] [PubMed] [Google Scholar]
- 27.Kuwana, R., M. Ikejiri, S. Yamamura, H. Takamatsu, and K. Watabe. 2004. Functional relationship between SpoVIF and GerE in gene regulation during sporulation of Bacillus subtilis. Microbiology 150:163-170. [DOI] [PubMed] [Google Scholar]
- 28.Lu, S., R. Halberg, and L. Kroos. 1990. Processing of the mother-cell sigma factor, sigma K, may depend on events occurring in the forespore during Bacillus subtilis development. Proc. Natl. Acad. Sci. USA 87:9722-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller, J. P., G. Bukusoglu, and A. L. Sonenshein. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J. Bacteriol. 174:4361-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantopoulos, K., and M. W. Hentze. 1998. Activation of iron regulatory protein-1 by oxidative stress in vitro. Proc. Natl. Acad. Sci. USA 95:10559-10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pantopoulos, K., and M. W. Hentze. 1995. Rapid responses to oxidative stress mediated by iron regulatory protein. EMBO J. 14:2917-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato, T., Y. Samori, and Y. Kobayashi. 1990. The cisA cistron of Bacillus subtilis sporulation gene spoIVC encodes a protein homologous to a site-specific recombinase. J. Bacteriol. 172:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schalinske, K. L., O. S. Chen, and R. S. Eisenstein. 1998. Iron differentially stimulates translation of mitochondrial aconitase and ferritin mRNAs in mammalian cells. Implications for iron regulatory proteins as regulators of mitochondrial citrate utilization. J. Biol. Chem. 273:3740-3746. [DOI] [PubMed] [Google Scholar]
- 35.Schlegl, J., V. Gegout, B. Schlager, M. W. Hentze, E. Westhof, C. Ehresmann, B. Ehresmann, and P. Romby. 1997. Probing the structure of the regulatory region of human transferrin receptor messenger RNA and its interaction with iron regulatory protein-1. RNA 3:1159-1172. [PMC free article] [PubMed] [Google Scholar]
- 36.Serio, A. W. 2005. The role of aconitase, a bi-functional protein, in iron regulation and sporulation of Bacillus subtilis. Ph.D. thesis. Tufts University, Boston, Mass.
- 37.Serio, A. W., and A. L. Sonenshein. 2006. Expression of yeast mitochondrial aconitase in Bacillus subtilis. J. Bacteriol. 188:6406-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 39.Somerville, G., M. S. Chaussee, C. I. Morgan, J. Ross Fitzgerald, D. W. Dorward, L. J. Reitzer, and J. M. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somerville, G., C. A. Mikoryak, and L. Reitzer. 1999. Physiological characterization of Pseudomonas aeruginosa during exotoxin A synthesis: glutamate, iron limitation, and aconitase activity. J. Bacteriol. 181:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stragier, P., B. Kunkel, L. Kroos, and R. Losick. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507-512. [DOI] [PubMed] [Google Scholar]
- 42.Tang, Y., and J. R. Guest. 1999. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 145:3069-3079. [DOI] [PubMed] [Google Scholar]
- 43.Tang, Y., J. R. Guest, P. J. Artymiuk, R. C. Read, and J. Green. 2004. Post-transcriptional regulation of bacterial motility by aconitase proteins. Mol. Microbiol. 51:1817-1826. [DOI] [PubMed] [Google Scholar]
- 44.Tang, Y., M. A. Quail, P. J. Artymiuk, J. R. Guest, and J. Green. 2002. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology 148:1027-1037. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, T. J., N. Bertrand, J. L. Tang, J. X. Feng, M. Q. Pan, C. E. Barber, J. M. Dow, and M. J. Daniels. 1998. The rpfA gene of Xanthomonas campestris pathovar campestris, which is involved in the regulation of pathogenicity factor production, encodes an aconitase. Mol. Microbiol. 28:961-970. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]