Abstract
Databases of experimentally determined protein interactions provide information on binary interactions and on involvement in multiprotein complexes. These data are valuable for understanding the general properties of the interaction between proteins as well as for the development of prediction schemes for unknown interactions. Here we analyze experimentally determined protein interactions by measuring various sequence, genomic, transcriptomic, and proteomic attributes of each interacting pair in the yeast Saccharomyces cerevisiae. We find that dividing the data into two groups, one that includes binary interactions within protein complexes (stable) and another that includes binary interactions that are not within complexes (transient), enables better characterization of the interactions by the different attributes and improves the prediction of new interactions. This analysis revealed that most attributes were more indicative in the set of intracomplex interactions. Using this data set for training, we integrated the different attributes by logistic regression and developed a predictive scheme that distinguishes between interacting and noninteracting protein pairs. Analysis of the logistic-regression model showed that one of the strongest contributors to the discrimination between interacting and noninteracting pairs is the presence of distinct pairs of domain signatures that were suggested previously to characterize interacting proteins. The predictive algorithm succeeds in identifying both intracomplex and other interactions (possibly the more stable ones), and its correct identification rate is 2-fold higher than that of large-scale yeast two-hybrid experiments.
Keywords: domain signature, genomewide analysis, stable interaction, transient interaction, logistic regression
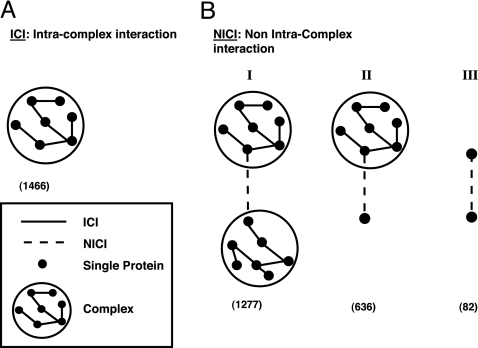
Protein interactions are central to almost all biological processes. Large-scale screens of protein–protein interactions (PPIs) in several organisms (1–4), together with PPI data from small-scale studies, have generated a large volume of experimental data that provides a partial picture of the cellular PPI networks. Previous studies that analyzed PPIs characterized their sequence domains and cellular properties (5–12) and provided insight into their evolution and regulation (13–23). At present, the richest information on PPIs is available for the yeast Saccharomyces cerevisiae, including documentation on experimentally determined binary interactions (1, 2, 24–26) as well as participation of proteins in the same complex (24, 27, 28). Intersection of these two data sources divides the binary interactions into those that occur within larger protein complexes [intracomplex interactions (ICIs)] and those that were not documented as belonging to complexes [non-intracomplex interactions (NICIs)]. The latter include interactions between proteins in different complexes, interactions between a noncomplexed protein and a protein in a complex, and interactions between two noncomplexed proteins (Fig. 1). A possible distinction between the ICIs and NICIs is the nature of the interactions: NICI interactions are likely to be transient whereas those between complex subunits, ICIs, are more stable. Separate analyses of the interactions in the ICI and NICI data sets allow better characterization of these interactions with regard to their various sequence, genomic, transcriptomic, and proteomic attributes. Indeed, our study shows that these two types of interactions differ in the examined characteristics, supporting our approach to distinguish rather than unify them while studying protein interactions. This separate characterization has an additional implication: by identifying NICIs with properties similar to those of ICIs, mistakes in the experimentally based annotations can be identified. It is possible that an NICI with properties similar to ICIs was misassigned because of incomplete experimental data, and our analysis may suggest reassigning it. This possibility is particularly intriguing in view of the current data, where most NICIs involve at least one protein from a complex (Fig. 1).
Fig. 1.
Two types of interactions, ICIs and NICIs. (A). ICIs: binary interactions within complexes (solid line). (B) NICIs: binary interactions not within complexes (dashed line). I, NICI between two complexes. II, NICI between a complex and a free protein. III, NICI between two free proteins. The counts of the various types of interacting pairs based on reliable data sets are shown in parentheses (for generation of data sets, see Fig. 2 and Materials and Methods).
Results
Attribute Assignment.
We selected nine attributes that may characterize pairs of physically interacting proteins (Table 1). Values for those nine attributes were assigned for each of all possible (≈6,0002/2) ≈1.8 × 107 pairs in the yeast proteome. Note that the attributes were defined at the pair level and not at the single-protein level. As Table 1 shows, the information about most attributes is incomplete, including both the computationally derived attributes (attributes 1–5) and the experimentally based attributes (attributes 6–9). Also, assignment of the computationally derived attributes depends on the stringency of the criteria used in the analysis. For example, following our criteria for fusion events (Tables 2 and 3, which are published as supporting information on the PNAS web site), this attribute was assigned to a very small fraction of all possible protein pairs (consistent with ref. 19). It is possible that with less stringent criteria, more pairs could be assigned. Using the stringent criteria, however, guarantees a higher quality of the data.
Table 1.
Data sources
| No. | Attribute abbreviation | Property of single protein | Proteome coverage, %* | Attribute of protein pair | No. of pairs with attribute† | Data source |
|---|---|---|---|---|---|---|
| 1 | DD | Domain signature | 65 | A domain–domain signature combination that appears in interacting protein pairs more often than expected at random | 454,714 | Our analysis (5) using InterPro database (51); learned from the data and assigned by 3-fold cross-validation |
| 2 | Fold | Protein fold | 26 | A combination of folds that appears in interacting protein pairs more often than expected at random | 177,895 | Our analysis using protein fold assignments of Hegyi et al. (52); learned from the data and assigned by 3-fold cross-validation |
| 3 | FE | NA‡ | NA‡ | Gene fusion event | 486 | Our analysis following Marcotte et al. (11) and Enright et al. (12) |
| 4 | PP | Phylogenetic profile | 100 | Consistent phylogenetic profiles | 822,789 | Our analysis following Pellegrini et al. (13) |
| 5 | GN | NA‡ | NA‡ | Conservation of gene neighborhood | 5,755 | von Mering et al. data (19) |
| 6 | Loc | Cellular localization | 72 | Colocalization | 3,497,490 | YPD (53) and Huh et al. (37) |
| 7 | Proc | Cellular process | 59 | Shared cellular process | 634,302 | YPD (53) |
| 8 | Exp | mRNA expression pattern | 100 | Coexpression | 94,370 | Based on clustering of Ihmels et al. (54) |
| 9 | Reg | Transcriptional regulation | 43.3 | Coregulation | 270,272 | YPD (53) and Lee et al. (55) |
*Fraction of proteins in S. cerevisiae that are annotated by this feature.
†No. of pairs with attributes among all possible ≈1.8 × 107 pairs in S. cerevisiae. Pairs with missing data were treated as not showing the attribute.
‡NA, not applicable.
Comparison of Attributes in the Different Data Sets.
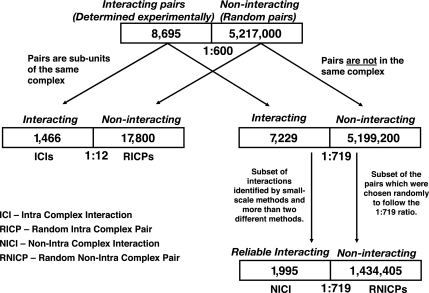
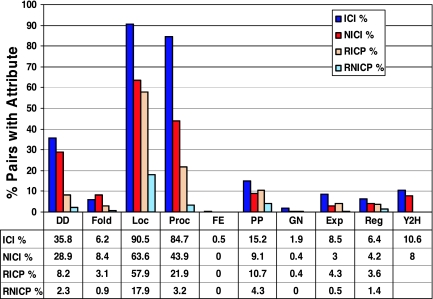
As described in Materials and Methods and in Fig. 2, we constructed two reliable data sets: 1,466 interacting pairs within complexes (ICIs) and 1,995 interacting pairs not within complexes (NICIs). For comparison with noninteracting protein data sets, we also constructed the corresponding random data sets: RICPs and RNICPs. Fig. 3 describes the fraction of pairs showing the various attributes in each data set under study. As seen in Fig. 3, for all attributes, except for the fold combinations, the fraction of pairs showing the attribute is higher in the set of ICIs compared with the set of NICIs (P ≤ 0.04 by a χ2 test with a Bonferroni correction). The fold attribute appeared more frequently in the set of NICIs, but this result was not statistically significant. We compared these fractions with the fraction of known ICIs and NICIs that the large-scale Y2H experiments detect (1, 2) (Fig. 3), and we found that four of the attributes are more sensitive than Y2H: domain–domain signatures, cellular colocalization, participation in a common cellular process, and consistent phylogenetic profiles. Interestingly, the Y2H detects a higher fraction of ICIs compared with NICIs (10.6% compared with 8%; P ≤ 0.01). We also examined the attribute frequencies in the respective sets of noninteracting proteins, RICPs and RNICPs, and we found that all attributes (except for the fusion event attribute) appeared less frequently than in the respective data sets of PPIs (Fig. 3).
Fig. 2.
Creating data sets of ICIs and NICIs and the corresponding data sets of noninteracting pairs. For the set of 8,695 interacting pairs, we generated a random set of 5,217,000 pairs. We chose this number to follow the rate of 1:600, based on the estimation of 30,000 interacting pairs and a total of 1.8 × 107 possible protein pairs in S. cerevisiae. The interacting pairs included 1,466 pairs in complexes. The random pairs included 17,800 pairs where both proteins participate in the same complex. The latter were used as the data set of protein pairs in complexes that are not known to interact [random intracomplex pairs (RICPs)]. The rest of the interacting pairs included 7,229 pairs, and the rest of the random pairs included 5,199,200 pairs (a ratio of 1:719). In the analysis, because we used only 1,995 known interacting pairs that were the most reliable, we also reduced the random set accordingly to keep the same ratio of 1:719 between interacting and noninteracting pairs. As a result, the set of noninteracting protein pairs not in complexes included 1,434,405 pairs that were derived randomly from the rest of 5,199,200 random pairs [random non-intracomplex pairs (RNICPs)].
Fig. 3.
Attribute coverage of the various data sets. Fractions of protein pairs with an attribute among the total number of pairs in a data set are shown [fractions of pairs revealed by the yeast two-hybrid (Y2H) method are shown for comparison]. ICI, 1,466 physically interacting protein pairs within complexes (blue); NICI, 1,995 physically interacting pairs not in complexes, identified by reliable methods (red); RICP, 17,800 random pairs (noninteracting) within complexes (light orange); and RNICP, 1,434,405 random pairs not in complexes (light blue). For a description of the data sets, see Fig. 2. For attribute abbreviations, see Table 1.
Prediction of Interacting Pairs.
Although each attribute distinguishes to some extent between interacting and noninteracting pairs, the integration of all attributes, appropriately weighted, is expected to provide better discrimination, and thus it could potentially be used for the development of a predictive algorithm. To incorporate the nine attributes into a predictive scheme, we used logistic regression (LR). Similar to linear regression, LR provides the best fitting function between a dependent variable and a set of independent variables. LR provides a function that incorporates the relative contributions of the independent variables (here, attributes) to compute the probability of an event (here, interaction between two proteins) (ref. 29; see also Supporting Methods, which is published as supporting information on the PNAS web site). It is possible then to choose a probability threshold above which a pair of proteins is determined as putatively interacting.
Our study shows that there are differences in the attribute distributions between ICIs and NICIs, suggesting that it would be more appropriate to treat them separately rather than unifying them as one data set. Therefore, we turned to developing two separate LR models for each type of interaction. However, the low specificity values of the NICI attributes (fractions of interacting pairs having an attribute among all pairs with that attribute), caused by the huge size of the data set of noninteracting pairs following our 1:600 rate estimation (Fig. 2), did not enable sufficient distinction between NICIs and RNICPs. Thus, the LR model developed on the set of noncomplexed proteins provided unsatisfactory predictions. The LR model developed by using the ICI and RICP data sets, however, looked much more promising, as described below. This LR model enables the identification of new ICIs based on their attributes.
The LR was carried out on the ICI and RICP data sets with 5-fold cross-validation (choosing 80% of the pairs for training and 20% for testing), showing consistent results and consistent attribute coefficients in all five tests (see Table 4, which is published as supporting information on the PNAS web site). Based on these results, the LR analysis was applied to the whole data sets of ICIs and RICPs, resulting in one set of attribute coefficients to be used in further predictions (see Table 5, which is published as supporting information on the PNAS web site). The overall model fit as estimated by the likelihood ratio was highly statistically significant (P < 0.0001). The coefficients of the domain–domain signatures, colocalization, and shared cellular process deviated significantly from zero (P < 0.0001). Interestingly, recently Lu et al. (30) also found that four strong features are sufficient for satisfactory predictions of co-complexed proteins, two of which regarded participation in a common cellular process.
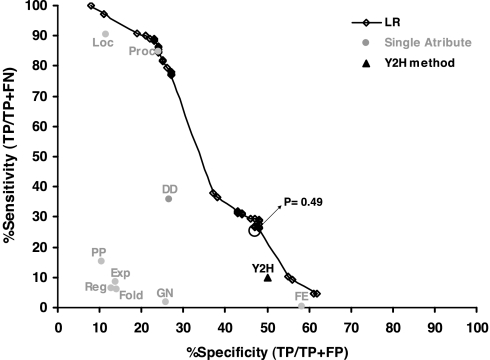
Fig. 4 describes the sensitivity/specificity values of the predictions for different probability thresholds, above which a pair is determined as interacting. The sensitivity is the fraction of correctly predicted pairs among the known interacting pairs. The specificity is the fraction of correctly predicted pairs among all predicted pairs. For comparison, we show also the sensitivity/specificity values of the interactions identified by Y2H and by the individual attributes. The combination of parameters by the LR provides better sensitivity and/or specificity than do most individual attributes and the Y2H method. For a specificity of ≈50%, similar to that of the genomewide Y2H, the LR achieves >2-fold higher sensitivity: we correctly predict 26% of ICIs, whereas the Y2H method identifies only 10.6% of them. When applied to the set of NICIs, the LR-based predictions succeed in identifying 11% of these interactions correctly (224 of 1,995 NICI interactions).
Fig. 4.
Performance of the predictive scheme based on the LR. For each probability threshold of the LR, the sensitivity vs. specificity is plotted (the connected line was drawn only for illustration). The threshold of 0.49 that ensures ≈50% specificity is marked on the graph. For comparison, the specificity/sensitivity values of the Y2H (triangle) and all other nine attributes (gray circles) are shown. For attribute abbreviations, see Table 1.
Our ability to identify interactions between free proteins and complexes by using the LR-based scheme that was trained on ICIs suggests that these interactions may have been missed from the complexes. Indeed, for some of these NICI interactions we managed to find support in the literature for their involvement in complexes. For space limitations, only one such example is described here. This example regards the heme activator protein HapI and the proteins that regulate its activity. It is known that the proteins Hsp82/Hsc82, Ydj1, and Sro9 are associated with HapI in the absence of heme (31, 32), and it was also shown that the proteins Ssa1 and Ssa2 play a major role in HapI repression (32). In our data, Ssa1 and Ssa2 are not documented as included in the complex with the other proteins, but our predictive scheme still succeeds in identifying the (known) interactions Ssa1–Hsp82 and Ssa2–Hsp82, suggesting that Ssa1 and Ssa2 participate in the complex. Likewise, we identify interactions between complexes, such as the interactions between the coat protein complex COPII with the soluble N-ethylmaleimide-sensitive fusion attachment receptor complexes t-SNARE and v-SNARE, which play a role in cellular vesicle transport (33).
When the algorithm was applied to all possible ≈1.8 × 107 pairs in the yeast proteome (excluding the known interacting pairs) we predicted 29,181 putative interactions (with a cutoff of 0.49, which yields ≈50% specificity in prediction as in Y2H). Of these putative interactions, 1,946 are between subunits of the same complex, i.e., predicted ICIs, and the rest are predicted NICIs. Among these predicted NICIs, 87% involved at least one complexed protein, and 13% involved two free proteins, probably regarding relatively stable interactions. Among the predicted interactions, there are interactions that were revealed by large-scale screening methods that are considered as less reliable, and our study supports them (see Table 6, which is published as supporting information on the PNAS web site). One such example regards the proteins Lst8 and Sec13, which belong to different complexes and play a role in protein transport. It was suggested that these two proteins may function as components of a post-Golgi secretory vesicle coat (34). The relationship between Sec13 and Lst8, which was determined previously only as a genetic interaction (34), was strongly supported by our analysis, and it was predicted as a PPI with a probability of 0.85. The pair Sec13–Lst8 showed six of the nine attributes: domain–domain signature, fold combination, colocalization, shared cellular process, fusion event, and consistent phylogenetic profiles. Remarkably, both Sec13 and Lst8 contain the WD-40 repeat that consists of five to eight tandem repeats, each containing a central Trp–Asp motif. Because the WD-40 repeat is known to be involved in PPI (35), it is possible that it also mediates the interaction between Sec13 and Lst8.
Discussion
The intersection of the information on pairwise interactions in the yeast S. cerevisiae with that on the involvement in a protein complex revealed an intriguing picture. In a data set of reliable interactions, 42% of the known pairwise interactions resided within multiprotein complexes. Among the rest of the interactions, 96% involved at least one pair-mate that participates in a protein complex (Fig. 1). This very high fraction may either imply that the current experimental methods that determine binary interactions are biased toward proteins in complexes or that most interactions in the cell occur either within or between complexes. It is also conceivable that interactions between free proteins and complexes or between different complexes may be interactions that occur only under specific conditions, and therefore they were not identified as belonging to one of the complexes. Do the ICIs and NICIs differ in their properties? In the current analysis we show that the ICIs are better characterized [larger fractions of pairs showing each of the attributes (Fig. 3)] and that by using their characteristics, additional interactions can be predicted within complexes, between complexes, and between free proteins and complexes.
Characterization of Interactions.
To characterize the various types of interactions, we examined their association with nine attributes of protein pairs (Table 1 and Supporting Methods). These attributes can be divided into two major classes: one class includes two attributes that relate directly to the physical interactions, the domain–domain signatures and fold combinations. Both attributes were learned from the database of experimentally determined PPIs, and they seem relevant to the actual physical interactions between proteins (it is widely acknowledged that there are special domains that participate in the physical interaction, and it is conceivable that there are preferred folds that are more suitable for interaction with each other). The other class includes attributes that may be indicative of functional relationships between proteins but not necessarily of their physical interactions. These attributes were derived from sources of information that are independent of the data sets of interacting proteins. These attributes can be also divided into different types. Colocalization and participation in a shared cellular process are implicit for PPIs (both ICIs and NICIs), and their presence or absence mostly testifies to the current state of annotation. The attributes of coexpression and coregulation are relevant to the coordinated transcription of the interacting proteins, and as such they are expected to provide insights into ICIs and NICIs. The attributes of fusion events, consistent phylogenetic profiles, and conserved gene neighborhoods have evolutionary implications, and they are expected to provide valuable insight into possible differences between ICIs and NICIs in this regard. Here we discuss the main findings regarding the various attributes.
Domain–domain signatures.
This attribute was more prominent in ICIs than in NICIs, and in general it was more abundant in PPIs than in noninteracting pairs. It was also assigned a high coefficient by the LR, suggesting that it is a good discriminator between interacting and noninteracting pairs. It should be emphasized that the domain–domain signatures were not derived directly from all interacting pairs, but rather they were learned from one part of the data and assigned to the other part (see Supporting Methods). Several other studies demonstrated the value of this attribute for PPI prediction by using different data sets of PPIs (6–9). These findings substantiate the biological meaning of domain–domain signatures and support the suggestion that they are characteristic of physically interacting proteins (5–9). Indeed, a literature and database inspection of the overrepresented pairs of domain signatures in interacting proteins (5) showed that 56% of the domain signature pairs for which information is available are involved in physical interactions (E.S. and H.M., unpublished results). Interestingly, pairs of identical domain signatures were found in 15% of the ICIs and in 7.5% of the NICIs. Among the interactions predicted on the full proteome, 13.5% had the same domain signatures in the two pair-mates, in general agreement with the suggestion that identical interfaces are often used for interaction (for review, see ref. 36).
Colocalization and shared cellular process.
Intuitively, involvement of two proteins in a complex implies that they should be found at the same cellular compartment in localization experiments (37). We found, however, that within complexes, interacting pairs are significantly more often documented as colocalized than other pairs, which suggests that noninteracting subunits within the complex may be either spurious or present transiently (depending on the cellular condition). The latter might complicate their detection as colocalized by independent studies. This possibility implies that the protein composition of multiprotein complexes changes dynamically, as indeed was found in the large-scale studies in which these complexes were discovered (27, 28). Thus, there are complexes that have identical core proteins, and different proteins may join them for specific tasks. Similar arguments hold for the shared cellular process attribute.
Because colocalization and shared cellular process are implicit for interacting proteins, their higher association with ICIs compared with NICIs indicates only that the annotation is incomplete. With complete annotation, we will be able to learn whether there are different cellular compartments that are preferred for the different interactions.
mRNA coexpression.
mRNA coexpression of pair-mates was more prominent among the interacting pairs in complexes than in other pairs in complexes. Dezso et al. (38) showed that there are protein pairs in complexes that exhibit these three properties (colocalization, coexpression, and shared process), and they suggested that these pairs constitute the cores of the complexes. Our results support this conjecture and emphasize that the protein pairs making up these cores were identified experimentally as physically interacting. mRNA coexpression was also relatively more pronounced in ICIs compared with NICIs, perhaps to ensure the right stoichiometry of subunits in complexes. Similar findings regarding the association of coexpression and coregulation with ICIs were also reported in other studies (39, 40).
Phylogenetic profile.
This attribute, which reflects the consistent presence/absence of the two pair-mates in different organisms, was more frequent among the ICIs than among the NICIs or RICPs. This observation may suggest that the stable interactions within complexes make up the cores that are responsible for basic, evolutionary conserved mechanisms, whereas the other protein pairs in the complexes or the transient pairs comprise interactions that are organism-specific and play a role in more specialized processes.
Prediction of Interacting Pairs.
Attempts to predict physical interactions have been reported with varying degrees of success (6–9, 41–43). Some of the attributes, such as coexpression or fusion events, were used previously for predictions of co-complexed proteins (44–46) or of functional relations (10–13). These predictions identify associations between protein pairs which are not necessarily physical interactions, although for the identified fusion events, many of the predicted relations correspond to actual interactions (as seen also in Fig. 4). In general, such attributes can support putative binary interactions, but they cannot determine them directly. Also, the domain–domain signatures were used by several groups for predicting PPIs (6–9). However, there are two limitations of using only domain–domain signatures for predictions. First, it is clear that not every two proteins showing the domain–domain signatures interact; and second, not all interacting proteins can be classified by the currently annotated domains (7). Therefore, combining the domain–domain signatures with other attributes in a predictive scheme allows for more reliable predictions.
An attempt to develop a predictive scheme based on the whole data set (ICIs and NICIs together) was not successful, nor did an attempt to develop a predictive scheme for NICIs based on their attributes succeed. In both cases, one major reason for the failure probably involves the very large differences in size between the interacting and noninteracting pairs. We believe that the sizes of these data sets reflect the actual situation in the cell, where, among millions of possible PPIs, only a very small fraction of pairs actually interact [≈30,000 based on several estimations (47)]. The well characterized ICIs and the smaller size of the RICP set allow the development of a reasonable predictive scheme based on the current attributes, which shows relatively good performance for this set. It is likely that for identifying NICIs among so many possible pairs, the studied attributes are not sufficient, and additional attributes of different types are required. Some additional attributes may be measures of essentiality (30) and coordinated protein levels (48, 49), which were shown recently to correlate with PPI and with functionally related proteins. Still, the predictive scheme that was trained on the ICIs succeeded in identifying 11% of the NICIs. This success rate is ≈60-fold higher than the chance probability for detecting these interactions (0.17%), and it is 1.4-fold higher than their identification rate by the Y2H method (using the same specificity rate as in the Y2H in the predictions). Most of the NICIs that were correctly identified by the LR model involve at least one protein in a complex. These interactions may have been missed from the complexes because they occur only under certain conditions, and now they are identified by us as putative components of the complexes.
The LR model provides the probability of interaction for each protein pair. In our analysis, we determined 0.49 as the threshold probability above which a pair is determined to be interacting (resulting in a specificity of ≈50%). We examined the attribute combinations that lead to probabilities above this threshold, and we found in the data set of interacting proteins as well as when the analysis was applied to the whole yeast proteome that only pairs that showed the three attributes of domain–domain signatures, colocalization, and shared cellular process passed the probability threshold. As discussed above, the first two characteristics are indeed prerequisite for an interaction to occur, which implies that with the current state of annotation, only proteins that are documented as colocalized, participating in a common cellular process, and containing appropriate domain signatures can be predicted to interact. However, what if such information is unavailable or if we are interested in more reliable prediction (requiring higher specificity)? Theoretically, there are additional attribute combinations that pass the threshold (for details, see Table 7, which is published as supporting information on the PNAS web site), and when the data annotation will be more complete, protein pairs with such combinations may be revealed as well.
Our predictive model provides a sensitivity that is >2-fold higher than the widely used Y2H method for the same specificity level. It is clear, however, that the sensitivity and specificity provided by the LR-based predictive scheme should be further improved. One possible improvement involves the attribute assignment. Our predictive scheme uses attributes that are all based on documentation available in the various biological databases. For many proteins in our data sets there is no information for some of these attributes (Table 1). For example, many of the yeast proteins are not yet characterized by domain signatures, and therefore they cannot be classified by this attribute. Thus, it is conceivable that when the documentation becomes complete, the associations learned by the LR model will be more precise and will lead to improved predictions. Also, additional attributes that are characteristic of physical interactions in general and of less stable interactions in particular should lead to improvement. Furthermore, using this scheme for initial screening and then applying a structure-based approach as proposed by Aloy et al. (43) may also be a promising direction, leading to a more accurate picture of both intra- and intercomplex interactions.
Materials and Methods
Data Sets of Interacting Protein Pairs.
The data of S. cerevisiae interacting protein pairs were collected from three public databases, MIPS (24), DIP (25), and BIND (26), and from compilations of genomewide Y2H assays (1, 2). After exclusion of redundancies and homodimers, the database included a total of 8,695 binary interactions, involving 4,136 proteins. From this database, two data sets were derived (Fig. 2): (i) a data set of 1,466 ICIs, which included pair-mates that were identified as participating in a complex of at least three proteins; the complex collection included a nonredundant set of curated complexes from MIPS (24) as well as large-scale compilations of tandem affinity purifications (27) and high-throughput mass spectrometric protein complex identification (28); (ii) a reliable set of 1,995 NICIs, which included protein pairs that were not identified in complexes. This set included interactions of relatively high confidence (50) that were identified experimentally by non-genomewide methods (coimmunoprecipitation, cross-linking, small-scale Y2H, etc.) and interactions that were identified by at least two different methods. Interactions that were identified as genetic only or by a large-scale Y2H screen only were not included.
Data Sets of Noninteracting Protein Pairs.
The proteome size of S. cerevisiae, which includes ≈6,000 proteins, implies that there are ≈1.8 × 107 possible protein pairs. Current estimations of the yeast interactome (the number of PPIs) range from 12,000 (50) to >30,000 (47). Taking the more frequently used estimation of 30,000 interactions implies that the ratio between interacting and noninteracting pairs in yeast is ≈1:600 (30,000:≈1.8 × 107). In our analyses, we follow this ratio of 1:600 between the known interacting pairs and the random pairs (representing the noninteracting pairs). For the 8,695 known binary interactions in our database, we generated 5,217,000 random pairs from all ≈6,000 yeast proteins, and from those pairs, we extracted the RICPs and RNICPs (see Fig. 2).
Data Sources of Attributes and Representation.
Table 1 provides a brief summary of the attributes and their data sources (for more detail, see Supporting Methods). Each yeast protein pair is represented as a vector of nine entries, where each entry represents an attribute, and the value indicates whether the attribute was found to characterize this particular interaction. An entry is assigned a value of 1 if the protein pair shows the attribute and a value of 0 if it does not show the attribute or if there is no information about the attribute.
Supplementary Material
Acknowledgments
We thank Joel Sussman for hosting E.S. in Rehovot and Ophry Pines, Mario Baras, Norman Grover, Shmuel Sattath, Ariel Jaimovich, and David Sprinzak for helpful discussions. We thank Amit Fliess for help in data mining and our group members for useful comments on the manuscript. This study was supported by Israeli Science Foundation (founded by the Israel Academy of Sciences and Humanities) Grant 558/01 (to H.M.) and by European Union Grant QLRI-CT-2001-00015. E.S. was supported by the Horowitz Foundation.
Abbreviations
- PPI
protein–protein interaction
- ICI
intracomplex interaction
- NICI
non-intracomplex interaction
- RICP
random intracomplex pair
- RNICP
random non-intracomplex pair
- Y2H
yeast two-hybrid
- LR
logistic regression.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Frontiers in Bioinformatics: Unsolved Problems and Challenges,” held October 15–17, 2004, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA. Papers from this Colloquium will be available as a collection on the PNAS web site. See the introduction to this Colloquium on page 13355 in issue 38 of volume 102. The complete program is available on the NAS web site at www.nasonline.org/bioinformatics.
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprinzak E, Margalit H. J Mol Biol. 2001;311:681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- 6.Gomez SM, Rzhetsky A. Pac Symp Biocomput. 2002;7:413–424. [PubMed] [Google Scholar]
- 7.Deng M, Mehta S, Sun F, Chen T. Genome Res. 2002;12:1540–1548. doi: 10.1101/gr.153002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng SK, Zhang Z, Tan SH. Bioinformatics. 2003;19:923–929. doi: 10.1093/bioinformatics/btg118. [DOI] [PubMed] [Google Scholar]
- 9.Gomez SM, Noble WS, Rzhetsky A. Bioinformatics. 2003;19:1875–1881. doi: 10.1093/bioinformatics/btg352. [DOI] [PubMed] [Google Scholar]
- 10.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 12.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 13.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandekar T, Snel B, Huynen M, Bork P. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 15.Ge H, Liu Z, Church GM, Vidal M. Nat Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 16.Grigoriev A. Nucleic Acids Res. 2001;29:3513–3519. doi: 10.1093/nar/29.17.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Mol Cell Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 18.Kemmeren P, van Berkum NL, Vilo J, Bijma T, Donders R, Brazma A, Holstege FC. Mol Cell. 2002;9:1133–1143. doi: 10.1016/s1097-2765(02)00531-2. [DOI] [PubMed] [Google Scholar]
- 19.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 20.Yeger-Lotem E, Margalit H. Nucleic Acids Res. 2003;31:6053–6061. doi: 10.1093/nar/gkg787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeger-Lotem E, Sattath S, Kashtan N, Itzkovitz S, Milo R, Pinter RY, Alon U, Margalit H. Proc Natl Acad Sci USA. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader JS, Chaudhuri A, Rothberg JM, Chant J. Nat Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 23.Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Proc Natl Acad Sci USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader GD, Betel D, Hogue CW. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 28.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 29.Hosmer D, Lemeshow S. Applied Logistic Regression. New York: Wiley; 2000. [Google Scholar]
- 30.Lu LJ, Xia Y, Paccanaro A, Yu H, Gerstein M. Genome Res. 2005;15:945–953. doi: 10.1101/gr.3610305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Hach A, Wang C. Mol Cell Biol. 1998;18:3819–3828. doi: 10.1128/mcb.18.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hon T, Lee HC, Hach A, Johnson JL, Craig EA, Erdjument-Bromage H, Tempst P, Zhang L. Mol Cell Biol. 2001;21:7923–7932. doi: 10.1128/MCB.21.23.7923-7932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato K, Nakano A. Nat Struct Mol Biol. 2005;12:167–174. doi: 10.1038/nsmb893. [DOI] [PubMed] [Google Scholar]
- 34.Roberg KJ, Bickel S, Rowley N, Kaiser CA. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enninga J, Levay A, Fontoura BM. Mol Cell Biol. 2003;23:7271–7284. doi: 10.1128/MCB.23.20.7271-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nooren IM, Thornton JM. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 38.Dezso Z, Oltvai ZN, Barabasi AL. Genome Res. 2003;13:2450–2454. doi: 10.1101/gr.1073603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jansen R, Greenbaum D, Gerstein M. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonis N, Van Helden J, Cohen GN, Wodak SJ. Genome Biol. 2004;5:R33. doi: 10.1186/gb-2004-5-5-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu L, Arakaki AK, Lu H, Skolnick J. Genome Res. 2003;13:1146–1154. doi: 10.1101/gr.1145203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valencia A, Pazos F. Methods Biochem Anal. 2003;44:411–426. [PubMed] [Google Scholar]
- 43.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, et al. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 44.Jansen R, Lan N, Qian J, Gerstein M. J Struct Funct Genomics. 2002;2:71–81. doi: 10.1023/a:1020495201615. [DOI] [PubMed] [Google Scholar]
- 45.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 46.Zhang LV, Wong SL, King OD, Roth FP. BMC Bioinformatics. 2004;5:38. doi: 10.1186/1471-2105-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grigoriev A. Nucleic Acids Res. 2003;31:4157–4161. doi: 10.1093/nar/gkg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraser HB, Hirsh AE, Wall DP, Eisen MB. Proc Natl Acad Sci USA. 2004;101:9033–9038. doi: 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lithwick G, Margalit H. Nucleic Acids Res. 2005;33:1051–1057. doi: 10.1093/nar/gki261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sprinzak E, Sattath S, Margalit H. J Mol Biol. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 51.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hegyi H, Lin J, Greenbaum D, Gerstein M. Proteins. 2002;47:126–141. doi: 10.1002/prot.10078. [DOI] [PubMed] [Google Scholar]
- 53.Csank C, Costanzo MC, Hirschman J, Hodges P, Kranz JE, Mangan M, O'Neill K, Robertson LS, Skrzypek MS, Brooks J, et al. Methods Enzymol. 2002;350:347–373. doi: 10.1016/s0076-6879(02)50973-3. [DOI] [PubMed] [Google Scholar]
- 54.Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Nat Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- 55.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.