Abstract
Neuromelanin (NM) isolated from the substantia nigra region of the human brain was studied by scanning probe and photoelectron emission microscopies. Atomic force microscopy reveals that NM granules are comprised of spherical structures with a diameter of ≈30 nm, similar to that observed for Sepia cuttlefish, bovine eye, and human eye and hair melanosomes. Photoelectron microscopy images were collected at specific wavelengths of UV light between 248 and 413 nm, using the spontaneous-emission output from the Duke OK-4 free electron laser. Analysis of the data establishes a threshold photoionization potential for NM of 4.5 ± 0.2 eV, which corresponds to an oxidation potential of −0.1 ± 0.2 V vs. the normal hydrogen electrode (NHE). The oxidation potential of NM is within experimental error of the oxidation potential measured for human eumelanosomes (−0.2 ± 0.2 V vs. NHE), despite the presence of a significant fraction of the red pigment, pheomelanin, which is characterized by a higher oxidation potential (+0.5 ± 0.2 V vs. NHE). Published kinetic studies on the early chemical steps of melanogenesis show that in the case of pigments containing a mixture of pheomelanin and eumelanin, of which NM is an example, pheomelanin formation occurs first with eumelanin formation predominantly occurring only after cysteine levels are depleted. Such a kinetic model would predict a structural motif with pheomelanin at the core and eumelanin at the surface, which is consistent with the measured surface oxidation potential of the ≈30-nm constituents of NM granules.
Keywords: free electron laser, oxidative stress, photoelectron imaging, substantia nigra, Parkinson's disease
Neuromelanin (NM) is a naturally occurring pigment in the human central nervous system that has received much attention in recent years because of its role in the selective neuronal loss associated with Parkinson's disease (PD). NM is found in highest concentration in catecholaminergic neurons of the substantia nigra (SN) and locus coeruleus regions of the human midbrain (1–3). An observable amount of NM begins to appear in humans at ≈3–5 years of age and increases steadily thereafter (4, 5). With the onset and progression of PD, the concentration of NM decreases rapidly as pigmented dopaminergic neurons are selectively degraded while nonpigmented neurons are mostly spared (6). A direct, causal link between NM concentration and neuronal vulnerability has not yet been demonstrated, although several hypotheses exist (7–9). In the regions of highest neuronal loss, namely the SN, studies have shown an increase in iron levels in the late stages of PD (10). Based on these observations, NM's role in avidly chelating redox-active metal ions, toxic organic compounds, and free radicals would serve as a protective role (11–13). Neurons that produce catecholamines (dopamine, norepinephrine, etc.) are known to experience a high oxidative load because of the synthetic pathway of their respective signaling molecules (14, 15). As a result, the synthesis of NM is hypothesized to be the result of the cell's defense mechanism against high oxidative stress. On the other hand, NM could become the source of free radicals and cytotoxic compounds upon degradation of highly pigmented neurons, thereby perpetuating a cycle of oxidative stress and neuronal loss (16, 17).
This complex duality concerning NM's physiological functions forms the basis of our investigation into the oxidative capacity of NM. Several groups have shown through chemical degradation analyses that NM is composed of both eumelanic and pheomelanic components (18–21). In addition to pigment, NM isolated from SN contains an aliphatic component and a peptide component (3); together, these form the large, aggregated structures observed in electron micrographs. The melanic component has been shown to be composed of two classes of molecules in specific proportions: one is a benzothiazine-based molecule characteristic of pheomelanin that is formed through the incorporation of cysteine with dopamine that forms 20–25% of the total melanic component of NM; the other is an indole-based molecule characteristic of eumelanin that is believed to be formed through the oxidation of dopamine, forming the remainder of the melanic component of NM. The identification of dopamine and cysteinyl-dopamine as building blocks of NM lend support to its detoxifying role, preventing an otherwise toxic intraneuronal accumulation of these compounds.
The molecular architecture of a pigment containing both pheomelanin and eumelanin is not understood. In vitro studies of the rate constants associated with the initial steps of eu- and pheomelanogenesis reveal that formation of pheomelanin dominates as long as cysteine is present (22–27). Once the supply of cysteine is depleted, eumelanin would be produced. Given that 20–25% of the pigment in NM is pheomelanin, such a model would suggest that the molecular structure of the NM pigment has a pheomelanin core and a surface that is predominantly eumelanin in character.
The morphological organization of naturally occurring pigments from sources other than the human brain has been investigated in recent years. These pigments include melanin from the ink sac of the cuttlefish Sepia officinalis (28), melanosomes isolated from human red and black hair (29), and melanosomes isolated from bovine and human eyes (30, 31). All of these melanosomes show a substructure of spherical constituents with diameters on the order of 30 nm, even though the size and shape of the fully aggregated melanosomes vary from sample to sample. Herein, our data confirm a similar substructure for NM.
In this study, we used photoelectron emission microscopy (PEEM) coupled to a free electron laser (FEL) as the light source to measure the ionization threshold for the surface of NM. There is a linear relationship between the measured ionization threshold and the surface oxidation potential (32). One of the powerful aspects of this technique is its ability to distinguish between pheomelanin and eumelanin pigments. Using human hair melanosomes as a standard, we have established that eumelanosomes have a surface oxidation potential of −0.2 ± 0.2 V vs. the normal hydrogen electrode (NHE) (33). Pheomelanosomes from human hair contain a mixture of eumelanin and pheomelanin, and this is reflected by two threshold potentials, −0.2 ± 0.2 and +0.5 ± 0.2 V, corresponding to the two pigments, respectively. Because FEL-PEEM is a surface technique, with penetration depths on the order of a few nanometers into organic materials, the pheomelanosome result indicates that both pigments can be found on or near the surface of the melanosomes.
Based on kinetic experiments discussed in detail later, there is good reason to hypothesize that the synthesis of NM will result in spatially heterogeneous structures comprised of a pheomelanin core that is encapsulated in eumelanin. FEL-PEEM can directly test this concept through its ability both to probe only the region near the surface of the granule and to distinguish eumelanin from pheomelanin.
Results
Imaging.
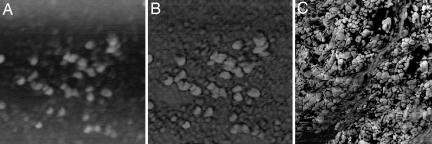
Isolated, dried NM samples of the SN from a human brain were imaged by using both scanning electron microscopy (SEM) and atomic force microscopy (AFM). Two types of structures are observed: small spherical particles, ≈30 nm in diameter, and larger NM granules, with an average diameter of ≈350 nm. The small particles were found in deposits of varying dimensions and were also seen on the surface of the larger granules, suggesting that these are aggregates of such smaller structures. In, Fig. 1, an SEM image shows aggregated NM granules. The ≈350-nm NM granules were rarely observed in isolation and appeared to have substructure that could not be adequately resolved by the electron microscope. We used AFM imaging to observe this substructure. AFM confirmed the SEM results but provided spatially resolved images from which the substructure of the NM granule could be seen (Fig. 2). The ≈350-nm granules, as well as aggregated NM deposits, appear to be comprised of the ≈30-nm-diameter entities. Such building blocks for pigments have been previously reported for many natural melanins, including human hair melanosomes, bovine and human eye melanosomes, and Sepia granules. Fig. 2 shows the height and phase AFM images resolving the NM substructure, measured as 30 ± 10 nm in diameter.
Fig. 1.
SEM image of dried NM pigment isolated from the SN region of human brain tissue. The image illustrates a commonly observed, large, amorphous aggregate (>10 μm in size) composed of smaller NM molecules, roughly spherical in shape and varying widely (200–500 nm) in diameter. The image is 6.75 μm × 4.50 μm.
Fig. 2.
AFM images of SN NM. The small substructures of NM, ≈30 nm in diameter, are observed both in isolation [A (height) and B (phase)] and in the highly aggregated structures [C (phase)]. The images in A and B are 950 nm × 950 nm; the image in C is 1.57 μm × 1.57 μm.
Threshold Photoionization Potentials from UV FEL-PEEM.
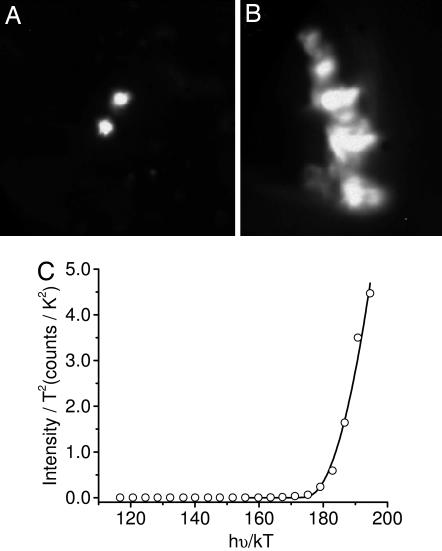
Wavelength-dependent PEEM images of isolated NM from the SN region of the human midbrain deposited on a silicon substrate were collected by using a tunable UV FEL light source. Fig. 3A and B shows PEEM images of isolated ≈350-nm granules and larger deposits of NM pigment. The integrated intensity (S) of the PEEM images was determined as a function of excitation wavelength for isolated granules and various regions within larger NM deposits. Fig. 3C shows data for an isolated granule, and the best fit of Eq. 1 gives an ionization threshold of ≈4.5 ± 0.2 eV. The threshold potential is independent of whether individual granules, large pigment deposits, or small regions within a pigment deposit are probed.
Fig. 3.
PEEM image of isolated (A) and aggregated (B) NM upon UV excitation by an FEL at 248 nm, and wavelength-dependent FEL-PEEM data plotted and fit according to Eq. 1 (C). The best fit to the experimental data gives a photoionization threshold of 4.5 ± 0.2 eV (275 nm) for both isolated and aggregated deposits. Data are shown for the isolated granule. The image in A is 2.3 μm × 2.3 μm; the image in B is 1.1 μm × 1.5 μm.
Discussion
The AFM, SEM, and PEEM images demonstrate that the SN NM pigment is stable and retains consistent morphology under the diverse experimental conditions. The AFM data are collected under ambient conditions, whereas the SEM and PEEM data on the dried NM pigment is collected under high vacuum. Therefore, the measured threshold photoionization potential is referenced to the vacuum level. This value is linearly related to the electrochemical oxidation potential; the potential of an electron at rest in a vacuum corresponds to −4.44 V vs. NHE (34). Accordingly, the determined threshold potential of 4.5 ± 0.2 eV for NM corresponds to an electrochemical oxidation potential of −0.1 ± 0.2 V vs. NHE. It is important to stress that the data are completely described by a single threshold potential and that the value is within experimental error of that measured for black human hair eumelanosomes, −0.2 ± 0.2 V vs. NHE (33). It is interesting to contrast this result to that for human hair pheomelanosomes, which exhibit two potentials, −0.2 ± 0.2 V and +0.5 ± 0.2 V vs. NHE. Chemical analysis shows these melanosomes to contain both eumelanin and pheomelanin. The −0.2 ± 0.2 V and +0.5 ± 0.2 V potentials are therefore ascribed to eumelanin and pheomelanin, respectively, and the values themselves affirm that pheomelanin is more reductive than eumelanin.
It is interesting to note that the chemical analyses of NM and human hair pheomelanosomes are similar in that both contain appreciable amounts of both eumelanin and pheomelanin. Specifically in the case of NM, cysteinyl-dopamine, the precursor of pheomelanin constituents, accounts for ≈25% of the melanic component of NM (18, 19). However, in contrast to the human hair pheomelanosome data, only a single threshold potential is found for the NM pigment, which is characteristic of the black pigment eumelanin. In fact, the synthesis of melanin in hair melanosomes is tyrosinase-dependent, whereas NM synthesis is not, and the synthetic pathway of NM is quite unknown.
This result suggests that pheomelanin molecules are not on or near (within a few nanometers) the surface of the NM granule. This is true for both the isolated granular structures, ≈350 nm in diameter, and the large aggregated deposits composed of ≈30-nm substructures. These results are self-consistent because the ≈350-nm granules are also aggregates of these smaller ≈30-nm constituents.
This result is of interest in terms of the melanogenesis of the ≈30-nm structures of NM. Several in vitro kinetic studies establish that the addition of cysteine to dopaquinone is kinetically favored over eumelanogenesis as long as the cysteine concentration is >1 μM (22–27). This process is further favored by the fact that dopaminequinone cyclizes ≈100-fold more slowly than does dopaquinone (15, 24). The production of eumelanin competes with the production of pheomelanin once the cysteine and 5-S-cysteinyldopa concentrations are sufficiently low. If the ≈30-nm structures are the products of the melanogenesis process, then these kinetic arguments would suggest that 5-S-cysteinyl-dopamine products form the core, or center, of the structure and that eumelanin encapsulates this core, giving rise to a final pigment that is approximately a 3:1 eumelanin/pheomelanin mixture. This type of molecular architecture for mixed melanins has been previously proposed based on biochemical studies of mixed melanogenesis and solubility properties of synthetic model systems (35, 36). This model for the structure is consistent with the measured surface oxidation potential, which only reflects the presence of eumelanin. The mechanism by which the ≈30-nm structures assemble into larger pigment granules is not known. Also, in cultures we showed that midbrain dopaminergic neurons produce the first NM granules 8–11 days after l-DOPA treatment (17).
It is not possible at this time to assign the electrochemical potential to a specific chemical species on the surface of NM because the molecular structure of these pigments remains to be determined. Several other molecular constituents in addition to melanin are found in NM, e.g., proteins, lipids, and metal ions (3). The electrochemical potential determined by the PEEM experiment is attributed to the pigment and not to these other constituents. Lipids and proteins have ionization thresholds in excess of 5.0 eV based on control experiments. Also, using Sepia granules, the surface potential is independent of Fe(III) concentration up to saturation and remains unchanged after treatment with EDTA, in which all metals are removed (37).
From a thermodynamic perspective, an oxidation potential of −0.2 V vs. NHE for the surface of NM is not sufficiently reductive to generate a high level of oxidative stress. Therefore, NM's surface electrochemical potential is less reductive than might be anticipated given NM's complicated but unresolved role in the selective loss of pigmented neurons of the SN associated with PD. NM can be oxidized and degraded by H2O2 (18), and this process likely occurs also in the human brain during PD, where an extensive depletion of NM occurs after neuronal death. Degradation may also result in exposure of the pheomelanic core. The oxidation potential of pheomelanosomes is +0.5 V vs. NHE (33), which is sufficient so that the reduction of oxygen is thermodynamically favorable. In fact, synthetic pheomelanins can reduce oxygen in the absence of light, a feature that was not observed with eumelanins (38). Further research on the role of NM in oxidative stress in SN neurons would help in gaining insight into the underlying mechanisms that govern the progression of PD.
Methods
Isolation of Human NM.
NM was isolated from the SN pars compacta region of the human midbrain of neurologically normal adult individuals as described in ref. 4. Briefly, the dissection of the SN from the frozen human midbrain was performed on a cold plate at −10°C within 36 h of death and was homogenized in distilled H2O (0.03 g/ml) in a glass-Teflon homogenizer, followed by centrifugation at 12,000 × g for 10 min. The resulting pellet was washed twice with 50 mM phosphate buffer (pH 7.4), incubated in 50 mM Tris buffer (pH 7.4) containing 0.5 mg/ml SDS at 37°C for 3 h, and incubated for another 3 h at 37°C in the same buffer containing 0.2 mg/ml proteinase K. The resulting pellet was washed twice with NaCl (9 mg/ml), three times with distilled water, and once each with methanol (2 ml) and hexane (1 ml). Finally, it was dried under vacuum and stored protected from light. The procedure described herein is the same used in a previous study on the characterization of pheomelanin and eumelanin content in NM (18). Reproducibility of pheomelanin and eumelanin composition for the granules studies herein was confirmed by elemental analysis of carbon, hydrogen, nitrogen, sulfur, and oxygen.
Sample Preparation.
Isolated SN NM was stored in a dessicator and protected from light. Samples for PEEM analysis were prepared as follows. Sections of (100)-oriented silicon wafers [N-type, P-doped, low resistivity of 0.005–0.01 Ω-cm, (Virginia Semiconductor, Fredericksburg, VA)] were cut into 9 × 9 mm squares by using a diamond-tipped knife and were cleaned by the standard (RCA) wet chemical procedure before deposition of NM on the surface (39). The hydrofluoric acid step was not included in our preparation because we desired a thin oxide layer for optimal sample deposition; therefore, this procedure results in a surface terminated with an ≈1-nm-thick silicon oxide layer. All chemicals for the silicon wafer RCA cleaning procedure (1 M HCl, 30% H2O2, 30% NH4OH, and 18 M H2SO4) were purchased from Fisher Chemical (Fairlawn, NJ) and were of the highest purity available. The wafers were dried over lens paper in a sterile Petri dish while flowing argon gas over them. NM samples were subsequently deposited onto the hydrophilic surface of the freshly RCA-cleaned silicon wafer by micropipetting 0.5 μl of a NM suspension in distilled water (>18.2 MΩ) obtained from a Simplicity system (Millipore, Billerica, MA). Samples were air-dried while being protected from light for <1 h before experimentation. This preparatory process was repeated immediately before each new experiment.
Photoelectron Emission Microscopy.
The Duke OK-4 UV FEL and PEEM are described in detail in ref. 40. We detected the photoelectron emission intensity of our NM samples within the spectral range of 248–413 nm (5.0–3.0 eV), using the spontaneous-emission mode of the FEL with an energy full width at half maximum of ±0.1 eV. All PEEM images were acquired by using a DVC-1312M digital camera (DVC Company, Austin, TX). The resolution of the digital camera is 1,300 × 1,030 pixels × 12 bits. The DVCView program was used to view/save images. After sample preparation (as described above), samples were always immediately transferred under ultrahigh vacuum into the PEEM chamber for the collection of data, with minimal exposure to UV radiation. We typically imaged large NM aggregates at fields of view in the PEEM of 150 μm, and smaller NM assemblies were imaged at a field of view of 5 or 1.5 μm. The focusing of the FEL was optimized for each wavelength used, and the FEL spot size on the sample was ≈30 × 100 μm.
Image Analysis.
Details of the methods of PEEM image analysis were published recently by us (32). In these experiments, the appropriate software gain values were chosen to avoid saturation of the images, and the wavelength-dependent integrated brightness was determined. Because this integrated brightness (S) also depends on the power at each wavelength, the incident power was measured at the input port of the PEEM and converted into a light fluence. However, the experimental setup limited the determination of light fluence to the input port only. Therefore, the wavelength-dependent data reveal relative, instead of absolute, photoionization quantum yields. The integrated brightness, S(λ), is taken to be proportional to the photocurrent collected and is corrected with the background as well as normalized for the software gain and the incident photon flux at each wavelength. Considering the temperature-related effects, S(λ), and the excitation energy, hν, should satisfy the following equation (41):
where A is a proportionality constant, T is the temperature at which the experiments are conducted, k is the Boltzmann constant, χ is the photoionization threshold, and
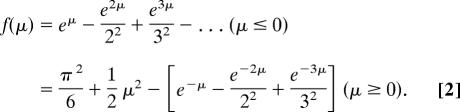 |
All data were plotted as integrated intensity divided by temperature squared, S/T2, as a function of relative energy, hν/kT, resulting in a linear relationship. The threshold was then determined by fitting the normalized data with Eq. 1 by using IGOR Pro software (WaveMetrics, Lake Oswego, OR).
Scanning Electron Microscopy.
Samples were prepared for SEM by using the same procedure as that used for PEEM samples (as described above). Once prepared, the silicon wafers containing a dried NM film were mounted on stainless-steel pegs by using double-stick copper tape before being transferred to the SEM chamber. The mounted samples were coated with gold/palladium mist under argon plasma for 4 min at 10 mA by using a Hummer V sputter coater (Anatech, Springfield, VA). A Philips FEI XL30 SEM-FEG (FEI, Portland, OR) equipped with a back-scattering secondary electron detector and a resolution of ≈3 nm was used to examine the samples in ultrahigh-resolution mode. The typical electron-beam conditions were 3–5 kV, with a spot size of 3.0 and a working distance of 4.0–6.0 mm. SEM images were captured and analyzed by using analySIS XL docu software (Soft Imaging Systems, Lakewood, CO). All images were saved as TIFF files.
Atomic Force Microscopy.
NM samples in a double-distilled water suspension were pipetted (0.5 μl) onto freshly cleaved mica and allowed to air dry in the dark. The instrument and method of imaging is described in detail in ref. 28. Briefly, AFM height and phase images were collected by using a NanoScope IIIa BioScope AFM (Digital Instruments, Santa Barbara, CA) operated in tapping mode. The AFM head was mounted on a Zeiss (Thornwood, NY) Axiovert S100 TV inverted optical microscope allowing visual observation of the sample.
Acknowledgments
J.D.S., G.S.E., and R.J.N. thank the Air Force Office of Scientific Research for support of this work through the Medical Free Electron Laser Program. F.A.Z., A.A., and L.Z. thank the Italian Fund for Basic Science (FIRB-MIUR Project RBNE03PX83_002) for support of this work and Ms. Chiara Bellei for technical assistance.
Abbreviations
- AFM
atomic force microscopy
- FEL
free electron laser
- NHE
normal hydrogen electrode
- NM
neuromelanin
- PD
Parkinson's disease
- PEEM
photoelectron emission microscopy
- SEM
scanning electron microscopy
- SN
substantia nigra.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 14647.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Graham DG. Arch Pathol Lab Med. 1979;103:359–362. [PubMed] [Google Scholar]
- 2.Zecca L, Stroppolo A, Gatti A, Tampellini D, Toscani M, Gallorini M, Giaveri G, Arosio P, Santambrogio P, Fariello RG, et al. Proc Natl Acad Sci USA. 2004;101:9843–9848. doi: 10.1073/pnas.0403495101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zucca FA, Giaveri G, Gallorini M, Albertini A, Toscani M, Pezzoli G, Lucius R, Wilms H, Sulzer D, Ito S, et al. Pigment Cell Res. 2004;17:610–617. doi: 10.1111/j.1600-0749.2004.00201.x. [DOI] [PubMed] [Google Scholar]
- 4.Zecca L, Fariello R, Riederer P, Sulzer D, Gatti A, Tampellini D. FEBS Lett. 2002;510:216–220. doi: 10.1016/s0014-5793(01)03269-0. [DOI] [PubMed] [Google Scholar]
- 5.Fenichel GM, Bazelon M. Neurology. 1968;18:817–820. doi: 10.1212/wnl.18.8.817. [DOI] [PubMed] [Google Scholar]
- 6.Gibb WR. Brain Res. 1992;581:283–291. doi: 10.1016/0006-8993(92)90719-p. [DOI] [PubMed] [Google Scholar]
- 7.Zecca L, Zucca FA, Costi P, Tampellini D, Gatti A, Gerlach M, Riederer P, Fariello RG, Ito S, Gallorini M, Sulzer D. J Neural Transm. 2003;65(Suppl):145–155. doi: 10.1007/978-3-7091-0643-3_8. [DOI] [PubMed] [Google Scholar]
- 8.Enochs WS, Sarna T, Zecca L, Riley PA, Swartz HM. J Neural Transm Parkinson's Dis Dementia Sect. 1994;7:83–100. doi: 10.1007/BF02260963. [DOI] [PubMed] [Google Scholar]
- 9.D'Amato RJ, Lipman ZP, Snyder SH. Science. 1986;231:987–989. doi: 10.1126/science.3080808. [DOI] [PubMed] [Google Scholar]
- 10.Zecca L, Zucca FA, Toscani M, Adorni F, Giaveri G, Rizzio E, Gallorini M. J Radioanal Nucl Chem. 2005;263:733–737. [Google Scholar]
- 11.Kimura S, Kurasaki M, Saito T, Ito K, Hosokawa T, Okabe M, Shiraishi K, Niioka T. Trace Elem Electrolytes. 2004;21:55–59. [Google Scholar]
- 12.Fornstedt B. J Neural Transm Parkinson's Dis Dementia Sect. 1989;1:279–295. doi: 10.1007/BF02263482. [DOI] [PubMed] [Google Scholar]
- 13.Mars U. Pigment Cell Res. 1999;12:266–274. doi: 10.1111/j.1600-0749.1999.tb00760.x. [DOI] [PubMed] [Google Scholar]
- 14.Tse DCS, McCreery RL, Adams RN. J Med Chem. 1976;19:37–40. doi: 10.1021/jm00223a008. [DOI] [PubMed] [Google Scholar]
- 15.Hawley MD, Tatawawadi SV, Piekarski S, Adams RN. J Am Chem Soc. 1967;89:447–450. doi: 10.1021/ja00978a051. [DOI] [PubMed] [Google Scholar]
- 16.Zecca L, Zucca FA, Wilms H, Sulzer D. Trends Neurosci. 2003;26:578–580. doi: 10.1016/j.tins.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Sulzer D, Bogulavsky J, Larsen KE, Behr G, Karatekin E, Kleinman MH, Turro N, Krantz D, Edwards RH, Greene LA, et al. Proc Natl Acad Sci USA. 2000;97:11869–11874. doi: 10.1073/pnas.97.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakamatsu K, Fujikawa K, Zucca FA, Zecca L, Ito S. J Neurochem. 2003;86:1015–1023. doi: 10.1046/j.1471-4159.2003.01917.x. [DOI] [PubMed] [Google Scholar]
- 19.Ito S, Wakamatsu K. Pigment Cell Res. 1998;11:120–126. doi: 10.1111/j.1600-0749.1998.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 20.Zecca L, Mecacci C, Seraglia R, Parati E. Biochim Biophys Acta. 1992;1138:6–10. doi: 10.1016/0925-4439(92)90144-c. [DOI] [PubMed] [Google Scholar]
- 21.Odh G, Carstam R, Paulson J, Wittbjer A, Rosengren E, Rorsman H. J Neurochem. 1994;62:2030–2036. doi: 10.1046/j.1471-4159.1994.62052030.x. [DOI] [PubMed] [Google Scholar]
- 22.Marmol V, Ito S, Bouchard B, Libert A, Wakamatsu K, Ghanem G, Solano F. J Invest Dermatol. 1996;107:698–702. doi: 10.1111/1523-1747.ep12365591. [DOI] [PubMed] [Google Scholar]
- 23.Ozeki H, Ito S, Wakamatsu K, Ishiguro I. Biochim Biophys Acta. 1997;1336:539–548. doi: 10.1016/s0304-4165(97)00068-8. [DOI] [PubMed] [Google Scholar]
- 24.Land EJ, Riley PA. Pigment Cell Res. 2000;13:273–277. doi: 10.1034/j.1600-0749.2000.130409.x. [DOI] [PubMed] [Google Scholar]
- 25.Ito S. Pigment Cell Res. 2003;16:230–236. doi: 10.1034/j.1600-0749.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- 26.Land EJ, Ito S, Wakamatsu K, Riley PA. Pigment Cell Res. 2003;16:487–493. doi: 10.1034/j.1600-0749.2003.00082.x. [DOI] [PubMed] [Google Scholar]
- 27.Jameson GNL, Zhang J, Jameson RF, Linert W. Org Biomol Chem. 2004;2:777–782. doi: 10.1039/b316294j. [DOI] [PubMed] [Google Scholar]
- 28.Clancy CMR, Simon JD. Biochemistry. 2001;40:13353–13360. doi: 10.1021/bi010786t. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Simon JD. Pigment Cell Res. 2003;16:1–13. doi: 10.1034/j.1600-0749.2003.00009.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Hong L, Wakamatsu K, Ito S, Adhyaru BB, Cheng CY, Bowers CR, Simon JD. Photochem Photobiol. 2005;81:510–516. doi: 10.1562/2004-10-19-RA-345.1. [DOI] [PubMed] [Google Scholar]
- 31.Hong L, Garguilo J, Anzaldi L, Edwards GS, Nemanich RJ, Simon JD. Photochem Photobiol. 2006 May 12; doi: 10.1562/2006-03-14-RA-846. [DOI] [PubMed] [Google Scholar]
- 32.Samokhvalov A, Garguilo J, Yang WC, Edwards GS, Nemanich RJ, Simon JD. J Phys Chem B. 2004;108:16334–16338. [Google Scholar]
- 33.Samokhvalov A, Hong L, Liu Y, Garguilo J, Nemanich RJ, Edwards GS, Simon JD. Photochem Photobiol. 2005;81:145–148. doi: 10.1562/2004-07-23-RC-245. [DOI] [PubMed] [Google Scholar]
- 34.Graetzel M. Nature. 2001;414:338–344. doi: 10.1038/35104607. [DOI] [PubMed] [Google Scholar]
- 35.Agrup G, Hansson C, Rorsman H, Rosengren E. Arch Dermatol Res. 1982;272:103–115. doi: 10.1007/BF00510400. [DOI] [PubMed] [Google Scholar]
- 36.Ito S, Wakamatsu K, Ozeki H. Pigment Cell Res. 2000;13(Suppl 8):103–109. doi: 10.1034/j.1600-0749.13.s8.19.x. [DOI] [PubMed] [Google Scholar]
- 37.Simon JD. Pigment Cell Res. 2006;19:527–528. doi: 10.1111/j.1600-0749.2006.00330.x. [DOI] [PubMed] [Google Scholar]
- 38.Ye T, Hong J, Garguilo J, Pawlak A, Edwards GS, Nemanich RJ, Sarna T, Simon JD. Photochem Photobiol. 2006;82:733–737. doi: 10.1562/2006-01-02-RA-762. [DOI] [PubMed] [Google Scholar]
- 39.Senturia SD. Microsystem Design. Boston: Kluwer Academic; 2001. p. 34. [Google Scholar]
- 40.Ade H, Yang W, English SL, Hartman J, Davis RF, Nemanich RJ, Litvinenko VN, Pinayev IV, Wu Y, Madey JMJ. Surf Rev Lett. 1998;5:1257–1268. [Google Scholar]
- 41.Fowler RH. Phys Rev. 1931;38:45–56. [Google Scholar]