Abstract
We previously showed that the calcium-binding protein S100A4 is overexpressed during the progression of prostate cancer (CaP) in humans and in the TRAMP (transgenic adenocarcinoma of the mouse prostate) mouse model. We tested a hypothesis that the S100A4 gene plays a role in the invasiveness of human CaP and may be associated with its metastatic spread. We observed that siRNA-mediated suppression of the S100A4 gene significantly reduced the proliferative and invasive capability of the highly invasive CaP cells PC-3. We evaluated the mechanism through which the S100A4 gene controls invasiveness of cells by using a macroarray containing 96 well characterized metastatic genes. We found that matrix metalloproteinase 9 (MMP-9) and its tissue inhibitor (TIMP-1) were highly responsive to S100A4 gene suppression. Furthermore, S100A4 suppression significantly reduced the expression and proteolytic activity of MMP-9. By employing an MMP-9-promoter reporter, we observed a significant reduction in the transcriptional activation of the MMP-9 gene in S100A4-siRNA-transfected cells. Cells overexpressing the S100A4 gene (when transfected with pcDNA3.1-S100A4 plasmid) also significantly expressed MMP-9 and TIMP-1 genes with increased proteolytic activity of MMP-9 concomitant to increased transcriptional activation of the MMP-9 gene. S100A4-siRNA-transfected cells exhibited a reduced rate of tumor growth under in vivo conditions. Our data demonstrate that the S100A4 gene controls the invasive potential of human CaP cells through regulation of MMP-9 and that this association may contribute to metastasis of CaP cells. We suggest that S100A4 could be used as a biomarker for CaP progression and a novel therapeutic or chemopreventive target for human CaP treatment.
Keywords: extracellular matrix, metastasis, biomarker
Approximately 27,350 prostate cancer (CaP)-related deaths are predicted during this year alone in the United States, and despite recent improvements in diagnostic and therapeutic techniques, the survival rate of CaP patients is poor because of the posttreatment recurrence of disease (1, 2). The lack of effective therapies for advanced CaP is related to a large extent to poor understanding of the molecular mechanisms underlying the progression of disease toward invasion and metastasis (3). Thus, the identification of new predictive biomarkers, especially those that are indicative of invasiveness of the disease, which could serve as targets for establishing effectiveness of therapeutic and chemopreventive interventions, will improve clinical management of CaP.
S100A4 (also known as mts1), a calcium-binding protein associated with invasion and metastasis of cancer cells, has been reported to be frequently overexpressed in metastatic tumors, normal cells with uninhibited movement (such as macrophages), and transformed cells, and in various cancer types such as breast, ovary, thyroid, lung, esophageal squamous cell carcinoma, gastric, and colon (4–6). Studies have shown that breast cancers expressing high levels of S100A4 have a significantly worse prognosis than breast cancers negative for S100A4 (ref. 7 and references therein). These studies have also shown that overexpression of S100A4 may be sufficient to induce a metastatic phenotype in human mammary tumor cells (7). Additional studies indicated that overexpression of S100A4 correlates with the in vitro invasive potential of glioma cells and breast cancer, conferring enhanced metastatic ability (refs. 7 and 8 and references therein). Recently, we showed that S100A4 is overexpressed during progression of CaP in humans and in transgenic adenocarcinoma of the mouse prostate (TRAMP) mice, an autochthonous transgenic model that develops CaP in a manner similar to human disease (9, 10). In the present study, we provide evidence to support the hypothesis that S100A4 plays a role in invasiveness of human CaP through the transcriptional regulation of matrix metalloproteinase (MMP)-9.
Results
Effect of siRNA on S100A4 Expression in PC-3 Cells.
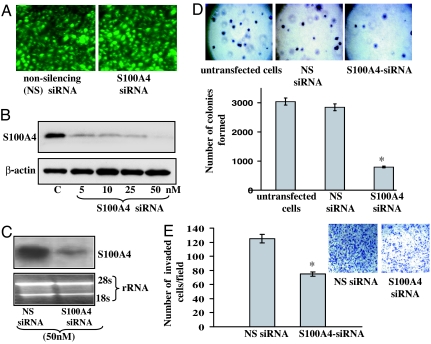
As shown in Fig. 1A, high (60–90%) transfection efficiency of siRNAs was observed in PC-3 cells. As determined by Western blot analysis, cells transfected with S100A4 siRNA displayed a dose-dependent reduction in the expression levels of S100A4 protein (Fig. 1B). Nonsilencing siRNA did not exhibit any effect on protein levels of S100A4 (Fig. 1B). Similarly, cells transfected with lamin A/C siRNA (which is used as a positive control for siRNA transfection) significantly reduced the expression of lamin A/C protein (data not shown). Further, the suppression of S100A4 by siRNA in cells was confirmed by Northern blot analysis. Cells transfected with S100A4 siRNA exhibited a significant reduction in mRNA level of S100A4 (Fig. 1C). These data confirmed the suppression effect of siRNA and established the efficiency of siRNA transfection.
Fig. 1.
S100A4 gene knockdown by siRNA transfection in PC-3 cells. (A) Photomicrographs showing transfection of fluorescein-labeled siRNA in PC-3 cells. (Magnification: ×100.) (B and C) Representative images showing expression of S100A4 protein and mRNA in nonsilencing siRNA control and S100A4-siRNA-transfected cells as analyzed by Western (B) and Northern (C) blotting. Equal loading of protein was confirmed by stripping the blots and reprobing with β-actin antibody, and equal loading of RNA was confirmed by measuring the 28S rRNA. Densitometric measurements of the bands in Western and Northern blot analyses were performed by using the digitizing software UN-SCAN-IT (Silk Scientific, Orem, UT). (D) Photomicrographs showing soft agar colony formation and histogram showing number of colonies formed by PC-3 cells transfected with siRNA. (E) Representative photomicrographs (Inset) and histogram showing invasive capability of PC-3 cells transfected with S100A4 siRNA in a chemoinvasion chamber. (Magnification: ×100.) Each bar represents mean ± SE; ∗, P < 0.05. All experiments were repeated three times with similar results.
Effect of S100A4 Gene Knockdown on Colony Formation.
To investigate the effect of S100A4 gene suppression on the growth of PC-3 cells, we performed a soft agar colony-formation assay. As shown in Fig. 1D, S100A4-siRNA-transfected cells formed a significantly reduced (P < 0.01) number of colonies compared with untreated and control siRNA-treated cells, suggesting that S100A4 might have growth-promoting effects on CaP cells and that suppression of this gene reduces the proliferative property of CaP cells.
Effect of S100A4 Gene Knockdown on Invasive Capability of PC-3 Cells.
The S100A4 gene is reported to confer invasive characteristics to various cancer cells; however, its role in the invasion and metastasis of CaP is yet unknown (11). Next, we analyzed the effect of S100A4 gene suppression on the invasive capability of highly invasive and metastatic CaP cells PC-3 by employing an in vitro chemoinvasion assay. As shown in Fig. 1E, suppression of the S100A4 caused a >50% reduction (P < 0.05) in the number of cells that traversed the membrane versus nonsilencing control. These data suggest that the S100A4 gene controls the motility and invasion of cancerous cells during the metastasis of human CaP.
Effect of S100A4 Gene Knockdown on Other Metastatic Genes.
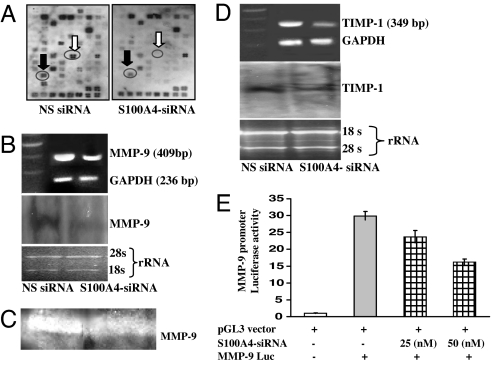
To determine the mechanism (such as interaction and regulation of other metastatic genes) through which S100A4 controls the invasion and metastasis of CaP cells, we performed a cDNA macroarray analysis of 96 well characterized metastatic genes. Suppression of the S100A4 gene caused a reduction in the expression level of various metastatic genes (Fig. 2A). The list of genes that were observed to exhibit changes in their expression in response to S100A4 gene knockdown is presented in Table 1. Several genes that were responsive to S100A4 gene suppression are known to be associated with degradation of the extracellular matrix (ECM) and are highly expressed during metastasis. Most notably, we observed that suppressing the S100A4 gene in PC-3 cells caused a significant reduction (>95%) in the expression of MMP-9 and tissue inhibitor of matrix metalloproteinase (TIMP)-1 (Fig. 2A). Because MMP-9 and TIMP-1 were highly responsive to S100A4 gene suppression, we selected these two genes for further studies.
Fig. 2.
Effect of S100A4 gene knockdown on 96 well characterized metastatic genes and on the expression level, gelatinolytic activity, and transcriptional activation of MMP-9 and TIMP-1. (A) Autoradiographic image of control (Left) and S100A4-siRNA-transfected cells (Right). Encircled tetra spots indicate the position of genes. Black arrow, TIMP-1; white arrow, MMP-9. (B) Representative images showing the expression of MMP-9 mRNA as determined by RT-PCR and Northern blot analysis. (C) Representative image showing gelatinolytic activity of MMP-9 in cells. Densitometric measurements of the bands in RT-PCR and Northern blot analysis were performed by using the digitizing software UN-SCAN-IT. (D) Representative images showing expression of TIMP-1 as determined by RT-PCR and Northern blot analysis. Equal loading of PCR products and RNA was confirmed by using constitutively expressed GAPDH and by measuring the 28S rRNA. All experiments were repeated three times with similar results. (E) Histogram showing the activity of MMP-9 promoter (in terms of relative luciferase activity) in transfected cells.
Table 1.
List of candidate metastasis-associated genes modulated by S100A4 gene
| Gene symbol | Gene description | Gene status after S100A4 siRNA |
|---|---|---|
| MMP-9 | Matrix metalloproteinase 9 (gelatinase B) | Down-regulated* |
| TIMP-1 | Tissue inhibitor of matrix metalloproteinase 9 | Down-regulated* |
| ERBB2 | V-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 | Down-regulated** |
| ICAM5 | Intercellular adhesion molecule 5 | Down-regulated** |
| MAP 2 K4 | SAPK/Erk kinase 1 | Down-regulated** |
| MGEA5 | Meningioma-expressed antigen 5 (hyaluronidases) | Down-regulated** |
| MMP-1 | Matrix metalloproteinase 1 | Down-regulated** |
| MMP-13 | Matrix metalloproteinase 13 | Down-regulated** |
| MMP-14 | Matrix metalloproteinase 14 | Down-regulated** |
| MMP-15 | Matrix metalloproteinase 15 | Down-regulated** |
| MUC1 | Mucin 1 | Down-regulated** |
| NGFB | Nerve growth factor | Down-regulated** |
| PDGFA | Platelet-derived growth factor α polypeptide A | Down-regulated** |
| PLAUR | Human urokinase-type plasminogen activator receptor (uPAR) | Down-regulated** |
| CAV 1 | Caveolin 1 | Up-regulated* |
| CDH 1 | E-cadherin (cadherin 1) | Up-regulated* |
| COL 4A2 | Collagen, type IV α2 | Up-regulated* |
| MDM 2 | Mouse double minute 2 (human homolog of p53-binding protein) | Up-regulated* |
| PTEN | Phosphatase and tensin homolog | Up-regulated* |
| TIMP-2 | Tissue inhibitor of matrix metalloproteinase 2 | Up-regulated* |
*, >70%; **, >35–60%.
Effect of S100A4 Gene Knockdown on MMP-9 Expression and Its Enzymatic Activity and Transcriptional Activation.
Increased MMP activity is considered important for the increased capability of cancerous cells to traverse the membrane and invade and metastasize to distant sites (12). Next, we analyzed the effect of S100A4 gene suppression on the expression and activity of MMP-9. PC-3 cells transfected with S100A4 siRNA exhibited a significant reduction in the expression of MMP-9 mRNA (Fig. 2B). These data were consistent with cDNA macroarray data, suggesting a possible link between the S100A4 gene and MMP-9 during the progression of human CaP. We also evaluated the effect of S100A4 gene suppression on proteolytic activity of MMP-9 protein by performing zymography with a gelatin gel. We observed that S100A4 gene suppression significantly decreased MMP-9 enzymatic activity in PC-3 cells (Fig. 2C). We suggest that an S100A4 gene knockdown-mediated decrease in the level of MMP-9 mRNA resulted in the decreased level of MMP-9 protein, hence its decreased activity.
Effect of S100A4 Gene Knockdown on TIMP-1.
TIMP-1, an endogenous inhibitor of MMP-9, is often found complexed to MMP-9 (13). Because TIMP-1 was observed to be the most responsive gene (along with MMP-9) to S100A4 gene suppression, we next determined the effect of S100A4 gene knockdown on the mRNA expression levels of TIMP-1 gene. We observed that S100A4 gene suppression significantly reduced expression of TIMP-1 mRNA (Fig. 2D).
Effect of S100A4 Gene Knockdown on MMP-9 Transcriptional Activation.
Because S100A4 gene suppression caused a reduction in expression of MMP-9 mRNA, we determined whether S100A4 regulates the activity of MMP-9 gene by cotransfecting a pGl3-MMP-9-luc-promoter plasmid with S100A4 siRNA in PC-3 cells. S100A4 gene suppression significantly decreased the promoter activity of MMP-9 in a dose-dependent manner, indicating the involvement of S100A4 in the regulation of the MMP-9 gene (Fig. 2E) without any effect on basal activity of pGL3 vector (data not shown).
Effect of S100A4 Overexpression on PC-3 Cells.
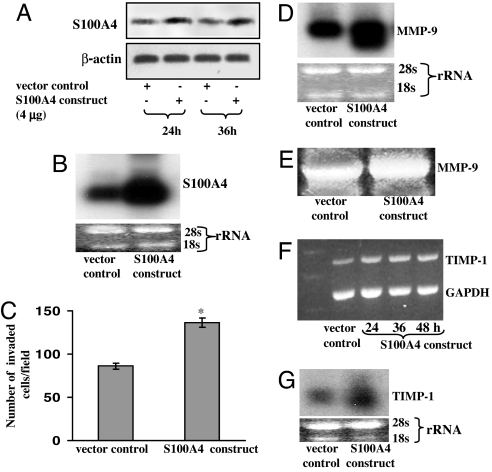
PC-3 cells transfected with pc.DNA3.1-S100A4 plasmid displayed a significant increase in the expression levels of S100A4 as compared with vector control (Fig. 3A). The overexpression of S100A4 was confirmed by performing Northern blot analysis 36 h after transfection (Fig. 3B). Because S100A4 expression was observed to be very high 36 h after transfection, we selected this time point for further studies.
Fig. 3.
Effect of S100A4 overexpression by transfection of S100A4-pcDNA3.1 plasmid on MMP-9, its tissue inhibitor, and on the invasive capability of PC-3 cells. (A and B) Representative images showing expression of S100A4 in vector (pcDNA3.1) and pcDNA3.1-S100A4 transfected cells as analyzed by Western (A) and Northern (B) blot analysis. (C) Histogram showing invasive capability of transfected cells. Each bar represents mean ± SE (n = 3); ∗, P < 0.05. (D) Autoradiographic images showing the expression of MMP-9 mRNA in transfected cells as determined by Northern blot analysis. (E) Representative image showing gelatinolytic activity of MMP-9 in transfected cells. (F and G) Representative images showing expression of TIMP-1 in transfected cells as determined by RT-PCR (F) and Northern blot (G) analysis. Equal loading of protein was confirmed by stripping the blots and reprobing with anti-β-actin antibody. Equal loading of PCR products and RNA was confirmed by using constitutively expressed GAPDH and by measuring the 28S rRNA, respectively. Densitometric measurements of the band in RT-PCR and Northern blot analysis were performed by using the digitizing scientific software UN-SCAN-IT. All experiments were repeated three times with similar results.
Effect of S100A4 Overexpression on Invasive Capability.
We analyzed the effect of S100A4 overexpression on the invasive capability of PC-3 cells. As shown in Fig. 3C, overexpression of S100A4 significantly increased (P < 0.05) the number of invasive cells. These data further support our hypothesis that S100A4 confers the invasive characteristics to cells during human CaP development.
Effect of S100A4 Overexpression on MMP-9 Expression and Its Enzymatic Activity.
Next we determined the effect of S100A4 overexpression on the expression and activity of MMP-9. PC-3 cells transfected with an S100A4 construct exhibited a significant increase in the expression level of MMP-9 mRNA (Fig. 3D). We found that S100A4 gene overexpression significantly increased the proteolytic activity of MMP-9 protein (Fig. 3E).
Effect of S100A4 Overexpression on Tissue Inhibitor of MMP-9.
We also measured the effect of S100A4 overexpression on TIMP-1 mRNA expression. Transfection with pcDNA3.1-S100A4 plasmid caused a significant increase in expression of TIMP-1 mRNA (Fig. 3 F and G).
Effect of S100A4 Overexpression on MMP-9 Transcriptional Activation.
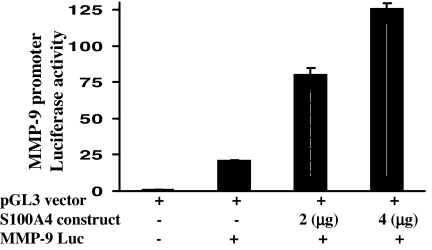
Transfection with pcDNA3.1-S100A4 plasmid significantly increased MMP-9 promoter activity in a dose-dependent manner in PC-3 cells (Fig. 4) without any effect on basal activity of pGL3 vector (data not shown).
Fig. 4.
Effect of S100A4 overexpression on transcriptional activation of MMP-9 in PC-3 cells. Histogram represents the relative luciferase activity in cells cotransfected with pGL3-MMP9-Luc, pcDNA3.1, or pcDNA3.1-S100A4.
Effect of S100A4 Gene Knockdown on Tumorigenicity in Athymic Nude Mice.
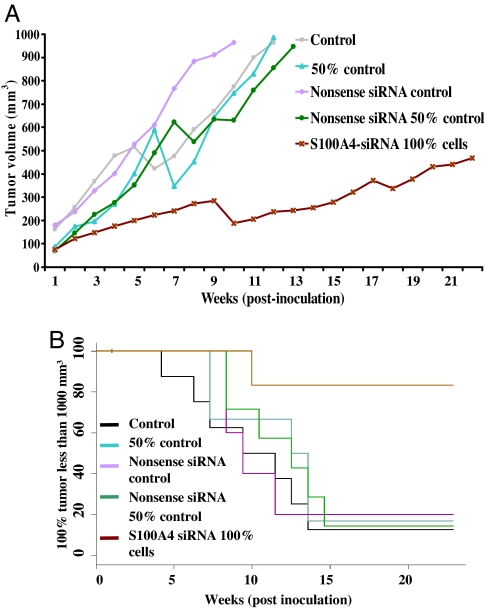
Because suppression of the S100A4 gene reduced PC-3 cell invasion in vitro, we next examined whether these in vitro data have in vivo relevance. To accomplish this goal, PC-3 cells transfected with S100A4 siRNA were grown as s.c. xenografts in nude mice to form tumors. Implantation of PC-3 cells onto nude mice produced visible tumors, with a mean latent period of 20 days. The average volume of tumors in mice implanted with control cells increased as a function of time and reached a preset end point of 1,000 mm3 in 10–11 weeks; however, mice implanted with S100A4-siRNA-transfected cells exhibited a significant reduction in tumor volume and did not reached the preset end point even after 21 weeks (Fig. 5A). Linear regression analysis showed that the rate of tumor growth was significantly low for S100A4-siRNA-transfected cells (data not shown). Next, we performed a Kaplan–Meier analysis to determine whether S100A4 gene suppression enhances tumor-free survival or delays the growth of tumors in nude mice. We found a significant increase (P < 0.001) in tumor-free survival in animals implanted with S100A4 gene-silenced cells (Fig. 5B). Furthermore, log–rank analysis showed that the average number of weeks to attain 1,000-mm3 tumor volume increased significantly from 11 to >21 weeks (P < 0.001) (data not shown). From these data we conclude that the S100A4 gene plays an important role in the CaP development in humans and its suppression significantly inhibits the tumor formation. Cells that grew in vivo despite S100A4 gene knockdown could be those cells that escaped the siRNA transfection.
Fig. 5.
Effect of S100A4 gene knockdown in PC-3 cells on the tumorigenecity in a nude mouse xenograft model. (A) Growth of tumors of control and S100A4-siRNA-transfected cells in terms of average volume of tumors as a function of weeks on test. (B) Tumor-free survival as assessed by Kaplan–Meier plot.
Discussion
Overexpression of S100A4 is reported to confer invasive potential to glioma and breast cancer cells and to cause a malignant phenotype in rat and human cancer cells; however, the role of S100A4 in human CaP metastasis is yet to be elucidated (14–18). In the present study, we observed that the S100A4 gene controls the invasiveness and growth of human CaP cells under in vitro and in vivo conditions through the transcriptional regulation of MMP-9. This report demonstrates that S100A4 regulates MMP-9 and its inhibitor TIMP-1 genes in human CaP cells.
In this study, we show that overexpression of the S100A4 gene increases the invasiveness of CaP cells and that its suppression reverses this effect. We also show that suppression of S100A4 gene reduces the growth and proliferative potential of human CaP cells. These data provide evidence that the S100A4 gene may be associated with proliferation, invasion, and metastatic spread of cancerous cells during progression of human CaP.
CaP cell invasion of distant sites causes metastatic disease and is the major cause of CaP-related deaths in humans (17). Degradation of ECM is required by tumor cells to invade distant tissues, and recently metastatic effects of S100A4 have been linked with ECM destruction in some cancer types (18). MMPs are known to degrade the ECM by proteolysis, and a positive correlation has been shown to exist between MMP expression levels and Gleason scores in CaP patients (19). Recently we have shown that MMP-9 is up-regulated during the progression of CaP in TRAMP mice (20). It has been reported that the S100A4 gene may induce changes in the expression of MMPs in osteosarcoma cells, resulting in a malignant phenotype (21). In the present study, we observed that S100A4 gene suppression significantly decreased the expression of MMP-9 and that overexpression of the S100A4 gene significantly increased MMP-9 expression. We also observed that suppression of the S100A4 gene caused a decrease in the proteolytic activity of MMP-9 protein, whereas overexpression of S100A4 caused a reverse effect. Furthermore, we observed that S100A4 gene knockdown decreased, and its overexpression increased, MMP-9 promoter activity, suggesting that transcriptional activation of MMP-9 is regulated by the S100A4 gene in CaP cells.
TIMP-1 expression has been reported to be increased in several cancer types, such as neck, breast, colorectal, bladder, and gastric cancers (22–24). TIMP expression in the cells has been shown to be driven by the expression of MMP, and overall, the balance between their activities determines net proteolytic activity (20). Extracellular S100A4 protein has been reported to stimulate invasive growth of mouse endothelial cells through modulation of TIMP activity (18). We observed that S100A4 gene suppression decreased, and its overexpression increased, the expression of TIMP-1 in CaP cells. We suggest that the S100A4 gene influences the ratio of MMP-9 to TIMP-1, leading to ECM degradation.
Finally, S100A4-siRNA-transfected CaP cells displayed a significantly slower tumor growth. Interestingly, the control cells (100% cells) and even 50% of control cells reached a volume of 1,000 mm3 around week 10, but the S100A4-siRNA-transfected cells did not reach a similar volume even after 21 weeks. Based on our observations that S100A4-siRNA-transfected cells proliferated more slowly under in vitro and in vivo conditions, we suggest that suppression of the S100A4 gene may have resulted in the loss of its proliferative property. These data strengthen our notion that the S100A4 gene plays a role during the progression of human CaP.
In summary, we demonstrated that the S100A4 gene controls proliferation, invasion, and tumorigenicity of human CaP cells at least in part through the transcriptional regulation of MMP-9 gene. We suggest that control of proliferation, invasion, and metastasis through suppression of the S100A4 gene may contribute to a novel therapeutic approach against CaP. This approach could be realized through development of specific S100A4 inhibitors or use of a gene therapy approach.
Methods
Cell Culture.
Human CaP PC-3 cells (American Type Culture Collection, Manassas, VA) were cultured in RPMI medium 1640 supplemented with 10% FBS and 1% penicillin–streptomycin.
siRNA Transfection.
siRNAs were commercially purchased from Qiagen (Valencia, CA). The sequence of selected regions to be targeted by siRNAs was 5′-AACGAGGTGGACTTCCAAGAG-3′ for S100A4, 5′-AACTGGACTTCAGAACA-3′ for constitutively expressed lamin A/C gene, and 5′-AATTCTCCGAACGTGTCTCGT-3′ for a nonsilencing siRNA (control). By using a DharmaFECT transfection kit (Lafayette, CO), cells were transfected with fluorescein-labeled siRNAs; i.e., nonsilencing siRNA (50 nM), lamin A/C siRNA (50 nM), and S100A4 siRNA (5–50 nM). Cells were harvested after 24 h and analyzed for expression of S100A4 and lamin A/C.
Western Blot Analysis.
Forty micrograms of protein was resolved over Tris–glycine, pH 8.6, 18% (for S100A4) and 12% (for lamin A/C) polyacrylamide gels and then transferred onto the nitrocellulose membranes. Expression levels of proteins were analyzed as described in ref. 10.
pcDNA3.1-S100A4 Plasmid Construction and Transfection.
pUC-S100A4 plasmid was procured from Genescript (Piscataway, NJ), and S100A4-cDNA was isolated from pUC18 S100A4 by using HindIII restriction endonuclease. The digested fragment was then ligated into pcDNA3.1 vector (Invitrogen, Carlsbad, CA) previously digested with HindIII and treated with calf intestinal alkaline phosphatase. The resulting construct was verified by direct sequencing. For transfection studies, PC-3 cells were plated at a density of 1 × 106 cells per well in six-well plates and incubated for 24 h in complete medium. The cells were then transfected with 2–4 μg of the S100A4 construct by using an Amaxa transfection kit (Gaithersburg, MD). For controls, the same amount of empty vector, pcDNA3.1, and GFP vector (as positive control for transfection) was also transfected.
MMP-9-Promoter-Driven Luciferase Plasmid Transformation.
The human MMP-9 promoter luciferase plasmid (pGL3-MMP-9-luc) was procured from the laboratory of Dougles D. Boyden (M. D. Anderson Cancer Center, Houston, TX). The details of pGL3-MMP-9-luc have been described previously (25).
pcDNA3.1-S100A4 Plasmid and pGL3-MMP-9-luc Cotransfection.
PC-3 cells plated at a density of 5 × 104 cells per well were cotransfected with pGL3-MMP-9-luc (200 ng per well) along with S100A4 siRNA (25 and 50 nM) or the S100A4-pcDNA3.1 construct (2–4 μg) for 24 h. Renilla luciferase (20 ng per well, pRL-TK; Promega, Madison, WI) was used as an internal control. In addition, for controls, the same amount of empty vectors, pGL3 and pcDNA3.1, was transfected into cells. The cells were then harvested, and luciferase activity was measured in quadruplicate by using a Dual-Luciferase reporter assay system (Promega). Relative luciferase activity was calculated with the values from the pGL3 vector alone group with or without the cotransfected group.
Colony Formation Assay.
Cells were transfected with S100A4 siRNA and nonsilencing siRNA. Cells were harvested 36 h after transfection and mixed with RPMI medium 1640 containing 10% FCS and 0.5% agar (bottom layer). Cells (5,000 cells per well) in 20% FCS/0.7% agarose (top layer) were plated and incubated at 37°C for 14 days. After incubation, plates were stained with 0.005% crystal violet for more than 2 h, and colonies were counted in two colony grids per well by using a microscope.
cDNA Macroarray.
Briefly, 5 μg of RNA was reverse transcribed, and a cDNA probe was synthesized by labeling the cDNAs with biotin-16-dUTP as per the vendor's protocol (SuperArray, Frederick, MD). The denatured probes were hybridized with GEArray Q series nylon membrane printed with cDNAs of 96 genes and detected by chemiluminescence followed by data analysis with the help of a GEArray analyzer (SuperArray).
Semiquantitative PCR and Northern Blot Analysis of S100A4, MMP-9, and TIMP-1.
PCRs were carried out by using forward and reverse primer combinations for S100A4 (forward 5′-TCAGAACTAAAGGAGCTGCTGACC-3′, reverse 5′-TTTCTTCCTGGGCTGCTTATCTGG-3′), MMP-9 (forward 5′-TACCACCTCGAACTTTGACAGCGA-3′, reverse 5′-AAAGGCACAGTAGTGGCCGTAGAA-3′), TIMP-1 (forward 5′-GACCTACACTGTTGGCTGTGAG-3′, reverse 5′-AAGAAAGATGGGAGTGGGAACAGG-3′), and GAPDH (forward 5′-AATCCCATCACCATCTTCCAGGAG-3′, reverse 5′-GCATTGCTGATGATCTTGAGGCTG-3′). The cDNA was amplified with an initial denaturation at 94°C for 3 min followed by the sequential cycles of denaturation at 94°C for 45 sec, annealing at 52°C for 1 min, and extension at 72°C for 1 min for 30 cycles, with final extension at 72°C for 5 min. For Northern blot analysis, 5 μg of RNA was subjected to electrophoresis and fixed to the nylon membrane by UV cross-linker. The membranes were hybridized with the respective [32P]dCTP-labeled probes for S100A4, MMP-9, and TIMP-1 followed by washing and detection by autoradiography.
Gelatin Zymography.
An equal number of cells (1 × 106) were transfected with S100A4 siRNA and pcDNA3.1-S100A4 plasmid, and 24–36 h after transfection, the conditioned media were harvested, concentrated, and electrophoresed (10 μg of protein) under nonreducing conditions. The gelatinolytic activity of MMP-9 was determined by employing a zymography kit (Invitrogen) as per the vendor's protocol.
In Vitro Chemoinvasion Assay.
Twenty-four hours after transfection with S100A4 siRNA or pcDNA3.1-S100A4 plasmid, cells were collected, resuspended in culture medium, and incubated in a chemoinvasion chamber kit containing polycarbonate filter coated with Matrigel (BD Biosciences, Bedford, MA) for 5 h. The invasive capability of cells was measured as per vendor's protocol. Cells were counted under a microscope in five predetermined fields at a magnification of ×200.
Tumorigenicity Studies in Nude Mice.
After growth to subconfluency, transfected (S100A4 siRNA or nonsilencing siRNA) and nontransfected cells were trypsinized and harvested. A total of 1 × 106 cells were suspended in 50 μl of medium and 50 μl of Matrigel. For s.c. tumor growth, the cell suspension was inoculated s.c. into the right flank on 6-week-old male nude (athymic) mice by using a 27-gauge needle. To account for transfection efficiency, an additional control group composed of animals implanted with 50% S100A4-siRNA-transfected cells (500,000 cells) was also included. Every week, tumor volume was measured as described earlier (26).
Statistical Analyses.
Student's t test for independent analysis was applied to evaluate differences between the transfected and nontransfected cells with respect to in vitro invasive properties and expression of MMP-9 and TIMP-1. Kaplan–Meier analysis was used to estimate tumor-free survival, and differences in survival were analyzed by log–rank analysis. Linear regression analysis was performed to estimate tumor volume as a function of time. Statistical analyses were carried out by using S-PLUS (Insightful, Seattle, WA) and Excel (Microsoft, Redmond, WA), where P < 0.05 was considered significant.
Acknowledgments
We thank Dr. Dougles D. Boyden (M. D. Anderson Cancer Center, Houston, TX) and Dr. Motoharu Seiki (Institute of Medical Science, University of Tokyo, Tokyo, Japan) for providing pGL3-MMP-9-luc-reporter plasmid. This work was supported by U.S. Public Health Service Grants R01 CA 78809, R01 CA 101039, and P50 DK 65303.
Abbreviations
- CaP
prostate cancer
- MMP
matrix metalloproteinase
- TIMP
tissue inhibitor of matrix metalloproteinase.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Klein EA, Thompson IM. Curr Opin Urol. 2004;14:143–149. doi: 10.1097/00042307-200405000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Saleem M, Adhami VM, Zhong W, Longley BJ, Lin CY, Dickson RB, Reagan-Shaw S, Jarrard DF, Mukhtar H. Cancer Epidemiol Biomarkers Prev. 2006;5:217–227. doi: 10.1158/1055-9965.EPI-05-0737. [DOI] [PubMed] [Google Scholar]
- 4.Heizmann CW. Methods Mol Biol. 2002;172:69–80. doi: 10.1385/1-59259-183-3:069. [DOI] [PubMed] [Google Scholar]
- 5.Taylor S, Herrington S, Prime W, Rudland PS, Barraclough R. Br J Cancer. 2002;86:409–416. doi: 10.1038/sj.bjc.6600071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett SC, Varney KM, Weber DJ, Bresnick AR. J Biol Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- 7.de Silva Rudland S, Martin L, Roshanlall C, Winstanley J, Leinster S, Platt-Higgins A, Carroll J, West C, Barraclough R, Rudland P. Clin Cancer Res. 2006;12:1192–1200. doi: 10.1158/1078-0432.CCR-05-1580. [DOI] [PubMed] [Google Scholar]
- 8.Camby I, Nagy N, Lopes MB, Schafer BW, Maurage CA, Ruchoux MM, Murmann P, Pochet R, Heizmann CW, Brotchi J, et al. Brain Pathol. 1999;9:1–19. doi: 10.1111/j.1750-3639.1999.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Hussain T, MacLennan GT, Fu P, Patel J, Mukhtar H. J Clin Oncol. 2003;21:106–112. doi: 10.1200/JCO.2003.03.024. [DOI] [PubMed] [Google Scholar]
- 10.Saleem M, Adhami VM, Ahmad N, Gupta S, Mukhtar H. Clin Cancer Res. 2005;11:147–154. [PubMed] [Google Scholar]
- 11.Mathisen B, Lindstad RI, Hansen J, El-Gewely SA, Maelandsmo GM, Hovig E, Fodstad O, Loennechen T, Winberg JO. Clin Exp Metastasis. 2003;20:701–711. doi: 10.1023/b:clin.0000006819.21361.03. [DOI] [PubMed] [Google Scholar]
- 12.Aalinkeel R, Nair MP, Sufrin G, Mahajan SD, Chadha KC, Chawda RP, Schwartz SA. Cancer Res. 2004;64:5311–5321. doi: 10.1158/0008-5472.CAN-2506-2. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Y, Muschel RJ. Cancer Res. 2002;62:1910–1914. [PubMed] [Google Scholar]
- 14.Hernan R, Fasheh R, Calabrese C, Frank AJ, Maclean KH, Allard D, Barraclough R, Gilbertson RJ. Cancer Res. 2003;63:140–148. [PubMed] [Google Scholar]
- 15.Ke Y, Jing C, Barraclough R, Smith P, Davies MP, Foster CS. Int J Cancer. 1997;71:832–837. doi: 10.1002/(sici)1097-0215(19970529)71:5<832::aid-ijc22>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Bjornland K, Winberg JO, Odegaard OT, Hovig E, Loennechen T, Aasen AO, Fodstad O, Maelandsmo GM. Cancer Res. 1999;59:4702–4708. [PubMed] [Google Scholar]
- 17.Satariano WA, Ragland KE, Van Den Eeden SK. Cancer. 1998;83:1180–1188. doi: 10.1002/(sici)1097-0142(19980915)83:6<1180::aid-cncr18>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt-Hansen B, Ornas D, Grigorian M, Klingelhofer J, Tulchinsky E, Lukanidin E, Ambartsumian N. Oncogene. 2004;23:5487–5495. doi: 10.1038/sj.onc.1207720. [DOI] [PubMed] [Google Scholar]
- 19.Stearns M, Stearns ME. Oncol Res. 1996;8:69–75. [PubMed] [Google Scholar]
- 20.Adhami VM, Siddiqui IA, Ahmad N, Gupta S, Mukhtar H. Cancer Res. 2004;64:8715–8722. doi: 10.1158/0008-5472.CAN-04-2840. [DOI] [PubMed] [Google Scholar]
- 21.Bjornland K, Bratland A, Rugnes E, Pettersen S, Johansen HT, Aasen AO, Fodstad O, Ree AH, Maelandsmo GM. J Pediatr Surg. 2001;36:1040–1044. doi: 10.1053/jpsu.2001.24735. [DOI] [PubMed] [Google Scholar]
- 22.Maatta M, Santala M, Soini Y, Talvensaari-Mattila A, Turpeenniemi-Hujanen T. Tumour Biol. 2004;25:188–192. doi: 10.1159/000081101. [DOI] [PubMed] [Google Scholar]
- 23.Culhaci N, Metin K, Copcu E, Dikicioglu E. BMC Cancer. 2004;4:42. doi: 10.1186/1471-2407-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida S, Yokota T, Ujiki M, Ding XZ, Pelham C, Adrian TE, Talamonti MS, Bell RH, Jr, Denham W. Biochem Biophys Res Commun. 2004;323:1241–1245. doi: 10.1016/j.bbrc.2004.08.229. [DOI] [PubMed] [Google Scholar]
- 25.Peters TJ, Albieri A, Bevilacqua E, Chapman BM, Crane LH, Hamlin GP, Seiki M, Soares MJ. Cell Tissue Res. 1999;295:287–296. doi: 10.1007/s004410051235. [DOI] [PubMed] [Google Scholar]
- 26.Saleem M, Kweon MH, Yun JM, Adhami VM, Khan N, Syed DN, Mukhtar H. Cancer Res. 2005;65:11203–11213. doi: 10.1158/0008-5472.CAN-05-1965. [DOI] [PubMed] [Google Scholar]