Abstract
There is controversy and uncertainty on how far north there were glacial refugia for temperate species during the Pleistocene glaciations and in the extent of the contribution of such refugia to present-day populations. We examined these issues using phylogeographic analysis of a European woodland mammal, the bank vole (Clethrionomys glareolus). A Bayesian coalescence analysis indicates that a bank vole population survived the height of the last glaciation (≈25,000–10,000 years B.P.) in the vicinity of the Carpathians, a major central European mountain chain well north of the Mediterranean areas typically regarded as glacial refugia for temperate species. Parameter estimates from the fitted isolation with migration model show that the divergence of the Carpathian population started at least 22,000 years ago, and it was likely followed by only negligible immigration from adjacent regions, suggesting the persistence of bank voles in the Carpathians through the height of the last glaciation. On the contrary, there is clear evidence for gene flow out of the Carpathians, demonstrating the contribution of the Carpathian population to the colonization of Europe after the Pleistocene. These findings are consistent with data from animal and plant fossils recovered in the Carpathians and provide the clearest phylogeographic evidence to date of a northern glacial refugium for temperate species in Europe.
Keywords: climate change, colonization, northern refugia, phylogeography, Quaternary
Climate change has had major impacts on geographic distributions, demography, and, thus, evolution of species. It is now well established that in the northern hemisphere many temperate species retreated from large continental areas during the height of the last glaciation (≈25,000–10,000 years B.P.) and were only able to survive in sheltered refugia, which provided suitable conditions. However, controversy and uncertainty remain regarding the number and location of glacial refugia that contributed to modern populations. Temperate-adapted woodland species would have been particularly strongly affected because the unglaciated areas at this time in northern and central Europe, Asia, and North America were largely treeless. Numerous distributional and phylogeographic patterns of fauna and flora are consistent with the process of colonization from refugia at lower latitudes after the Pleistocene (1–3). In Europe, in particular, there has been much emphasis on the Mediterranean peninsulas of Iberia, Italy, and the Balkans as possible locations for glacial refugia (4, 5). However, there is evidence that refugia for temperate species may also have existed further north in central and western Europe (6), locations described as “northern refugia” by Stewart and Lister (7). We follow the terminology of Stewart and Lister. The most compelling evidence supporting the existence of northern refugia comes from the vicinity of the Carpathians, a mountain chain >1,500 km long in central Europe. In particular, plant pollen and macrofossils recovered from various places in the Carpathians showed that coniferous and broad-leaved trees were part of the local full-glacial environment (8, 9). This finding is supported by evidence from animal fossils, which provide records of woodland species of small mammals in deposits from the last glaciation in the Carpathians (10, 11). Furthermore, divergent mtDNA sequences from extant Carpathian populations of several temperate vertebrate species, including amphibians (12, 13) and fish (14, 15), indicate the maintenance of separate lineages that may have persisted in the Carpathians during the last glaciation. Various lines of evidence thus point to the existence of a glacial refugium for temperate species in the Carpathians. As a result, modern populations of species in central and northern Europe might have not been derived exclusively from southern refugial populations, but also from populations that survived in much more northerly regions. A proper realization of the importance of northern refugia thus has the potential to change dramatically the way that postglacial colonization of Europe is viewed, providing a different perspective on the communities of temperate species that currently occur there.
This phylogeographic study examines the genetic evidence for a Carpathian refugium in a European woodland mammal, the bank vole (Clethrionomys glareolus). The bank vole is a small rodent that is widely distributed in Europe between the British Isles and northern Spain in the west to central Siberia in the east. It is found abundantly in a wide variety of woodland habitats, such as temperate broad-leaved and mixed forests (16), and its glacial distribution most likely tracked this type of environment (11). The bank vole is an excellent model to test the relative contribution of traditionally recognized refugia and those that may have existed farther north, because its current wide distribution includes all three Mediterranean peninsulas. The bank vole has already been the subject of phylogeographic analyses that suggested that glacial refugia located in central and eastern Europe, as opposed to the Mediterranean refugia, made a major contribution to the modern population of this species in Europe (6, 17). The Carpathian Mountains have been proposed as a possible refugial area for this species based on the fossil record. Bank vole fossils have been identified in deposits from the last glaciation in the Carpathian Mountains together with typical glacial assemblages and suggest that the species may have had a continuous presence in this region (10, 11, 18). We have collected mtDNA sequences from the Carpathians and adjacent regions (Fig. 1) and present the clearest evidence yet that a bank vole population survived in a central European refugium through the height of the last glaciation. Together with recent findings in North America (19), our results give substantial credence to the occurrence of glacial refugia for temperate species further north than traditionally recognized.
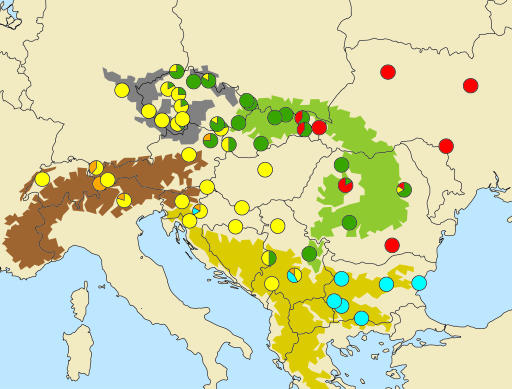
Fig. 1.
Map showing the collection sites and the distribution of haplotype lineages. Pie charts show the proportions of haplotypes belonging to each lineage within each population. The colors equate to the clades identified in Fig. 2. Approximate distribution of major mountain regions is shown in green for the Carpathians, in gray for the Bohemian Massif, in brown for Alps, and in yellow for the Balkans.
Results and Discussion
Phylogenetic Analyses.
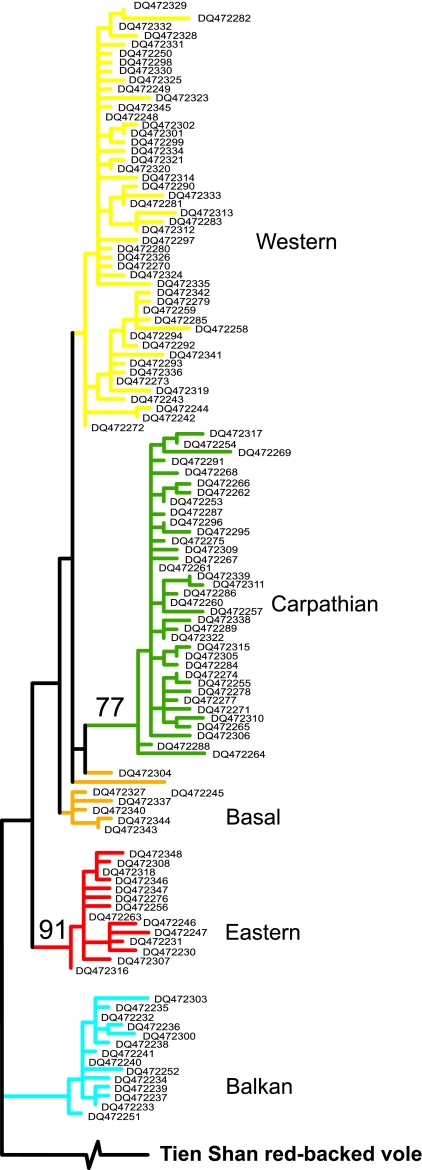
The reconstruction of phylogenetic relationships among the 119 haplotypes identified in 224 bank voles demonstrates multiple divergent clades (Fig. 2). Three of these clades were described in an earlier range-wide phylogeographic survey that revealed five European C. glareolus clades but failed to resolve finer scale divisions (17). With the enhanced sampling and longer sequences, we discovered a “Carpathian clade” defined by four synonymous nucleotide changes (0.0086–0.0192 net divergence from other clades), which was supported by a bootstrap value of 77% (Fig. 2) and had a localized geographic distribution. The 36 haplotypes in this clade were recovered only from sites in the Carpathians and their close vicinity, and the majority of bank voles from the Carpathians carried haplotypes from this clade (Fig. 1). The remaining 83 haplotypes in our study represented the western, eastern, and Balkan clades identified earlier. There were also seven distinct haplotypes that fell outside these major clades, basal to the Carpathian and western clades and geographically embedded within the distribution of the western clade (Figs. 1 and 2). Despite the dense sampling, the major clades did not show extensive geographic overlap except where their distributions come in contact. These results thus demonstrate that the Carpathian clade is a geographically localized and monophyletic lineage, indicating that ancestors of bank voles from this clade have been isolated in the past from the ancestors of the other clades.
Fig. 2.
Maximum likelihood estimate of the phylogeny of 119 mtDNA haplotypes. The tree has been rooted with the sequence of the Tien Shan red-backed vole (C. centralis) representing an outgroup to the bank vole sequences. Haplotypes are identified by GenBank accession numbers.
Population Divergence Analysis.
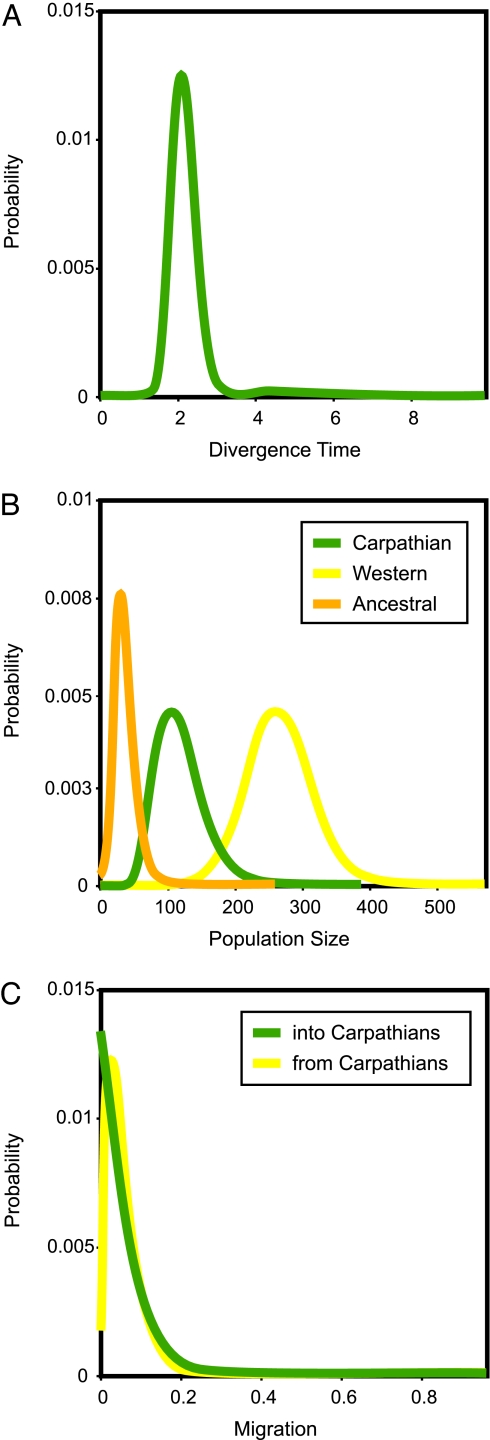
If bank voles of the Carpathian clade descended from a population that survived in a Carpathian refugium at the height of the last glaciation, the split from voles of the western population should have occurred earlier than that time. Fig. 3A shows the estimated posterior probability distribution of the divergence time between the two populations obtained from the isolation with migration (IM) model. The divergence time is clearly resolved, with posterior distribution that has a single narrow peak and bounds that fall within the prior distribution. Given the range of mutation rate estimates (see Materials and Methods), the position of the peak (t = 2.11) corresponds to 30,153–108,887 years. The IM analysis thus suggests that the divergence of the two populations occurred during the height of the last glaciation (≈25,000–10,000 years B.P.) or earlier, and the bank voles from the Carpathian clade must have survived in a separate refugium. If, on the contrary, the Carpathian population had been founded from the same southern refugial source as the western population and their divergence was postglacial, then the estimate of t should be much more recent. However, any divergence time < 22,000 years has very low probability in our IM analysis (Table 1), making such a scenario highly unlikely.
Fig. 3.
Posterior probability distributions (scaled by the mutation rate) estimated for divergence time (A), effective population sizes (B), and migration rates (C).
Table 1.
Maximum-likelihood estimates of gene flow and divergence parameters
| Parameter | Carpathian population | Western population |
|---|---|---|
| t | 2.11 (1.54–2.86) | |
| t, years | 30,153–108,887 (21,988–147,683) | |
| θ | 96.61 (59.26–172.44) | 259.24 (180.92–350.12) |
| θancestral | 20.10 (7.5–42.21) | |
| m | 0.0035 (0.0005–0.1435) | 0.0255 (0.0015–0.1145) |
| 2Nm | 0.169 | 3.305 |
The maximum likelihood values and 90% highest posterior density (HPD) intervals (in parentheses) are shown except for the population migration rate 2Nm= θ m/2, where 90% HPD intervals were not directly available. The time parameter was converted to the time scale in years using a range of plausible mutation rates (see Materials and Methods).
The effective population size for the Carpathian population is >2 times smaller than that for the western population, and the size of the estimated ancestral population was ≈5 and 15 times smaller than the two recent populations, respectively (Table 1 and Fig. 3B). This finding suggests that the Carpathian and western populations survived in separate refugia during the height of the last glaciation but most likely expanded from the same ancestral population at the end of an earlier glaciation.
The estimated rate of mtDNA gene flow into the Carpathian population after its separation is at or near zero, whereas gene flow from the Carpathians to the western population shows a clear nonzero peak at 2Nm = 3.3 (Table 1 and Fig. 3C). The IM analyses thus suggest that, after their divergence, the populations have been exchanging genes primarily from the Carpathians to the western population, further supporting the local origin of the Carpathian clade.
Although the McDonald–Kreitman test provided no evidence of selection on the cytochrome b (cytb) gene (P > 0.10), the possibility that adaptive divergence at a linked gene could retard mtDNA gene flow into the Carpathians cannot be excluded. In such a case, gene flow rates would be the result of demography and selection (rather then demography alone), but it would nevertheless underscore the ecological and evolutionary uniqueness of the Carpathian population and would stand in support of the Carpathian refugium hypothesis.
Our study of the bank vole provides the clearest phylogeographic evidence to date of a European glacial refugium for temperate species that was distinctly north of the traditionally recognized Mediterranean refugia. The Bayesian coalescent method generates a divergence time for the Carpathian population that predates the last glacial maximum. This genetic evidence is consistent with the fossil record supporting the persistence of bank voles and other woodland mammals during the height of the last glaciation in the Carpathians but not in more westerly areas (11). More broadly, the finding of our study is consistent with recent evidence suggesting that coniferous and broad-leaved trees were part of the local full-glacial environment in various parts of the Carpathians (8, 9, 20). Therefore, rather then migrating south to track favorable climate and habitat, some temperate species apparently tolerated the climate change at the glacial maximum and survived in sheltered northern refugia where moister conditions occurred and tree cover could develop.
The distribution and deep structure among the bank vole clades suggests a complex colonization history from multiple refugia. Our finding of gene flow out of the Carpathians points to the significance of the Carpathian refugium as a source for the colonization of other areas after the Pleistocene. Postglacial colonization from multiple refugia in central Europe was suggested for bank voles by our earlier range-wide phylogeographic study (17). For the western clade, the most southern localities are southern France, northwestern Italy, and the western Balkans. Therefore, it is possible that this lineage occupied one or more woodland refugia in the foothills of the Alps and/or the western Balkans (21, 22). These data suggest a complex pattern of colonization of central and northern Europe after the Pleistocene by bank voles from the Carpathians and other refugia.
Our phylogeographic study of the bank vole generates an impetus for similar detailed analyses on other temperate species in the Carpathians and in additional areas, e.g., the vicinity of the Alps, southern France, and southern parts of the Ural Mountains, which have been identified as potential northern refugia (e.g., refs. 23–25). If, as appears to be the case with the bank vole, some species colonized central and northern Europe both from refugia in relatively southern and relatively northern locations, it may have had an impact on the genetic and ecological properties of the species. From which type of refugium a particular population derives may have significance with regards to physiological traits such as cold-tolerance or dispersal capacity. If postglacial colonization from northern refugia was common, the concept that species responded to climate change mainly by undergoing large-scale distribution shifts to track suitable conditions might be erroneous. Instead, maintaining small isolated populations that persisted because of a locally favorable climate might be the major mechanism by which temperate species responded to the climate change (7, 26). Proper realization of the importance of northern refugia thus has the potential to change dramatically the way that postglacial colonization of Europe is viewed, providing a different perspective on how temperate species respond to changing environmental conditions.
Materials and Methods
Samples.
A total of 224 bank voles were collected from 56 localities across the Carpathians and adjacent geographic regions in central and southeastern Europe in Austria, Bulgaria, Czech Republic, Germany, Croatia, Hungary, Switzerland, Italy, Romania, Serbia and Montenegro, Slovakia, Slovenia, and Ukraine (Fig. 1 and Table 2, which is published as supporting information on the PNAS web site). A single individual of the Tien Shan red-backed vole (Clethrionomys centralis), phylogenetically a sister species to the bank vole, was sequenced and used as outgroup. Tissue samples of liver, spleen, or toe clips were obtained from mammalogists conducting fieldwork or were collected for the purpose of this study, and were stored in 95% ethanol at 4°C.
Molecular Biological Techniques.
Genomic DNA was extracted by the Qiagen (Valencia, CA) DNeasy Tissue kit. A 1,074-bp fragment of the mtDNA cytb gene was amplified by PCR by using primers located within the cytb (5′-CCCTCTAATCAAAATCATCAA-3′) and Thr tRNA genes (5′-TTTCATTTCTGGTTTACAAGAC-3′). The primers were designed on the basis of published cytb sequences (27), and sequences that were generated with the primer (5′-TGGTGGGGGAAGAGTCCTT-3′), designed within the Pro tRNA gene by using sequences published by Stacy et al. (28). The PCR conditions followed standard methods described for arvicolid rodents (17, 23). Negative extraction and PCR controls with no tissue and no template DNA, respectively, were used in each experiment. The resulting PCR products were purified by using the Qiagen QIAquick PCR Purification kit or the Millipore (Bedford, MA) Montage PCR centrifugal filter devices and were directly cycle-sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA), the sequencing primer LCLE2 (17), and a newly designed reverse sequencing primer (5′-GTTGGGTTGTTGGATCCTG-3′). The extension products were run on ABI 3730 automated sequencers. Sequences were aligned manually, and any ambiguity was resolved by sequencing the complementary strand.
Phylogenetic Reconstruction.
The phylogenetic relationships among the sequences were reconstructed by using the maximum likelihood optimality criterion. The analyses were performed by the algorithm in PHYML 2.4.4 that simultaneously adjusts tree topology and branch lengths to maximize tree likelihood (29), and using the HKY evolutionary model (30) with the base frequencies A, 0.31; C, 0.29; and G, 0.13; a transition/transversion ratio of 18.07; the proportion of invariable sites set at 0.68; and γ-distributed rates across sites with the shape parameter α equaling to 1.00. This model was determined to be the most appropriate for our dataset by the hierarchical likelihood ratio test of goodness of fit of 56 different nested models to the data, as implemented in Modeltest 3.06 (31). To quantify the confidence in the partitioning within the trees, we performed the nonparametric bootstrap test as applied to phylogenetics by Felsenstein (32) using 1,000 replications.
IM Model Analysis.
We analyzed the data under the IM model of population divergence (33, 34). The model assumes that an ancestral population splits into two descendant populations with gene flow possibly continuing between the diverging populations. To fit the IM model to the bank vole data, we used a Bayesian coalescent method that integrates over all possible genealogies by using a Markov chain Monte Carlo (MCMC) approach. This method estimates posterior probability distributions for six demographic parameters including divergence time, two-directional gene flow rates, and effective population sizes of two current populations and the ancestral population (34). We have taken the fossil evidence suggesting that the Carpathians provided glacial refugia for bank voles (11, 17, 18) as a hypothesis of the presence of a distinct and isolated population. The Carpathian population was defined as 43 bank voles collected in the Carpathians; this population excludes 14 individuals that represent recent admixture of the eastern clade (ref. 17; Fig. 1) to eliminate their negative effect on the parameter estimates (35). The western population included 138 bank voles from regions westerly adjacent to the Carpathians and principally characterized by the western clade but carrying also haplotypes of the Carpathian clade and haplotypes basal to both these clades (Figs. 1 and 2). Four bank voles carrying haplotypes of the highly divergent Balkan clade were excluded as representing admixture from the eastern Balkans (Fig. 1). We used the IM program (34) to run the MCMC simulations assuming the HKY model of sequence evolution and uniform prior distributions of parameter ranges, which were empirically determined to ensure that the posterior distributions fell completely within the prior distributions (36). The peaks of the posterior distributions were thus taken as maximum likelihood estimates of the parameters (33, 37). For credibility intervals we recorded for each parameter the 90% highest posterior density (HPD) interval, i.e., the shortest span that includes 90% of the probability density of a parameter. The analysis was done using several independent runs, each with 4 to 15 chains under the Metropolis coupling to improve mixing (36). Each chain was initiated with a burn-in period of 100,000 updates, and the total length of each analysis was between 5 and 30 million updates. The analysis was considered to have converged on a stationary distribution if the independent runs generated similar posterior distributions (38). To convert the parameter estimates scaled by the mutation rate to calendar years, we used a wide range of plausible divergence rates available for cytb of arvicolid rodents of 3.6% to 13% per million years (17, 39, 40). These divergence rates equate to 1.9 × 10−5 to 7.0 × 10−5 mutations per year for the gene region studied (1,074 base pairs). To explore whether natural selection at cytb may have acted to limit gene flow, we tested whether the bank vole variation conforms to expectations under selective neutrality using the McDonald–Kreitman test (41) implemented in DnaSP 4.10.8 (42). For this analysis, we used Fisher's exact test to compare the ratio of nonsynonymous to synonymous polymorphisms within the bank vole (overall and for each clade separately) to the ratio of the number of nonsynonymous to synonymous fixed differences between the bank vole and the Tien Shan red-backed vole.
Supplementary Material
Acknowledgments
We thank the many colleagues who assisted with field collections, including A. Belancic, V. Bjedov, I. Coroiu, J. Farkas, J. Hausser, G. Horvath, L. Choleva, J. Margaletić, S. Marková, N. Martínková, P. Miklós, E. Mikolášková, P. Mikulíček, A. Mishta, P. Munclinger, D. Murariu, P. Nová, D. Peshev, F. Sedláček, F. Spitzenberger, M. Stanko, P. Suchomel, M. Šandera, L. Tomović, P. Vogel, J. Uhlíková, V. Vohralík, and P. Zupančič. This work was funded by the Royal Society of London/North Atlantic Treaty Organization Postdoctoral Fellowship grant (to P.K. and J.B.S.), Czech Science Foundation Grant 206/05/P032 (to P.K.), and Academy of Sciences of the Czech Republic Grant IRP IAPG AV0Z 50450515 (to P.K.).
Abbreviation
- IM
isolation with migration.
Footnotes
References
- 1.Lessa EP, Cook JA, Patton JL. Proc Natl Acad Sci USA. 2003;100:10331–10334. doi: 10.1073/pnas.1730921100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hewitt GM. Philos Trans R Soc London Ser B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Runck AM, Cook JA. Mol Ecol. 2005;14:1445–1456. doi: 10.1111/j.1365-294X.2005.02501.x. [DOI] [PubMed] [Google Scholar]
- 4.Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF. Mol Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- 5.Hewitt GM. Biol J Linn Soc. 1999;68:87–112. [Google Scholar]
- 6.Bilton DT, Mirol PM, Mascheretti S, Fredga K, Zima J, Searle JB. Proc R Soc London Ser B; 1998. pp. 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart JR, Lister AM. Trends Ecol Evol. 2001;16:608–613. [Google Scholar]
- 8.Jankovská V, Chromy P, Nižnianská M. Acta Palaeobotanica. 2002;42:39–50. [Google Scholar]
- 9.Willis KJ, van Andel TH. Q Sci Rev. 2004;23:2369–2387. [Google Scholar]
- 10.Nadachowski A. Acta Theriologica. 1989;34:37–53. [Google Scholar]
- 11.Horáček I. Geolines. 2000;11:103–107. [Google Scholar]
- 12.Babik W, Branicki W, Crnobrnja-Isailovic J, Cogalniceanu D, Sas I, Olgun K, Poyarkov NA, Garcia-París M, Arntzen JW. Mol Ecol. 2005;14:2475–2491. doi: 10.1111/j.1365-294X.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- 13.Voros J, Alcobendas M, Martinez-Solano I, Garcia-Paris M. Mol Phylogenet Evol. 2006;38:705–718. doi: 10.1016/j.ympev.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Kotlík P, Berrebi P. Mol Phylogenet Evol. 2002;24:10–18. doi: 10.1016/s1055-7903(02)00264-6. [DOI] [PubMed] [Google Scholar]
- 15.Janko K, Culling MA, Ráb P, Kotlík P. Mol Ecol. 2005;14:2991–3004. doi: 10.1111/j.1365-294X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 16.Raczyński J. In: Ecology of the Bank Vole. Petrusewicz K, editor. Warsaw, Poland: Polish Scientific; 1983. pp. 3–10. [Google Scholar]
- 17.Deffontaine V, Libois R, Kotlík P, Sommer R, Nieberding C, Paradis E, Searle JB, Michaux JR. Mol Ecol. 2005;14:1727–1739. doi: 10.1111/j.1365-294X.2005.02506.x. [DOI] [PubMed] [Google Scholar]
- 18.Nadachowski A, Miekina B, Garapich A. In: Oblazowa Cave: Human Activity, Stratigraphy, and Palaeoenvironment. Valde-Nowak P, Nadachowski A, Madeyska T, editors. Krakow, Poland: Institute of Archaeology and Ethnology, Polish Academy of Sciences; 2003. pp. 134–140. [Google Scholar]
- 19.Rowe KC, Heske EJ, Brown PW, Paige KN. Proc Natl Acad Sci USA. 2004;101:10355–10359. doi: 10.1073/pnas.0401338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Björkman L, Feurdean A, Wohlfarth B. Rev Palaeobot Palynol. 2003;124:79–111. [Google Scholar]
- 21.Petit RJ, Brewer S, Bordacs S, Burg K, Cheddadi R, Coart E, Cottrell J, Csaikl UM, van Dam B, et al. For Ecol Manage. 2002;156:49–74. [Google Scholar]
- 22.Heuertz M, Funeschi S, Anzidei M, Pastorelli R, Salvini D, Paule L, Frascaria-Lacoste N, Hardy OJ, Vekemans X, et al. Mol Ecol. 2004;13:3437–3452. doi: 10.1111/j.1365-294X.2004.02333.x. [DOI] [PubMed] [Google Scholar]
- 23.Jaarola M, Searle JB. Mol Ecol. 2002;11:2613–2621. doi: 10.1046/j.1365-294x.2002.01639.x. [DOI] [PubMed] [Google Scholar]
- 24.Rendell S, Ennos RA. Mol Ecol. 2002;11:69–78. doi: 10.1046/j.0962-1083.2001.01413.x. [DOI] [PubMed] [Google Scholar]
- 25.Pinceel J, Jordaens K, Pfenninger M, Backeljau T. Mol Ecol. 2005;14:1133–1150. doi: 10.1111/j.1365-294X.2005.02479.x. [DOI] [PubMed] [Google Scholar]
- 26.Pearson RG. Trends Ecol Evol. 2006;21:111–113. doi: 10.1016/j.tree.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Cook JA, Runck AM, Conroy CJ. Mol Phylogenet Evol. 2004;30:767–777. doi: 10.1016/S1055-7903(03)00248-3. [DOI] [PubMed] [Google Scholar]
- 28.Stacy JE, Jorde PE, Steen H, Ims RA, Purvis A, Jakobsen KS. Mol Ecol. 1997;6:751–759. doi: 10.1046/j.1365-294x.1997.d01-470.x. [DOI] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 31.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 32.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen R, Wakeley J. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hey J, Nielsen R. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slatkin M. Mol Ecol. 2005;14:67–73. doi: 10.1111/j.1365-294X.2004.02393.x. [DOI] [PubMed] [Google Scholar]
- 36.Won Y-J, Sivasundar A, Wang Y, Hey J. Proc Natl Acad Sci USA. 2005;102:6581–6586. doi: 10.1073/pnas.0502127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Won Y-J, Hey J. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- 38.Hey J. PLoS Biol. 2005;3:1–11. doi: 10.1371/journal.pbio.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conroy CJ, Cook JA. J Mammal. 2000;81:344–359. [Google Scholar]
- 40.Galbreath KE, Cook JA. Mol Ecol. 2004;13:135–148. doi: 10.1046/j.1365-294x.2004.02026.x. [DOI] [PubMed] [Google Scholar]
- 41.McDonald JH, Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 42.Rozas J, Sanchez-Delbarrio JC, Messeguer X, Rozas R. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.