Abstract
Alterations in phosphorylation of cellular proteins are a hallmark of malignant transformation. Degradation of these phosphoproteins could generate cancer-specific class I MHC-associated phosphopeptides recognizable by CD8+ T lymphocytes. In a comparative analysis of phosphopeptides presented on the surface of melanoma, ovarian carcinoma, and B lymphoblastoid cells, we find 5 of 36 that are restricted to the solid tumors and common to both cancers. Differential presentation of these peptides can result from differential phosphorylation of the source proteins. Recognition of the peptides on cancer cells by phosphopeptide-specific CD8+ T lymphocytes validates the potential of these phosphopeptides as immunotherapeutic targets.
Keywords: tandem mass spectrometry, immobilized metal-affinity chromatography
Class I MHC molecules are cell surface proteins that bind and display a broad repertoire of peptides produced by degradation of cellular proteins to CD8+ T lymphocytes. Changes in protein expression and/or metabolism that accompany infection or cellular transformation alter the peptides that are displayed by these MHC molecules. CD8 T lymphocyte recognition of specific peptides on pathogen infected or cancerous cells leads to activation of cytolysis and cytokine secretion programs, resulting in inflammation and cellular destruction. Proteins degraded by the proteasome are a major source of peptides displayed on class I MHC molecules (1). Rapid protein degradation by the proteasome also is an important mechanism for regulating the activity of many transcription factors, cell growth modulators, signal transducers, and cell cycle proteins (2–4). Interestingly, this degradation often depends on E3 ubiquitin-ligases whose activity in turn depends on phosphorylation of the target protein (5–7). Malignant transformation leads to alterations in the protein kinase pathways regulating cell growth, differentiation, and cell death (8, 9). Deregulated signaling cascades often lead to increases in the extent of protein phosphorylation within the cell (10). We hypothesized that degradation of these proteins would generate phosphopeptides that are uniquely or differentially presented on malignant cells by class I MHC molecules. These phosphopeptides would be attractive candidates for cancer immunotherapy.
Results and Discussion
To define class I MHC-associated phosphopeptides derived from proteins potentially associated with malignant transformation, we analyzed mixtures of >10,000 peptides presented by HLA-A*0201 molecules on the surface of two melanoma cells, DM331 and SLM2, an ovarian carcinoma, COV413, and an Epstein–Barr virus-transformed B lymphoblastoid cell line (BLCL), JY. Peptides associated with immunoaffinity-purified HLA-A*0201 molecules were extracted with acid (11) and converted to methyl esters under acidic conditions with either d0- or d3-methanol (12). Samples with different isotopic labels from any two of the above cell lines were combined, enriched for phosphopeptides by immobilized metal-affinity chromatography and analyzed by the combination of nanoflow-HPLC and tandem MS on a hybrid linear-quadrupole ion-trap/Fourier-transform mass spectrometer (11, 13). In these experiments, signals for class I MHC phosphopeptides presented on the surface of both cell lines appear in the mass spectrum as doublets separated by 3 m/z units per carboxylate group in the molecule. Phosphopeptides that are unique to one or the other cell line appear as singlets. Reproducibility of the peptide extraction, derivatization, and analytical methodology was demonstrated by analyzing multiple d0- and d3-labeled samples prepared from the same or different lysates of JY BLCL. Identical doublet peaks were observed in each experiment (data not shown). Next, we analyzed samples prepared by mixing d0- and d3-labeled peptide methyl esters from multiple pairs of the above four cell lines. We detected and sequenced 36 phosphopeptides presented by HLA-A*0201 on one or more of the four cell lines (Table 1). Although relatively small in comparison to the number identified in cell extracts (14), the number of phosphopeptides detected is likely to be limited by the HLA-A*0201-binding motif (15) and the location of the phosphorylated residue (11). The observed peptides match to 35 source proteins in the human protein database (www.ncbi.nlm.nih.gov/BLAST).
Table 1.
Distribution and copies per cell of HLA-A2-associated phosphorylated epitopes on cancer cell lines
| Source protein | Sequence | DM331 | SLM2 | COV413 | JY |
|---|---|---|---|---|---|
| Cancer cell-specific phosphopeptides | |||||
| Insulin receptor substrate 2 | RVApSPTSGV | ++ | +++ | +++ | − |
| Tensin-3/tumor endothelial marker 6 | VMIGpSPKKV† | +++ | +++ | ++ | − |
| β-catenin | YLDpSGIHSGA | + | + | + | − |
| FLJ13725 | RTLpSHISEA | ++ | + | + | − |
| β-synemin/Desmuslin | RTFpSPTYGL | ++ | + | ++ | − |
| Jun-C/D | KLApSPELERL | ++ | − | + | − |
| Breast cancer anti-estrogen resistance 3 | IMDRpTPEKL† | +++ | + | − | − |
| TFIID transcription initiation factor subunit 13 | RLFpSKELRC‡ | ++ | ++ | − | − |
| Ribosomal protein S17 | KLLDFGSLpSNLQV | + | − | − | − |
| Ub-carboxyl terminal hydrolase 10 | KLLpSPSNEKL | + | − | − | − |
| Phosphoinositol 3-phosphate-binding protein 3 | SLQPRSHpSV | − | − | + | − |
| Shared phosphopeptides on cancer and BLCL | |||||
| CDC25b | GLLGpSPVRA | ++ | + | ++ | + |
| Human bromodomain containing protein-4 | AVVpSPPALHNA | − | ++ | ++ | + |
| FLJ10707 | LMFpSPVTSL§ | + | − | − | + |
| Ribosomal protein L4 | ILKpSPEIQRA | + | + | − | + |
| KIAA1328 protein | KLMpSPKADVKL§ | ++ | ++ | − | ++ |
| Interleukin enhancer binding factor 3 | KLFPDpTPLAL | ++ | + | + | ++ |
| Thyroid hormone receptor interacting protein 12 | SLLTpSPPKA | ++ | ++ | ++ | ++ |
| Trafficking protein particle complex 1 | RLDpSYVRSL¶ | ++ | + | + | ++ |
| Early mitotic inhibitor 1 | VMFRpTPLASV§ | + | + | − | + |
| ORF 17, chromosome 2 | RLSpSPLHFV | + | + | − | + |
| Heterogeneous nuclear ribonucleoprotein AO | AMAApSPHAV† | + | + | + | ++ |
| Adenosine monophosphate deaminase 2 | RQIpSQDVKL | ++ | ++ | + | +++ |
| Heat shock protein 27 | RQLpSSGVSEI | ++ | ++ | ++ | +++ |
| Unknown | RLLpSPLSSA‖ | ++ | ++ | ++ | +++ |
| Cell cycle checkpoint kinase 1 | KLIDIVpSSQKV | + | + | + | +++ |
| Testicular Receptor 2 (TR2) | RQDpSTPGKVFL | + | + | + | +++ |
| SRp46 splicing factor | SMpTRSPPRV† | ++ | ++ | − | +++ |
| Anaphase promoting complex subunit 1 | VLLpSPVPEL | + | − | + | ++ |
| FLJ22624 | TLApSPSVFKST | − | + | − | ++ |
| Mitochondrial escape 1-like 1 | RLQpSTSERL | − | + | − | ++ |
| BLCL-specific phosphopeptides | |||||
| Protein kinase D2 | RQApSLSISV | − | − | − | ++ |
| Lymphocyte-specific protein 1 | RQApSIELPSMAV††† | − | − | − | ++++ |
| Lymphocyte-specific protein 1 | KLIDRTEpSL | − | − | − | ++ |
| Premature ovarian failure, 1B | RTYpSGPMNKV† | − | − | − | ++ |
| Nedd4 binding protein 2 | KMDpSFLDMQL§ | − | − | − | ++ |
Protein sources were obtained by searching peptide sequences against the nr and refseq databases for human proteins (www.ncbi.nlm.nih.gov/BLAST). Bolded text indicates known phosphoproteins. Italicized text indicates that the protein is a member of a known phosphoprotein family but that phosphorylation status is undocumented. Underlined residues indicate known phosphosites (www.phosphosite.org). −, not detected; +, 0.5–5 copies per cell; ++, 5–50 copies per cell; +++, 50–500 copies per cell; ++++, >500 copies per cell.
†Met and Metox forms of the peptide were detected.
‡The Cys residue is either oxidized or cysteinylated.
§The Metox form of the peptide was detected.
¶C-terminal truncated versions of the peptide were found on several of the cell lines.
‖The peptide modified with +18 mass units on the second S was also identified on DM331 and COV413.
††The 10-mer (RQApSIELPSM) was found on JY BLCL.
Because it has been suggested that the main source of peptides presented by class I MHC molecules involves proteasomal degradation of defective ribosomal products or misfolded proteins (16), we examined the peptides for evidence that the observed phosphorylation sites result from normal protein phosphorylation processes. Of the 35 identified source proteins, 28 have described functions (Table 3, which is published as supporting information on the PNAS web site). Twenty-one of the identified source proteins are known phosphoproteins (Table 1); five others are members of known phosphoprotein families, but their phosphorylation status is presently undocumented. Of the seven source proteins with unknown functions, one (FLJ13725) has been shown to be phosphorylated (14). Of the 36 phosphorylation sites shown in Table 1, seven have been reported previously (14, 17–20). Presentation of the peptide, YLDpSGIHSGA (residues 30–39 of β-catenin) is particularly noteworthy. Phosphorylation of Ser33 is necessary for targeting β-catenin for ubiquitination and proteasome-mediated degradation (17, 21), and the proteasome is intimately involved in the creation of most class I MHC-associated epitopes. These observations suggest a link between this particular phosphorylation and the process of source protein degradation that leads to display of the phosphopeptide by HLA-A*0201. These results suggest that the observed class I MHC-associated phosphopeptides are derived from degradation of properly folded, biologically active proteins that are recognized and posttranslationally modified by their respective kinases.
Next, we evaluated whether a correlation exists between the phosphopeptides presented on each of the four cell lines and the annotated functions of the corresponding source proteins. Twenty phosphopeptides presented by HLA-A*0201 are detected on at least one cancer cell and on JY BLCL (Table 1). Fifteen of these phosphopeptides are derived from source proteins with known functions (Table 3). Seven phosphopeptides are involved in cell cycle regulation and represent all source proteins with this function that we identified. A likely explanation of this finding is that both JY BLCL and the cancer cells are actively proliferating, although JY BLCL is virally transformed and not derived from a tumor. Because cancer cells are more likely to be in cycle than untransformed cells, we believe that phosphopeptides in this category still have potential as immunotherapeutic targets.
Of the 36 phosphopeptides detected in association with HLA-A*0201, 11 are presented on one or more cancer cell lines, but not on JY BLCL (Table 1). Five of these peptides are of particular interest as immunotherapeutic targets because they are found on all three cancer cell lines. Source proteins for 10 of 11 phosphopeptides in this category have known functions (Table 3). Several of these proteins (IRS2, Tensin-3/TEM6, BCAR3, and β-catenin) are involved in cytoplasmic signaling. This group, as well as the transcriptional activator Jun-C/D, are known to either be up-regulated or more active in some cancer cells (22–29). Thus, the phosphopeptides displayed on cancer cells and not JY BLCL are preferentially derived from source proteins associated with cytoplasmic signaling pathways and cellular transformation.
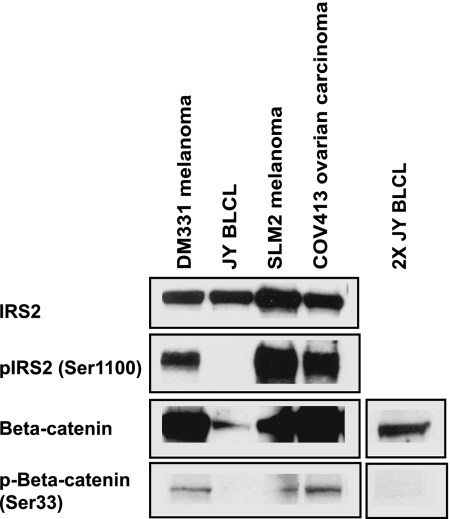
To understand the basis for differential display of phosphopeptides by HLA-A*0201 molecules on JY BLCL and the three cancer cell lines, Western blot analysis of cell lysates was used to determine the levels of the source proteins, IRS2 and β-catenin, and the corresponding phosphosites observed in the MHC-associated phosphopeptides. All four cell lines express IRS2 and β-catenin proteins (Fig. 1). However, we detected only IRS2 phosphorylated on Ser1100 and β-catenin phosphorylated on Ser33 in the three cancer cell lines (Fig. 1). Because the phosphopeptides RVApSPTSGV (residues 1097–1105 of IRS2) and YLDpSGIHSGA (residues 30–39 of β-catenin) are presented by HLA-A*0201 on the three cancer cell lines but not on JY BLCL (Table 1), we conclude that the differential presentation of these epitopes is due to differential phosphorylation of the corresponding source proteins. Interestingly, we have not been able to detect the nonphosphorylated form of the IRS2 epitope in the extracted peptides by MS (data not shown), indicating that it is either absent or present at levels at least 5-fold lower than that of the corresponding phosphopeptide. The nonphosphorylated IRS2 peptide does bind to purified HLA-A*0201 in vitro, but its affinity is lower by a factor of four relative to that of the phosphorylated form (Table 4, which is published as supporting information on the PNAS web site). Phosphorylation-augmented binding also was observed for TRAPPC1129–137 and CDC25b38–46 to a lesser extent, whereas phosphorylation diminished the binding of β-catenin30–39. However, all of the measured affinities are within the range observed for other naturally processed peptides known to be displayed. Thus, phosphorylation of Ser1100 in pIRS21097–1105 might promote display of this epitope by enhancing its binding to HLA-A*0201. Alternatively, phosphorylation may augment degradation and processing of the parent protein or prevent enzymatic cleavage of the peptide in the proteasome.
Fig. 1.
Differential phosphorylation of source proteins in cancer cells results in differential presentation of phosphorylated peptides by class I MHC. Shown are immunoblotting data for the expression of total IRS2 protein, phosphorylated IRS2 on Ser1100, total β-catenin, and phosphorylated β-catenin on Ser33. All blots were performed on 60 μg of protein, except in the case of the 2X JY BLCL, for which 120 μg of protein was probed. These results are representative of three independent experiments.
To demonstrate the potential of using class I MHC-restricted phosphopeptides as potential therapeutics against cancer, we generated CD8+ T lymphocytes that recognize the phosphopeptides presented by HLA-A*0201 from the source proteins, IRS2, β-catenin, and CDC25b. Mice expressing a transgenic recombinant HLA-A*0201 molecule (AAD) were immunized with activated bone-marrow-derived dendritic cells pulsed with either phosphorylated IRS2 (pIRS21097–1105), phosphorylated β-catenin (pβ-catenin30–39), or phosphorylated CDC25b (pCDC25b38–46) peptides. Three weeks later, CD8+ T lymphocytes were isolated and cultured in vitro in the presence of irradiated splenocytes pulsed with the same phosphopeptides (11). These CD8+ T lymphocytes secrete IFN-γ when they specifically recognize target cells that have been pulsed with synthetic peptides corresponding to the phosphorylated forms of these epitopes but not the nonphosphorylated homologues (Fig. 2A). We conclude that MHC phosphopeptides can be used to elicit CD8+ T lymphocytes that recognize only the phosphorylated form of the peptides.
Fig. 2.
pIRS21097–1105-, pβ-catenin30–39-, and pCDC25b38–46-specific CD8+ T lymphocytes recognize endogenously processed and presented phosphopeptides on the surface of cancer cell lines. (A) Recognition of C1RAAD stimulators pulsed with the indicated concentrations of synthetic phosphopeptide or its nonphosphorylated homolog by either pIRS21097–1105- or pCDC25b38–46-specific CD8+ T lymphocytes (Left) or pβ-catenin30–39-specific CD8+ T lymphocytes (Right). (B) Recognition of endogenously processed phosphopeptides on the indicated cell lines by pIRS21097–1105-, pβ-catenin30–39-, and pCDC25b38–46-specific CD8+ T cells.
Next, we evaluated whether these specific CD8+ T lymphocytes would recognize phosphopeptides generated from intracellular source proteins by the endogenous class I MHC antigen-processing pathway. CD8+ T lymphocytes specific for pIRS21097–1105 or pβ-catenin30–39 recognize all three cancer cell lines but not JY BLCL (Fig. 2B), consistent with their representation as determined by MS (Table 1). CD8+ T lymphocytes specific for pCDC25b38–46 recognize the three cancer cells and JY BLCL but with different efficiencies (Fig. 2B). Levels of IFN-γ secreted by CD8+ T lymphocytes that recognize the COV413 ovarian carcinoma and DM331 melanoma are approximately 10-fold higher then that released upon recognition of SLM2 melanoma and JY BLCL. These results are consistent with the levels of epitope detected in association with the HLA-A*0201 molecule by MS (Table 1).
To definitively show that phosphopeptide display and T cell recognition were due to expression and phosphorylation of the expected source protein, we transiently transfected the murine colorectal adenocarcinoma MC38AAD with a plasmid encoding human IRS2 (phIRS2-GFP) and evaluated its recognition by pIRS21097–1105-specific CD8+ T lymphocytes. Although we detected a low level of murine IRS2 in untransfected or mock-transfected cells by Western blot, MC38AAD cells transfected with the hIRS2-GFP plasmid showed a high level of human IRS2 that was also phosphorylated on Ser1100 (data not shown). pIRS21097–1105-specific CD8+ T lymphocytes only produced IFN-γ in response to MC38AAD cells that were transiently transfected with phIRS2-GFP or were exogenously pulsed with the RVApSPTSGV peptide and did not respond to MC38AAD cells that were untransfected or transfected with a GFP construct (Table 2). We conclude that recognition of MHC phosphopeptides endogenously expressed on cancer cells by specific CD8+ T lymphocytes is a direct result of both the level of expression and phosphorylation of the underlying source protein. Collectively, these results provide important validation of these peptides as potential targets for cancer immunotherapy.
Table 2.
CD8+ T cell recognition of endogenously processed and presented pIRS21097–1105 after transfection with the human IRS2 gene
| Cell line | Murine IFN-γ, pg/ml |
|---|---|
| SLM2AAD | 365.5 ± 103 |
| JY BLCL | 0 ± 0 |
| MC38AAD | |
| Mock (no DNA) | 0 ± 0 |
| pMAXGFP (2 μg) | 0 ± 0 |
| phIRS2-GFP (2 μg) | 14.1 ± 3.8 |
| phIRS2-GFP (4 μg) | 70.9 ± 5.7 |
| Mock + RVApSPTSGV peptide | 343.2 ± 51.2 |
Production of IFN-γ by pIRS21097–1105-specific CD8+ T cells was determined 24 h after incubation with the indicated stimulators by ELISA. Data are presented as means ± SD.
Methodologies used to discover class I MHC-restricted antigens displayed selectively on cancer cells frequently involve identification of the peptide target recognized by an already characterized T lymphocyte (30). Another approach relies on the ability to identify candidate source proteins, usually based on selective expression or overexpression and on the use of predictive algorithms to identify appropriate peptide sequences. Here, we employ a combination of isotopic labeling, enrichment of phosphopeptides by immobilized metal-affinity chromatography, and differential display MS to compare and sequence class I phosphopeptides presented by multiple cancer cell lines. The focus on the phosphate modification enables “sifting” of the complex mixture of MHC-associated peptides by MS to identify a small number of peptides with high relevance to cellular growth control processes. This modification provides the opportunity to rapidly assess the expression of the epitope and not just the source protein in multiple tumors of the same or different histological types with CD8+ T lymphocytes and phosphopeptide-specific antibodies. Our results using this approach establish a category of shared cancer antigens that are directly related to cellular growth control processes. These shared antigens are likely targets for immunotherapy.
Materials and Methods
Cell Lines and Transfectants.
All cell lines were grown at 37°C with 5% CO2 in growth medium consisting of RPMI medium 1640 supplemented with 10% FCS, 15 mM Hepes, and 2 mM l-glutamine. MC38 murine colorectal adenocarcinoma, COV413 (HLA-A2+ and HLA-B7+) ovarian carcinoma (31), and DM331 (HLA-A2+) (32) and SLM2 (HLA-A2+) melanomas were transfected with a chimeric class I MHC molecule (α1 and α2 domains of HLA-A*0201 plus an α3 domain of H-2Dd; AAD) and cultured under selection in growth medium supplemented with 600 μg/ml G418 (GIBCO/BRL, Carlsbad, CA). An AAD transfectant of the BLCL, C1R, was maintained in growth medium supplemented with 300 μg/ml G418. MC38AAD cells were transiently transfected with 2 or 4 μg of phIRS2-GFP (a generous gift from Lothar Vassen, Universitatsklinikum Essen, Essen, Germany) or 2 μg pMAXGFP plasmid using Nucleofector kit V (Amaxa Biosystems, Gaithersburg, MD) according to the manufacturer's directions. Cells were used to stimulate CD8+ T lymphocytes 48 h after transfection, as described below.
Antibodies.
An antibody specific for pSer1100 in IRS2 was generated by AnaSpec (San Jose, CA) by immunizing rabbits with a phosphopeptide conjugate of keyhole limpet hemocyanin corresponding to residues 1095–1108 of IRS2 (ARVApSPTSGVKRL). Other antibodies were obtained as follows: β-catenin phosphorylated on Ser33 (catalog no. sc-16743-R; Santa Cruz Biotechnology, Santa Cruz, CA), IRS2 protein (catalog no. 06-506; Upstate Biotechnology, Charlottesville, VA), and β-catenin (catalog no. sc-7199, Santa Cruz Biotechnology).
Isolation of HLA-Associated Peptides.
Class I MHC molecules were immunoaffinity-purified from the four cell lines, and their associated peptides were extracted as previously described (11). Cells (2 × 109 to 5 × 109) were lysed in a solution containing 20 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1% CHAPS, 1 mM PMSF, 5 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A. All protease inhibitors were purchased from Roche Applied Science (Indianapolis, IN). Phosphatase inhibitors (cocktails I and II from Sigma–Aldrich, St. Louis, MO) were added at a dilution of 1:100 to prevent dephosphorylation of peptides during extraction. The mixture was subjected to centrifugation, and the resulting supernatant was passed over an Econocolumn (Bio-Rad, Hercules, CA) containing the HLA-A2-specific Ab, BB7.2, bound to recombinant-protein A fast-flow Sepharose beads (Amersham Pharmacia Biotech, Piscataway, NJ) (33). Peptides were eluted from the purified class I MHC molecules with 0.2 M acetic acid. The concentration of acetic acid in the resulting solution was adjusted to 10%, and peptides were separated from β-microglobulin and MHC molecules by filtration through a 5-kDa cutoff (UFC4LCC25) filter (Millipore, Billerica, MA). Extracted peptides were stored at −80°C.
Liquid Chromatography/MS (LC/MS).
HLA-A*0201 peptides extracted from 1 × 108 cells were analyzed by nanoflow-HPLC/microelectrospray ionization coupled directly to a home-built Fourier-transform ion cyclotron resonance mass spectrometer (34). Briefly, samples were loaded onto a 7-cm, reversed-phase C18 (HPLC) column (360-μm o.d. and 50-μm i.d.; Polymicro Technologies, Phoenix, AZ), packed with 5-μm, C18 particles (YMC, Milford, MA), and gradient [gradient A: 0.1 M acetic acid (Sigma–Aldrich) in H2O; gradient B: 70% acetonitrile (Mallinckrodt, Phillipsburg, NJ) and 0.1 M acetic acid in H2O], eluted (0–60% gradient B in 20 min and 60–100% gradient B in 5 min) through a 2-μm diameter, laser-pulled electrospray tip (micropipette puller model P-2000; Sutter Instrument, Novato, CA) directly into the mass spectrometer with an 1100 series binary LC pump (Agilent, Palo Alto, CA) at a flow rate of ≈60 nl/min. Full-scan mass spectra (300 ≤ m/z ≤ 2,000) were collected at one scan per second, with a resolution of 5,000–10,000.
Phosphopeptide Enrichment.
Immunoaffinity-purified peptides were converted to methyl esters as previously described (35), except that the reaction was performed twice using anhydrous methanol and acetyl chloride (Alltech Biotechnology, Deerfield, IL). Samples were taken to dryness, reconstituted in 30 μl of 1:1:1 acetonitrile/methanol/aqueous acetic acid (0.01%), loaded onto an activated Fe3+-immobilized metal-affinity chromatography column (360-μm o.d. and 150-μm i.d.) packed with 7 cm of POROS 20 MC packing material (PerSeptive Biosystems, Framingham, MA) and eluted with 10 μl of 50 mM ascorbic acid (Sigma–Aldrich) onto a C18 microcapillary precolumn (360-μm o.d. and 75-μm i.d.) packed with 5 cm of C18 irregular reversed-phase packing material (5- to 20-μm diameter) (YMC) (36).
LC/MS/MS of Phosphopeptides.
A precolumn loaded with phosphopeptides was connected with polytetrafluoroethylene tubing [0.06-in o.d. and 0.012-in i.d. (1 in = 2.54 cm); Zeus Industrial Products, Orangeburg, SC] to the end of an analytical column (360-μm o.d. and 50-μm i.d.) packed with 7 cm of C18 RP packing material (5-μm diameter). Phosphopeptides were eluted with a 1-h gradient (0–60% gradient B in 60 min and 60–100% gradient B in 3 min) as described above, except that elution was to a hybrid, linear-quadrupole ion-trap/Fourier-transform mass-spectrometer (13) (Thermo Electron Corporation, Waltham, MA). The instrument was operated in a data-dependent mode and cycled through acquisition of a full-scan mass spectrum with the Fourier-transform mass spectrometer plus 10 MS/MS spectra recorded in the linear trap on the top 10 most abundant ions observed in the full-scan spectrum. Other experimental parameters included (i) one microscan per spectrum, (ii) ±1.5 Da precursor m/z, (iii) 35% collision energy, (iv) 30-ms ion activation, and (v) 40-s dynamic exclusion.
Comparative Analysis of MHC Phosphopeptides.
HLA-A*0201 peptide samples from each of the four different cell lines were divided in half and converted to methyl esters with either d0- or d4-methanol, as described above (12). Samples with different isotopic labels from any two of the cell lines were combined, enriched for phosphopeptides by immobilized metal-affinity chromatography, and analyzed by LC/MS as described above. In these experiments, signals for class I MHC phosphopeptides presented on the surface of both cell lines appear in the mass spectrum as doublets separated by 3 m/z units per carboxylate group in the molecule. Phosphopeptides that are unique to one or the other cell line appear as singlets. Data analysis was performed by using a three-dimensional visual representation of the chromatogram available in the Xcalibur software (Thermo Electron Corporation). Peptide sequences were determined by a combination of manual interpretation of MS/MS spectra, accurate mass measurements, and MASCOT Sequence Query (www.matrixscience.com/home.html). Sequences were confirmed by recording MS/MS spectra on the corresponding synthetic peptides. Synthetic peptides were prepared by standard Fmoc chemistry using a peptide synthesizer (model APEX 396; Advanced Automated Peptide Protein Technologies, Louisville, KY). Protein sources for confirmed peptide sequences were obtained by searching the nr and refseq databases for human proteins (www.ncbi.nlm.nih.gov/BLAST).
Western Blot Analysis.
Cell pellets were solubilized in lysis buffer (2 × 107 cells per milliliter) containing 50 mM Tris·HCl, pH 7.4, 1% IGEPAL (Sigma–Aldrich), 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1:100 dilutions of phosphatase inhibitor cocktails I and II. Lysates were centrifuged at 10,000 × g for 30 min at 4°C, and the total amount of protein was determined by using a DC protein assay (Bio-Rad). Samples were boiled in sample buffer for 5 min, and a total of 60 μg of protein was loaded and separated on a 4–20% gradient gel (ISC BioExpress, Kaysville, UT) by SDS/PAGE. Proteins were transferred to Nitropure nitrocellulose (Osmonics, Minnetonka, MN) and blocked with TBS containing 3% nonfat dry milk. Blots were probed overnight with phosphospecific antibodies, washed with TBS containing 0.1% Tween 20, probed with Goat anti-rabbit IgG horseradish peroxidase (Upstate Biotechnology) and developed by using enhanced chemiluminescence (ECL; Amersham Pharmacia). Blots were treated with Restore Western blot stripping buffer (Pierce) for 45 min at 37°C, checked for remaining primary antibody via incubation with goat anti-rabbit IgG horseradish peroxidase, and, if clear, reprobed with antibody to parent protein overnight at 4°C. Blots were then washed and probed with goat anti-rabbit IgG horseradish peroxidase and developed with ECL.
Generation of Phosphopeptide-Specific CD8+ T Lymphocytes and Analysis of Cytokine Production After Stimulation with Cancer Cell Lines.
CD8+ T lymphocytes specific for the indicated phosphopeptides were generated as described previously (11). Briefly, AAD-transgenic mice were immunized with phosphopeptide-pulsed, activated bone-marrow-derived dendritic cells (11). Separate bulk splenic T cell cultures were established in vitro with irradiated splenocytes that had been pulsed with the respective phosphorylated peptide at 1 μM. Cultures were restimulated with irradiated, phosphopeptide-pulsed splenocytes every 7–10 d in the presence of 10 Cetus units/ml IL-2 (Chiron, Emeryville, CA). To detect phosphopeptide-specific T cell reactivity, 50,000 CD8+ T lymphocytes were incubated with either 20,000 cancer cells or 20,000 C1RAAD antigen-presenting cells that had been pulsed with different doses of either phosphorylated or nonphosphorylated peptides for 1 h and then washed to remove free peptide. Supernatants were harvested after 24 h and analyzed by ELISA (eBioscience, San Diego, CA) according to the manufacturer's directions for murine IFN-γ produced by the activated CD8+ T lymphocytes.
Supplementary Material
Acknowledgments
This work was supported by U.S. Public Health Service Grants AI20963 (to V.H.E.) and AI33993 (to D.F.H.). A.L.Z. was supported by a fellowship from the Cancer Research Institute.
Abbreviations
- LC
liquid chromatography
- BLCL
B lymphoblastoid cell line.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 14649.
References
- 1.Rock KL, Goldberg AL. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW. Trends Cell Biol. 2002;12:104–107. doi: 10.1016/s0962-8924(01)02238-3. [DOI] [PubMed] [Google Scholar]
- 3.Reed SI. Nat Rev Mol Cell Biol. 2003;4:855–864. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- 4.Koepp DM, Harper JW, Elledge SJ. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 5.Ang XL, Wade Harper J. Oncogene. 2005;24:2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson KD. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 8.Evan GI, Vousden KH. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 9.Blume-Jensen P, Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 10.Easty DJ, Bennett DC. Melanoma Res. 2000;10:401–411. doi: 10.1097/00008390-200010000-00001. [DOI] [PubMed] [Google Scholar]
- 11.Zarling AL, Ficarro SB, White FM, Shabanowitz J, Hunt DF, Engelhard VH. J Exp Med. 2000;192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ficarro S, Chertihin O, Westbrook VA, White F, Jayes F, Kalab P, Marto JA, Shabanowitz J, Herr JC, Hunt DF, Visconti PE. J Biol Chem. 2003;278:11579–89. doi: 10.1074/jbc.M202325200. [DOI] [PubMed] [Google Scholar]
- 13.Syka JE, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, et al. J Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 14.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc Natl Acad Sci USA. 2004;101:12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelhard VH. Curr Opin Immunol. 1994;6:13–23. doi: 10.1016/0952-7915(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 16.Yewdell J. Mol Immunol. 2002;39:139–146. doi: 10.1016/s0161-5890(02)00097-4. [DOI] [PubMed] [Google Scholar]
- 17.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 18.Landry J, Lambert H, Zhou M, Lavoie JN, Hickey E, Weber LA, Anderson CW. J Biol Chem. 1992;267:794–803. [PubMed] [Google Scholar]
- 19.Huang CK, Zhan L, Ai Y, Jongstra J. J Biol Chem. 1997;272:17–19. doi: 10.1074/jbc.272.1.17. [DOI] [PubMed] [Google Scholar]
- 20.Sevilla A, Santos CR, Barcia R, Vega FM, Lazo PA. Oncogene. 2004;23:8950–8958. doi: 10.1038/sj.onc.1208015. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 22.Kornmann M, Maruyama H, Bergmann U, Tangvoranuntakul P, Beger HG, White MF, Korc M. Cancer Res. 1998;58:4250–4254. [PubMed] [Google Scholar]
- 23.Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cancer Res. 2001;61:6649–6655. [PubMed] [Google Scholar]
- 24.Bamberger AM, Milde-Langosch K, Rossing E, Goemann C, Loning T. J Cancer Res Clin Oncol. 2001;127:545–550. doi: 10.1007/s004320100255. [DOI] [PubMed] [Google Scholar]
- 25.Nagle JA, Ma Z, Byrne MA, White MF, Shaw LM. Mol Cell Biol. 2004;24:9726–9735. doi: 10.1128/MCB.24.22.9726-9735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai D, Iyer A, Felekkis KN, Near RI, Luo Z, Chernoff J, Albanese C, Pestell RG, Lerner A. Cancer Res. 2003;63:6802–6808. [PubMed] [Google Scholar]
- 27.Karim R, Tse G, Putti T, Scolyer R, Lee S. Pathology. 2004;36:120–128. doi: 10.1080/00313020410001671957. [DOI] [PubMed] [Google Scholar]
- 28.Boissan M, Beurel E, Wendum D, Rey C, Lecluse Y, Housset C, Lacombe ML, Desbois-Mouthon C. Am J Pathol. 2005;167:869–877. doi: 10.1016/S0002-9440(10)62058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nateri AS, Spencer-Dene B, Behrens A. Nature. 2005;437:281–285. doi: 10.1038/nature03914. [DOI] [PubMed] [Google Scholar]
- 30.Admon A, Barnea E, Ziv T. Mol Cell Proteomics. 2003;2:388–398. doi: 10.1074/mcp.R300004-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Los G, Verdegaal E, Noteborn HP, Ruevekamp M, de Graeff A, Meesters EW, ten Bokkel Huininkqq D, McVie JG. Biochem Pharmacol. 1991;42:357–363. doi: 10.1016/0006-2952(91)90723-i. [DOI] [PubMed] [Google Scholar]
- 32.Slingluff CL, Jr, Colella TA, Thompson L, Graham DD, Skipper JC, Caldwell J, Brinckerhoff L, Kittlesen DJ, Deacon DH, Oei C, et al. Cancer Immunol Immunother. 2000;48:661–672. doi: 10.1007/s002620050015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parham P, Brodsky FM. Hum Immunol. 1981;3:277–299. doi: 10.1016/0198-8859(81)90065-3. [DOI] [PubMed] [Google Scholar]
- 34.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Anal Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 35.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 36.Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. J Biol Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.