Abstract
Several pathogenic strains of Escherichia coli exploit type III secretion to inject “effector proteins” into human cells, which then subvert eukaryotic cell biology to the bacterium's advantage. We have exploited bioinformatics and experimental approaches to establish that the effector repertoire in the Sakai strain of enterohemorrhagic E. coli (EHEC) O157:H7 is much larger than previously thought. Homology searches led to the identification of >60 putative effector genes. Thirteen of these were judged to be likely pseudogenes, whereas 49 were judged to be potentially functional. In total, 39 proteins were confirmed experimentally as effectors: 31 through proteomics and 28 through translocation assays. At the protein level, the EHEC effector sequences fall into >20 families. The largest family, the NleG family, contains 14 members in the Sakai strain alone. EHEC also harbors functional homologs of effectors from plant pathogens (HopPtoH, HopW, AvrA) and from Shigella (OspD, OspE, OspG), and two additional members of the Map/IpgB family. Genes encoding proven or predicted effectors occur in >20 exchangeable effector loci scattered throughout the chromosome. Crucially, the majority of functional effector genes are encoded by nine exchangeable effector loci that lie within lambdoid prophages. Thus, type III secretion in E. coli is linked to a vast phage “metagenome,” acting as a crucible for the evolution of pathogenicity.
Keywords: bacterial pathogenesis, bacterial protein secretion, bioinformatics, genomics, virulence
Studies on Escherichia coli, and particularly on its relations with bacteriophages, laid the foundations of molecular biology. However, E. coli is more than just a model organism. As a widely distributed commensal, it colonizes organisms as diverse as humans, birds, and reptiles (1, 2). It is also a fearsome pathogen, causing a range of infections in humans and other animals (3). This ecological versatility is matched by a remarkable genomic diversity: as much as a quarter of the genome can vary from strain to strain. Bacteriophages account for one major source of diversity (4). Another is the acquisition of clusters of virulence genes en bloc by horizontal gene transfer, as so-called “pathogenicity islands” (5). Probably the most important pathogenicity island in E. coli is the locus for enterocyte effacement (or LEE). This locus encodes the Esc-Esp type III secretion system, which is crucial to the virulence of the human pathogens, enterohaemorrhagic and enteropathogenic E. coli (EHEC and EPEC). In particular, the LEE-encoded system confers on these bacteria the ability to elicit a characteristic response when applied to enterocytes: the attaching and effacing lesion (6).
Type III secretion systems (T3SSs) are complex multiprotein assemblages common to many plant and animal pathogens or symbionts that allow bacteria to subvert eukaryotic cell biology by injecting bacterial “effector proteins” into the host cell cytoplasm. Given its status as a pathogenicity island, it is tempting to assume that the LEE represents a self-contained unit, containing not only the genes for the secretion and translocation apparatus, but also for all of the effectors that might be secreted through the system. This assumption has been reinforced by the observation that the cloned LEE from EPEC is able to confer the attaching and effacing phenotype on E. coli K-12 and by the discovery and characterization of seven translocated effectors that are encoded within the LEE (7, 8).
Unlike proteins that are secreted through the Sec pathway, there is no signal peptide or any other known feature of T3SS effectors that can be used to identify them by sequence analysis alone. However, the effector repertoires of even distantly related type III-secreting bacteria often contain homologous proteins, suggesting that homology searches are likely to prove fruitful in finding new effectors (9, 10). In addition, effector genes, as recent arrivals in a bacterial genome, may exhibit a base composition distinct from the average for their host genome.
Recently, a handful of potential or proven non-LEE-encoded effectors have been identified, chiefly in the mouse pathogen, Citrobacter rodentium, which also harbors a LEE-encoded T3SS (8, 11). However, drawing on comparisons with other type III secreting organisms, such as the plant pathogen Pseudomonas syringae (12, 13), we speculated that the repertoire of E. coli effector genes might be much larger than currently recognized and that new effectors might be identified through a systematic genome-wide survey, which had yet to be carried out for any attaching and effacing strain. Because we were keen to examine a genome-sequenced strain with clear pathogenic potential in humans, we chose to investigate the effector repertoire of the RIMD 0509952 strain of EHEC (also known as the “Sakai strain”), which was responsible for >9,000 cases and 12 deaths in an outbreak in the Japanese city of Sakai (14, 15).
Results and Discussion
Prediction of Potential EHEC O157:H7 Effectors.
Initially, we exploited a bioinformatics approach to identify potential effectors encoded in the completed genome sequence of the Sakai strain (15). We performed homology searches with >300 effectors from type III-secreting organisms, including plant and animal pathogens and symbionts (Table 2 and Data Set 1, which are published as supporting information on the PNAS web site). This approach led to the identification of >60 putative effector genes in EHEC (Table 3, which is published as supporting information on the PNAS web site). Comparisons with the original protein-coding query sequences, and with other E. coli genomes suggest that 13 of these are probably pseudogenes, where a longer ancestral coding sequence has been disrupted by truncations and/or frameshifts that are likely to render any protein product nonfunctional (Table 4, which is published as supporting information on the PNAS web site). Thus, we conclude that there are 49 candidate effector genes in the Sakai genome that are potentially fully functional.
Experimental Confirmation of Predictions.
Next, we sought to confirm our predictions experimentally, using a variety of approaches (Fig. 1). Initially, we used a proteomics approach to identify type III secreted proteins. In particular, we exploited an EHEC ΔsepL mutant, which secretes effectors into the culture supernatant at far higher levels than the wild type (11). In addition, high-level expression of pchA from a multicopy plasmid, which encodes a transcriptional activator of the LEE, was used to enhance the expression of the Esc-Esp T3SS (16). By comparing the protein secretion profile of the ΔsepL strain with an isogenic non-type-III-secreting ΔsepL ΔescR mutant, we corroborated the status of 31 proteins from the bioinformatics survey as type-III-secreted effectors (Fig. 1). The presence of the Sakai orthologs of nine proteins already known to be translocated into eukaryotic cells (Map, EspB, EspF, EspG, EspH, EspZ TccP/EspFu, EspI/NleA, NleB, and NleE; refs. 17–25)) among our secreted effectors confirms the validity of our approach.
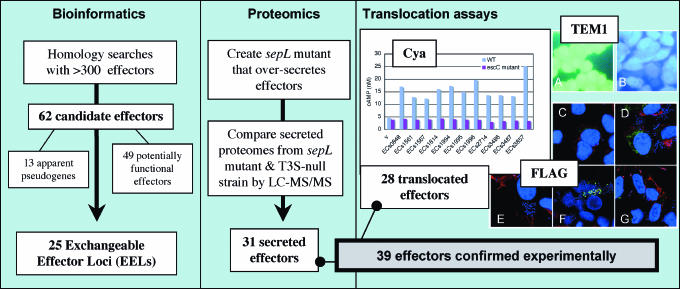
Fig. 1.
Experimental flow chart. BLAST searches with a comprehensive database of known and predicted effector sequences were used to identify 62 candidate effectors in 25 loci within the genome of EHEC O157:H7 Sakai. Using a proteomic approach, 31 candidate effectors were found to be secreted by T3S. Subsequently, three methods for measuring T3S-dependent translocation into eukaryote cells were used to identify 28 translocated effectors. In total, 39 candidate effectors were confirmed experimentally either by proteomics or by translocation assays or both methods. Representative translocation data (including controls) are provided for Cya, TEM1 and FLAG translocation assays. Cya: cAMP levels of Caco-2 cell extracts after infection with wildtype (WT) or escC− (T3S negative) E. coli carrying CyaA-effector fusion plasmids (v = vector only); TEM1: HeLa cell fluorescence was observed after infection with WT or escN− (T3S negative) E. coli carrying TEM1-effector fusion plasmids. Blue or green fluorescence indicates translocation or no translocation, respectively. (A) ECs1567 (escN− mutant). (B) ECs1567 (WT); FLAG: Caco-2 cell fluorescence was observed after infection with WT E. coli carrying FLAG-effector fusion plasmids. FLAG-effector fusion proteins, nucleus and F-actin were fluorescently stained green, blue, and red, respectively. Green fluorescence within Caco-2 cells indicates translocation. (C) ECs1567. (D) ECs1814. (E) Vector only. (F) ECs1994.(G) ECs3485. Further details of the experimental approach are described in Results and Discussion.
Subsequently, we confirmed the effector status of 28 of our candidates by showing T3SS-dependent translocation of effector fusion proteins into eukaryotic cells from adherent bacteria (Table 1). In the first rounds of translocation assays, we used two reporter systems to investigate proteins detected in the proteomics screen: translocation of CyaA-fusion proteins detected by an increase of intracellular cAMP concentration and translocation of FLAG-tagged protein detected by immunofluorescent staining. In later experiments, using a TEM1 β-lactamase fusion assay, we detected translocation of several additional candidate effectors, including some that had not been detected in the secretome of the sepL mutant. Taken together, the proteomics approach and the translocation assays confirmed the effector status of 39 EHEC proteins.
Table 1.
Summary of E. coli O157:H7 effectors
| Effector* | Sakai ID | Family† | Context‡ | Evidence§ |
|---|---|---|---|---|
| EspX1 | ECs0025 | PPR | O-I 1 | H |
| EspY1 | ECs0061 | SopD-N | C-I | B |
| EspY2 | ECs0073 | SopD-N | O-I 3 | H |
| EspY3 | ECs0472 | SopD-N; PRR | C-I | H |
| NleB2-1 | ECs0846 | NleB | Sp3 | H |
| NleC | ECs0847 | NleC | Sp3 | B |
| NleH1-1 | ECs0848 | NleH | Sp3 | SC |
| NleD | ECs0850 | NleD | Sp3 | B |
| EspX2 | ECs0876 | PPR | O-I 37 | B |
| EspF2-1′ | ECs1126 | EspF | Sp4 | H ψ |
| EspV′ | ECs1127 | AvrA | Sp4 | Neg ψ |
| EspX7 | ECs1560¶ | PPR; LRR | Sp6 | S |
| EspN | ECs1561¶ | CNF | Sp6 | S |
| NleB2-2′ | ECs1566 | NleB | Sp6 | H ψ |
| EspO1-1 | ECs1567 | OspE | Sp6 | SCFB |
| EspK | ECs1568 | LRR | Sp6 | SFB |
| NleG2-1′ | ECs1810/1 | NleG | Sp9 | S ψ |
| NleA | ECs1812 | NleA | Sp9 | S |
| NleH1-2 | ECs1814 | NleH | Sp9 | SCFB |
| NleF | ECs1815 | NleF | Sp9 | SB |
| EspO1-2 | ECs1821 | OspE | Sp9 | H |
| NleG | ECs1824 | NleG | Sp9 | SF |
| EspM1 | ECs1825 | IpgB | Sp9 | SF |
| NleG9′ | ECs1828 | NleG | Sp9 | H ψ |
| NleG2-2 | ECs1994‖ | NleG | Sp10 | SCF |
| NleG6-1 | ECs1995‖ | NleG | Sp10 | SCF |
| NleG5-1 | ECs1996‖ | NleG | Sp10 | SCF |
| EspR1 | ECs2073 | LRR | O-I 62 | H |
| EspR2′ | ECs2074/5 | LRR | O-I 62 | H ψ |
| NleG5-2 | ECs2154‖ | NleG | Sp11 | S |
| NleG6-2 | ECs2155‖ | NleG | Sp11 | S |
| NleG2-3 | ECs2156‖ | NleG | Sp11 | S |
| NleG7′ | ECs2226** | NleG | Sp12 | B ψ |
| NleG3′ | ECs2227/8†† | NleG | Sp12 | H ψ |
| NleG2-4′ | ECs2229 | NleG | Sp12 | H ψ |
| EspL1 | ECs2427 | AR | C-I | H |
| EspR3 | ECs2672 | LRR | C-I | H |
| EspR4 | ECs2674 | LRR | C-I | H |
| EspJ | ECs2714 | EspJ | Sp14 | SC |
| TccP | ECs2715 | EspF | Sp14 | S |
| EspM2 | ECs3485 | IpgB | Sp17 | SF |
| NleG8-2 | ECs3486 | NleG | Sp17 | SCF |
| EspW | ECs3487 | HopW | Sp17 | SC |
| NleG6-3′ | ECs3488†† | NleG | Sp17 | B ψ |
| EspL2 | ECs3855 | AR | SpLE3 | SF |
| NleB1 | ECs3857 | NleB | SpLE3 | SCF |
| NleE | ECs3858 | NleE | SpLE3 | SF |
| EspF1 | ECs4550 | EspF | LEE | S |
| EspB | ECs4554 | EspB | LEE | S |
| Tir | ECs4561 | Tir | LEE | B |
| Map | ECs4562 | IpgB | LEE | S |
| EspH | ECs4564 | EspH | LEE | SB |
| EspZ | ECs4571 | EspZ | LEE | SFB |
| EspG | ECs4590 | EspG | LEE | SB |
| EspL3′ | ECs4642/3†† | AR | O-I 152 | H ψ |
| EspY4 | ECs4653 | SopD-N | O-I 153 | B |
| EspX3′ | ECs4654/5 | PPR | O-I 153 | H ψ |
| EspY5′ | ECs4657 | SopD-N | O-I 153 | H ψ |
| EspL4 | ECs4935 | AR | C-I | H |
| EspX4 | ECs5021 | PPR | C-I | Neg |
| EspX5 | ECs5048 | PPR | C-I | H |
| EspX6 | ECs5295 | PPR | O-I 174 | H |
*Named according to a scheme based on the P. syringae scalable nomenclature for Hrp-dependent effectors (see Materials and Methods). TccP is also known as EspFu or EspF2-2 (this study).
†LRR, leucine-rich repeats; AR, ankyrin repeats; PPR, pentapeptide repeats; SopD-N, SopD N-terminal domain.
‡Prophage nomenclature as in Hayashi et al. (15); O-I, O-island, nomenclature as in Perna et al. (56); C-I, “coli island,” refers to gene clusters present in some or all E. coli genomes but absent in related species such as S. enterica.
§Confirmatory evidence. S, detected in secretome of ΔsepL mutant; C, translocation detected by using CyaA fusion; F, translocation detected using FLAG tag; B, translocation detected by using β-lactamase fusion; H, homologous to known effector but not confirmed by translocation assay; Neg, negative using β-lactamase fusion; ψ, predicted pseudogene.
¶Frame-shift present in EHEC O157:H7 EDL933.
‖Near identical copies in genome (>95% identity): ECs1994/ECs2156; ECs1995/ECs2155; ECs1996/ECs2154.
**Miscalled start codon. CDS length should be 651 nt.
††Not annotated as pseudogene in EHEC O157:H7 Sakai (15).
At the protein level, the EHEC effector sequences fall into >20 families (Table 1). Newly identified effector families were named according to the commonly accepted “Esp” (E. coli secreted protein) nomenclature as EspK-R, omitting the names EspP (already used) and EspQ (easily confused with EspO). Many of the effector families can be further divided on tightly defined phylogenetic grounds into two or more subfamilies. We have distinguished these subfamilies through the use of a numerical suffix, following a scheme based on the P. syringae nomenclature for Hrp-dependent effectors (26) (Table 1 and below). Where there are multiple members in the Sakai strain of a phylogenetically defined subfamily, we have used an additional numerical suffix to distinguish them (e.g., NleG2-1, NleG2-2, etc.). We made two exceptions to the need to define families and subfamilies according to the rules proposed for P. syringae effectors: all effectors with a WEX5F motif were lumped together in the EspY family, whereas all SopA-like effectors were placed in the EspX family.
The NleG Family.
The largest effector family encoded by EHEC O157:H7 is the NleG family, with 14 members in the Sakai strain alone, a surprising discovery given that all that was previously known of this family was a short peptide sequence assigned to a type-III-secreted protein from C. rodentium (11). A TBLASTN search with this peptide sequence against the unfinished C. rodentium genome sequence (www.sanger.ac.uk/Projects/C_rodentium) allowed us to identify the full-length protein-coding sequence that encompassed the peptide. Using this full-length sequence, we found many new NleG homologs, including the 14 in the Sakai strain, 13 in EPEC E22, together with additional homologs in Salmonella bongori and C. rodentium. We have shown that the majority of these putative proteins from the Sakai strain are translocated by the LEE-encoded T3SS (Table 1). It is clear that a remarkable expansion of the nleG gene family has taken place in the EHEC O157:H7 and EPEC E22 lineages, with several gene duplications followed on occasion by disruptions (Fig. 3, which is published as supporting information on the PNAS web site).
Following the approach adopted by the P. syringae research community (26), we exploited a phylogenetic approach to produce a scalable nomenclature for these proteins (Fig. 4, which is published as supporting information on the PNAS web site). Considerable sequence divergence is apparent within the protein family, but there is no apparent similarity to any proteins of known function. However, the presence of conserved patches of sequence centred on three kinds of potential active-site residues (His, Cys, Asp) makes it tempting to speculate that this family might be characterized by a conserved but as yet cryptic enzymatic activity (27) (Fig. 3).
Homologs of Effectors from Plant Pathogens.
%Several EHEC candidate effectors show homology to T3SS effectors from plant pathogens, strengthening the idea that effectors generally target ancient and conserved aspects of eukaryotic cell biology (28). Others have already commented on the weak similarity between NleD, HopPtoH, and botulinum toxin, which suggests that these effectors might be zinc metalloproteases (29). EspW, which we have shown to be translocated, shares 29% identity with the C-terminal domain of HopPmaA/HopW (Table 3), a protein of unknown function, identified in a genome-wide functional screen for effectors in P. syringae (30). EspV encodes a short (60 aa) peptide that shows significant similarity to the AvrA effector from P. syringae (31) (but not to the unrelated but confusingly named AvrA from Salmonella) (Table 1). Although ECs1127 appears to be an espV pseudogene in the Sakai strain, longer potentially functional AvrA-like coding sequences are discernable in the genomes of EPEC strains E22 and E110019, C. rodentium, and EHEC O84:H4 bacteriophage BP-4795 (32).
Three Families of Chromosomally Encoded Effectors Allied to Shigella Osp Proteins.
The EHEC effector repertoire includes chromosomally encoded homologs of several putative or poorly characterized plasmid-encoded effectors from Shigella. There are four homologs of the OspD proteins from Shigella flexneri. OspD3 has been previously characterized as an enterotoxin (SenA) (33), but no function has yet been ascribed to OspD1 and OspD2. In the Sakai strain, the OspD-like proteins fall into two subfamilies on the basis of genomic context and functional characteristics. EspL2 is encoded by the phage-like element SpLE3, is secreted by the sepL mutant and is translocated through the Esc-Esp T3SS (Table 1). All four OspD-like proteins from the Sakai strain share a similar domain structure: an N-terminal ShET enterotoxin domain, with C-terminal ankyrin-like repeats (Fig. 5, which is published as supporting information on the PNAS web site). Orthologs of the non-phage-encoded OspD-like proteins are encoded in some E. coli/Shigella genomes that lack the LEE, including strain K-12, where the ortholog of EspL4 is misannotated as a regulator of acetyl CoA synthetase (this hypothetical function has been discredited: A. Wolfe, personal communication).
EspO1-1 and EspO1-2 are homologs of the OspE1 and OspE2 proteins. Both OspE proteins are relatively short (115 aa) and differ by only a single residue (34). OspE2 has recently been shown to be required for the maintenance of cell architecture of Shigella-infected cells (35). We found EspO1-1 to be translocated in all of our assays (Table 1). Genes corresponding to EspO1-1 and EspO1-2 are present in the genome of EHEC strain EDL933 but have not been recognized in the annotation. Furthermore, homology searches with EspO1-1 revealed additional unannotated members of this family encoded in the genomes of several serovars of Salmonella and in variant LEE clusters from EHEC O103:H2 strain RW1374 and from a rabbit EPEC strain (Fig. 6, which is published as supporting information on the PNAS web site) (36, 37). NleH1-1 and NleH1-2 are homologs of the OspG protein, a protein kinase that interferes with the NF-κB signaling (38). We have shown that both are translocated by T3S (Table 1). Both EHEC sequences contain the conserved residues of the three protein kinase catalytic motifs that are implicated in autophosphorylation of OspG (38) (Fig. 7, which is published as supporting information on the PNAS web site).
A Plethora of Other Effectors.
The genome of the Sakai strain encodes easily identifiable homologs of the “non-LEE encoded” effectors (NleA–F), recently identified in C. rodentium (11), and of the seven LEE-encoded effectors (Map, Tir, EspB, EspF, EspG, EspH, and EspZ) (for recent review, see ref. 8). Our results are consistent with previous studies on the secretion and/or translocation of these effectors, but also provide evidence that NleF is translocated (Table 1). Intriguingly, we found three NleB homologs encoded within prophages in the EHEC genome, at least once of which (NleB1) appears to be translocated. Similarly, we found two additional members of the Map/IpgB family (EspM1 and EspM2), both of which are translocated.
We found that EspN, which is type-III-secreted in a sepL mutant, showed sequence similarity to part of cytotoxic necrotizing factor 1 from E. coli (39). However, this similarity excludes the C-terminal catalytic domain in CNF, suggesting that EspN has a significantly different function. Five of the proteins from the Sakai strain (EspY1–5) possess a N-terminal WEX5F domain that has been linked to type-III secretion and is conserved in several well characterized Salmonella effectors (40, 41) and in putative effectors from Edwardsiella and Sodalis (Fig. 8, which is published as supporting information on the PNAS web site). Two of these proteins proved positive in translocation assays, although none was detected in the survey of the secreted proteome (Table 1). Likewise, of the seven SopA/PipB homologs (EspX1–7) identified in the genome, two (EspX2 and EspX7) were found to be secreted or translocated (Table 1).
Distribution of Effector Loci.
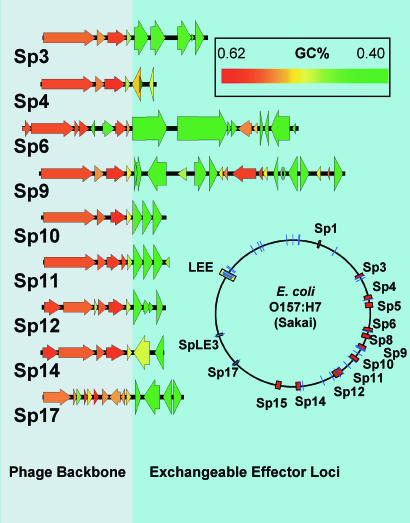
The genes encoding proven or predicted effectors in the Sakai strain occur in over twenty exchangeable effector loci (EELs) scattered throughout the chromosome (Fig. 2). The EELs fall into three groups: two pathogenicity islands (the LEE and SpLE3), nine EELs within lambdoid prophages (which encode the majority of functional effector genes) and 14 non-phage EELs. There are several distinctive features of the lambdoid prophage EELs: they are always located just downstream of the tail fiber genes, they always contain more than one effector gene (in the extreme case, the Sp9 EEL, there are eight effector genes encoded in a 13-kbp locus) and they stand out from their host phage backbone in possessing an extreme bias toward low GC content (Fig. 2).
Fig. 2.
The genome view shows lambdoid prophages in orange and effector loci in blue. The blow-ups of the terminal portions of lambdoid phages shows the clear distinction in GC content between effector genes and the phage backbone. Effector genes highlighted by increased height. For reasons of space, a cluster of insertion sequence remnants has been omitted from the end of Sp12. Sp5 and Sp15, the two Shiga toxin-encoding prophages, do not encode any T3SS effectors. In all cases, phage encoded effector genes fall within the prophage boundaries defined by Hayashi et al. (15).
The non-prophage EELs consist chiefly of lineage-specific insertions of one or a few genes. Interestingly, several of the non-prophage-encoded effector genes EELs also occur, often as pseudogenes, in the model strain E. coli K-12, even though it is considered nonpathogenic (Table 5). We speculate that these represent ancient phage remnants and/or encode substrates of a second T3SS, ETT2, which is now defunct in the Sakai strain. The ETT2 secretion locus is distributed in a broad range of E. coli lineages, including K-12, and is thought to have been acquired at a relatively early stage of E. coli diversification (42). However, we found that at least two non-phage-encoded candidates can be translocated through the LEE-encoded system (EspY1 and EspY4), confirming that they are (at least were) genuine effectors.
Conclusions
Our studies lead to a remarkable increase in the number of known translocated T3S effectors in E. coli and to some striking conclusions. First, through preliminary analyses of other genome-sequenced strains (Table 4), it is clear that E. coli strains show striking differences in the number, sequence diversity, and strain distribution of T3S effectors. Such differences in effector repertoire are likely to be reflected in differences in host range or other virulence phenotypes. Secondly, genes associated with T3S account for a significant component of the mobile gene pool in E. coli genomes: our calculations show that around a quarter of EHEC genes in the lowest fifth centile of G+C content are associated with T3S. Furthermore, it appears that the major function of lambdoid prophages in EHEC is to carry type III-secretion effectors, nine of 13 lambdoid prophages in the EHEC genome carry effector genes. When remnants of insertion sequences are excluded, putative or proven effector genes account for all but three of the 64 genes within the passenger compartments (or “morons”; ref. 43) of these nine prophages. In addition, three of the lambdoid prophages also encode pch genes, which regulate gene expression within the LEE (16). Thus, we conclude that type III secretion in E. coli is connected to a vast phage “metagenome,” which acts as crucible for the evolution of pathogenesis in this species.
Materials and Methods
Bioinformatics Search for New Effector Candidates.
Over 300 proven or predicted effectors were collated from recent T3S literature (Table 2), and the peptide sequences were used to search the E. coli O157:H7 Sakai genome and protein sequences using TBLASTN and BLASTP under default conditions (44). An E value <1e-05 was chosen as a cutoff value for significance. All newly identified effectors were then subjected to PSI-BLAST searches over the NCBI's NR peptide database to identify more distantly related E. coli Sakai homologs. Pseudogenes were identified on the grounds of partial matches to much longer homologous coding sequences, and where possible, evidence of frame shifts or truncations was gathered by comparing family members at the nucleotide level. The genomic context and G+C content of all candidate effector genes was examined by using the coliBASE resource (45). The distribution of effectors in other E. coli genomes was determined by using TBLASTN searches (again with an E value cutoff of <1e-05) and genome comparisons facilitated by the coliBASE genome comparison tool (45).
Proteomic Analysis of Proteins in Culture Supernatant.
The ΔsepL mutant (SKI1204) and the ΔsepL ΔescR mutant (SKI 1205) of EHEC O157:H7 Sakai (RIMD 0509952) were constructed by using method and plasmids of Datsenko and Wanner (46). To ensure maximal expression of the LEE, a positive regulator of LEE, PchA, was overexpressed in these strains. To create a pchA-overexpressing plasmid, pGEM-pchA, a DNA fragment corresponding to the pchA gene was synthesized by PCR from the chromosomal DNA of O157:H7 Sakai using specific primers (CACAGGAATATATCCGTACCC and AGTATGTGTCACTGGCCTATACGG) and cloned into pGEM-T (Promega). Proteins were harvested from the culture supernatant of EHEC strain SKI1204 (O157:H7 Sakai ΔsepL)/pGEM-pchA or SKI 1205 (O157:H7 Sakai ΔsepL ΔescR)/pGEMpchA. Bacteria were grown in Dulbecco's modified Eagle media (DMEM) to 1.2 OD600, and the supernatant was separated by centrifugation and filtration. Proteins in the supernatant were precipitated with 6% TCA and dissolved in SDS-sample buffer, then were separated by SDS/page. The gel was cut in 25 slices, and proteins in each gel slice were identified by using LC-MS/MS and the EHEC O157:H7 Sakai database as described (47). Proteins detected in the sample from SKI1204 but absent from the sample from SKI1205 were judged to be candidate effectors and investigated further.
Translocation Assays.
Three independent methods based on translational fusion plasmids were used to assay T3S-dependent translocation from E. coli into eukaryotic cells. Fusion plasmids for each gene were constructed from PCR products encompassing the full gene length or approximately the first 300 nt as determined by the E. coli O157:H7 Sakai gene predictions. In all cases, fusion plasmids without DNA inserts produced negative results. T3S-deficient mutants were also tested with each plasmid to ensure that any observed translocation depended on the T3SS. All effector candidates identified by the proteomics and bioinformatics screens were tested in one or more of the translocation assays, with occasional exceptions where there was published evidence confirming effector status (EspF, EspG, EspH, TccP, and NleA), there were difficulties in cloning (ECs1560 and ECs1561), additional bioinformatics analysis suggested that the candidate was a pseudogene, or other members of the same family had already proven positive in a translocation assay (e.g., only seven of the 14 NleG homologs were tested; only ECs1568 was tested from the LRR family, only ECs1567 was tested from the OspE family).
Cya Translocation Assay (48).
N-terminal translational fusions of CyaA to each gene were constructed by using pTB101-cyaA, which encodes the N-terminal part (1–412) of Bordetella pertusis CyaA toxin. The EPEC E. coli strain B171 and an isogenic T3S-negative mutant (ΔescC), each harboring each the cyaA-fusion plasmid, were grown in DMEM containing 0.2 mM IPTG to late exponential growth phase and used to infect Caco-2 cells. After incubating for 4 h, cells were washed and incubated in 2.5% perchloric acid for 10 min. Cell extract was obtained by centrifugation and cAMP concentration in the extract was measured by using the cAMP EIA Kit (Cayman Chemical, Ann Arbor, MI).
FLAG-Tagged Translocation Assay (49).
C-terminal FLAG fusions were constructed with pFLAG-CTC (Sigma, St. Louis, MO). The EHEC O157:H7 Sakai strain and an isogenic T3S-negative mutant (ΔescR) harboring FLAG-fusion plasmids were grown in DMEM containing IPTG for 2 h and used to infect Caco-2 cells. After washing off unattached bacteria, cells were further incubated for 2 h in fresh DMEM then washed and fixed with 4% paraformaldehyde. FLAG-tagged proteins were visualized with anti-FLAG antibody (Sigma) after attachment of Alexa Fuor 484-conjugated anti-mouse antibody (Molecular Probes, Eugene, OR). Nucleus and F-actin were also stained by DAPI and rhodamine-phalloidene, respectively.
β-Lactamase Translocation Assay (50).
N-terminal translational fusions with TEM-1 β-lactamase were constructed by using pCX340 (50) or its Gateway (Invitrogen, Carlsbad, CA) compatible derivative pCX340gw (this study). Enteropathogenic E. coli strain 2348/69 and an isogenic T3S-negative mutant (ΔescN) harboring the TEM-1 fusion plasmids were grown for 2.5 h in DMEM before being used to infect HeLa or Hep2 cells (1.5 h in DMEM with IPTG). After infection, cells were washed and loaded with the fluorescent substrate CCF2-AM. Cleavage of CCF2-AM by the TEM-1 β-lactamase was detected by blue fluorescence of eukaryotic cells after illumination at 409 nm, indicating translocation of the effector fusion protein. Conversely, green fluorescence due to the presence of uncleaved CCF2-AM was taken to indicate absence of translocated fusion protein within the eukaryotic cells.
Phylogenetic Analyses to Establish a Scalable Nomenclature.
For each effector family, homologous sequences were retrieved from the NCBI GenBank database, and where appropriate, the unfinished genomes of C. rodentium, E. coli, Shigella, and Salmonella species. Multiple alignments were prepared for each family (for further details, see Supporting Text, which is published as supporting information on the PNAS web site). Homologous families were divided into subfamilies by using the criteria defined for P. syringae Hop effectors by the P. syringae community (26), i.e., subfamily groups share <0.75 aa diversity and >0.75 between-group amino acid diversity (as measured by using the Jones–Taylor–Thornton substitution matrix). In exceptional cases, where the between-group diversity was close to the cutoff and more than one distinct clade with low intragroup diversity was apparent, the family was divided (e.g., NleB). Where more than one homolog from the same subfamily exists in the same genome, an additional distinguishing numerical suffix is added. Names are further extended with an apostrophe if the respective CDS are truncated by insertions/deletions.
Bioinformatics Analysis of G+C Content and Phage EELs.
The percentage G+C content was calculated for all genes in the Sakai genome, and then genes were sorted by rank order to calculate percentiles. For the purposes of calculation, “genes associated with T3S” were taken to include those in the LEE and genes for the non-LEE-encoded effectors described in Table 1. The lambdoid prophage passenger compartments or morons were defined as those portions of the prophage genomes lying between the last putative tail fiber gene (homologs of ECs0845) and the end of the phage (as defined by genomic comparisons; ref. 15).
Supplementary Material
Acknowledgments
Work in the M.J.P. laboratory was supported by Biotechnology and Biological Sciences Research Council Grants BBD0101951 and EGA16107. Work in the T.T. and T.H. laboratory was supported by Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a Grant-in-Aid from the Ministry of Health, Labor, and Welfare (H17-Sinkou-ippan-019). S.A.B. was supported by a Special Research Fellowship in Bioinformatics from the Medical Research Council (U.K.) to work in the M.J.P. laboratory and by a Howard Florey Centenary Fellowship from the National Health and Medical Research Council (Australia). R.Y. was supported was by Biotechnology and Biological Sciences Research Council (U.K.) Grant BB/D010195/1, S.A.M. was supported by a Medical Research Council studentship, and C.W.B. was supported by the Division of Immunity and Infection. Work in the G.F. laboratory was supported by the Medical Research Council and Wellcome Trust.
Abbreviations
- LEE
locus for enterocyte effacement
- EHEC
enterohaemorrhagic E. coli
- EPEC
enteropathogenic E. coli
- T3SS
type III secretion system.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Souza V, Rocha M, Valera A, Eguiarte LE. Appl Environ Microbiol. 1999;65:3373–3385. doi: 10.1128/aem.65.8.3373-3385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon DM, Cowling A. Microbiology. 2003;149:3575–3586. doi: 10.1099/mic.0.26486-0. [DOI] [PubMed] [Google Scholar]
- 3.Kaper JB. Int J Med Microbiol. 2005;295:355–356. doi: 10.1016/j.ijmm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi M, Kurokawa K, Hayashi T. Trends Microbiol. 2001;9:481–485. doi: 10.1016/s0966-842x(01)02173-4. [DOI] [PubMed] [Google Scholar]
- 5.Hacker J, Blum-Oehler G, Hochhut B, Dobrindt U. Acta Microbiol Immunol Hung. 2003;50:321–330. doi: 10.1556/AMicr.50.2003.4.1. [DOI] [PubMed] [Google Scholar]
- 6.Jerse AE, Yu J, Tall BD, Kaper JB. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDaniel TK, Kaper JB. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 8.Garmendia J, Frankel G, Crepin VF. Infect Immun. 2005;73:2573–2585. doi: 10.1128/IAI.73.5.2573-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pallen MJ, Beatson SA, Bailey CM. FEMS Microbiol Rev. 2005;29:201–229. doi: 10.1016/j.femsre.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Pallen MJ, Beatson SA, Bailey CM. BMC Microbiol. 2005;5:9. doi: 10.1186/1471-2180-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, et al. Proc Natl Acad Sci USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JH, Urbach JM, Law TF, Arnold LW, Hu A, Gombar S, Grant SR, Ausubel FM, Dangl JL. Proc Natl Acad Sci USA. 2005;102:2549–2554. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR. Trends Microbiol. 2002;10:462–469. doi: 10.1016/s0966-842x(02)02451-4. [DOI] [PubMed] [Google Scholar]
- 14.Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. Am J Epidemiol. 1999;150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, et al. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 16.Iyoda S, Watanabe H. Microbiology. 2004;150:2357–2571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- 17.Kenny B, Jepson M. Cell Microbiol. 2000;2:579–290. doi: 10.1046/j.1462-5822.2000.00082.x. [DOI] [PubMed] [Google Scholar]
- 18.Campellone KG, Robbins D, Leong JM. Dev Cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Garmendia J, Phillips AD, Carlier MF, Chong Y, Schuller S, Marches O, Dahan S, Oswald E, Shaw RK, Knutton S, Frankel G. Cell Microbiol. 2004;6:1167–1183. doi: 10.1111/j.1462-5822.2004.00459.x. [DOI] [PubMed] [Google Scholar]
- 20.Tu X, Nisan I, Yona C, Hanski E, Rosenshine I. Mol Microbiol. 2003;47:595–606. doi: 10.1046/j.1365-2958.2003.03329.x. [DOI] [PubMed] [Google Scholar]
- 21.Elliott SJ, Krejany EO, Mellies JL, Robins-Browne RM, Sasakawa C, Kaper JB. Infect Immun. 2001;69:4027–4033. doi: 10.1128/IAI.69.6.4027-4033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly M, Hart E, Mundy R, Marches O, Wiles S, Badea L, Luck S, Tauschek M, Frankel G, Robins-Browne RM, Hartland EL. Infect Immun. 2006;74:2328–2337. doi: 10.1128/IAI.74.4.2328-2337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanack KJ, Crawford JA, Tatsuno I, Karmali MA, Kaper JB. Infect Immun. 2005;73:4327–4337. doi: 10.1128/IAI.73.7.4327-4337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruenheid S, Sekirov I, Thomas NA, Deng W, O'Donnell P, Goode D, Li Y, Frey EA, Brown NF, Metalnikov P, et al. Mol Microbiol. 2004;51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 25.McNamara BP, Koutsouris A, O'Connell CB, Nougayrede JP, Donnenberg MS, Hecht G. J Clin Invest. 2001;107:621–669. doi: 10.1172/JCI11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindeberg M, Stavrinides J, Chang JH, Alfano JR, Collmer A, Dangl JL, Greenberg JT, Mansfield JW, Guttman DS. Mol Plant Microbe Interact. 2005;18:275–282. doi: 10.1094/MPMI-18-0275. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 28.Buttner D, Bonas U. Curr Opin Plant Biol. 2003;6:312–319. doi: 10.1016/s1369-5266(03)00064-5. [DOI] [PubMed] [Google Scholar]
- 29.Marches O, Wiles S, Dziva F, La Ragione RM, Schuller S, Best A, Phillips AD, Hartland EL, Woodward MJ, Stevens MP, Frankel G. Infect Immun. 2005;73:8411–8417. doi: 10.1128/IAI.73.12.8411-8417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 31.Napoli C, Staskawicz B. J Bacteriol. 1987;169:572–578. doi: 10.1128/jb.169.2.572-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creuzburg K, Recktenwald J, Kuhle V, Herold S, Hensel M, Schmidt H. J Bacteriol. 2005;187:8494–8498. doi: 10.1128/JB.187.24.8494-8498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nataro JP, Seriwatana J, Fasano A, Maneval DR, Guers LD, Noriega F, Dubovsky F, Levine MM, Morris JG., Jr Infect Immun. 1995;63:4721–4728. doi: 10.1128/iai.63.12.4721-4728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchrieser C, Glaser P, Rusniok C, Nedjari H, D'Hauteville H, Kunst F, Sansonetti P, Parsot C. Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 35.Miura M, Terajima J, Izumiya H, Mitobe J, Komano T, Watanabe H. Infect Immun. 2006;74:2587–2595. doi: 10.1128/IAI.74.5.2587-2595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tauschek M, Strugnell RA, Robins-Browne RM. Mol Microbiol. 2002;44:1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- 37.Jores J, Wagner S, Rumer L, Eichberg J, Laturnus C, Kirsch P, Schierack P, Tschape H, Wieler LH. Int J Med Microbiol. 2005;294:417–425. doi: 10.1016/j.ijmm.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. Proc Natl Acad Sci USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buetow L, Flatau G, Chiu K, Boquet P, Ghosh P. Nat Struct Biol. 2001;8:584–588. doi: 10.1038/89610. [DOI] [PubMed] [Google Scholar]
- 40.Miao EA, Miller SI. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brumell JH, Kujat-Choy S, Brown NF, Vallance BA, Knodler LA, Finlay BB. Traffic. 2003;4:36–48. doi: 10.1034/j.1600-0854.2003.40106.x. [DOI] [PubMed] [Google Scholar]
- 42.Ren CP, Chaudhuri RR, Fivian A, Bailey CM, Antonio M, Barnes WM, Pallen MJ. J Bacteriol. 2004;186:3547–3360. doi: 10.1128/JB.186.11.3547-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhuri RR, Khan AM, Pallen MJ. Nucleic Acids Res. 2004;32:D296–D299. doi: 10.1093/nar/gkh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murata Y, Doi T, Taniguchi H, Fujiyoshi Y. Biochem Biophys Res Commun. 2005;327:183–191. doi: 10.1016/j.bbrc.2004.11.154. [DOI] [PubMed] [Google Scholar]
- 48.Sory MP, Cornelis GR. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 49.Hopp TP, Prickett KS, Price KL, Libby RT, March CJ, Cerretti DP, Urdal DL, Conlon PJ. Biotechnology. 1988;6:1204–1210. [Google Scholar]
- 50.Charpentier X, Oswald E. J Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.