Abstract
How heterocyst differentiation is regulated, once particular cells start to differentiate, remains largely unknown. Using near-saturation transposon mutagenesis and testing of transposon-tagged loci, we identified three presumptive regulatory genes not previously recognized as being required specifically for normal heterocyst maturation. One of these genes has a hitherto unreported mutant phenotype. Two previously identified regulatory genes were further characterized.
Because cyanobacterial heterocysts are micro-oxic cells within oxygen-producing filaments (13, 42), they are hospitable to oxygen-sensitive processes, including nitrogen fixation and hydrogen production (12-14, 38). Which vegetative cells differentiate into heterocysts depends upon HetR (7), whose dimer binds to DNA but with unknown specificity (20), and upon a peptide, PatS (44), that inhibits differentiation. However, the regulation of heterocyst maturation remains largely undefined. Bacteria use signaling systems, often based on protein phosphorylation, to coordinate adaptive processes (1, 36). Several proteins that participate in the regulation of heterocyst differentiation and function are known: (i) canonical DNA-binding proteins, especially NtcA (17, 18) and the related protein DevH (16, 35); (ii) unorthodox DNA-binding proteins (20, 24); and (iii) components of phosphorylated two-component regulatory systems and a Ser/Thr kinase (8, 9, 27, 30, 32, 45, 46, 48). Other processes, e.g., transport (22), may also critically regulate differentiation. Despite these insights, ignorance of molecular participants in regulation hinders understanding of the process of heterocyst differentiation as a whole. Fox genes, by definition, are required specifically for fixation of dinitrogen in the presence of oxygen (10). We have identified additional regulatory Fox genes upon which heterocyst maturation depends.
Regulatory-family genes with Fox− mutant phenotypes.
We mutated Anabaena sp. with the transposon (Tn) Tn5-1063 and identified mutants that are capable of aerobic growth on the nitrate-containing medium AAN but not on (N-free) AA medium (19). Transposon insertions present in 1,076 Fox− mutants were localized in 491 open reading frames (ORFs) that included 83% of the previously known Fox genes but also genes known not to be Fox genes. We sought to determine, in part by complementation, whether transposon-mutated ORFs that were annotated (21) as members of a regulatory family and that had not previously been identified as playing a regulatory role in heterocyst formation were responsible for the corresponding mutant phenotypes. We did so by previously described methods (11, 19), testing all such genes for the ability of plasmids (Tables 1 and 2) to restore corresponding mutants to a Fox+ phenotype.
TABLE 1.
Fox− regulatory ORFs mutated by Tn5-1063, complementation, and predicted functions of their products
| ORF (name of gene) | No. of mutants with a Tn in that ORF | Mutant(s) studied | Chromosomal site (21) of Tn insertion (bp) | Complementing fosmid clone | Complementing replicating plasmid | Predicted function | Heterocyst envelope phenotypea |
|---|---|---|---|---|---|---|---|
| alr0117 (hepN) | 6 | FQ671 | 120135 | anc0709 | pRL2855a | Two-component sensory histidine kinase | Hgl+ Hep− |
| all0187 (conR) | 1 | FQ1062 | 201581 | anc1418 | pRL3055 | Transcriptional regulator | Hgl+ Hep+b |
| alr1086 (henR) | 11 | FQ621 | 1272510 | anc3515 | pRL3194 | Two-component response regulator | Hgl− Hep− (has fibrous layer) |
| all2760 (hepS) | 6 | FQ1487, FQ1641 | 3356965, 3357475 | anc1982 | pRL2858a | Serine/threonine kinase | Hgl+ Hep− |
| alr5348 | 3 | FQ1281 (11) | —c | — | — | Bears Spo0J-like domain | Hgl+ (11) Hep− |
Within the context of this study, Hgl+ and Hgl− phenotypes are, respectively, those in which heterocysts bear or do not bear a laminated layer of envelope glycolipids. More generally, an Hgl mutant can be one in which heterocyst envelope glycolipids are not synthesized (11) or are synthesized but not deposited in an organized array, as in an hglK mutant (2). Hep+, heterocysts bear a homogeneous envelope layer of compacted polysaccharide; Hep−, they bear no such layer.
In FQ1062, polar junctions of heterocysts to vegetative cells are usually abnormally little constricted.
—, see reference 11.
TABLE 2.
Cyanobacterial strains and plasmids used
| Strain or plasmid | Derivation and/or relevant characteristicsa |
|---|---|
| Strains of Anabaena sp. | |
| FQ621 | Bmr Nmr Smr; alr1086::Tn5-1063 |
| FQ671 | Bmr Nmr Smr; alr0117::Tn5-1063 |
| FQ1062 | Bmr Nmr Smr; all0187::Tn5-1063 |
| FQ1281 (11) | Bmr Nmr Smr; alr5348::Tn5-1063 |
| FQ1487 | Bmr Nmr Smr; all2760::Tn5-1063 |
| FQ1490 (11) | Bmr Nmr Smr; Tn5-1063 upstream from hglA; Hgl− |
| FQ1641 | Bmr Nmr Smr; all2760::Tn5-1063 |
| PCC 7120 | Wild type, from R. Haselkorn |
| Plasmids | |
| anc0709 | Cmr Emr; Anabaena sp. chromosomal DNA from bp 111372 to bp 129814 in BamHI site of pRL838 (21) |
| anc1418 | Cmr Emr; Anabaena sp. chromosomal DNA from bp 197711 to bp 215670 in BamHI site of pRL838 (21) |
| anc1982 | Cmr Emr; Anabaena sp. chromosomal DNA from bp 3353565 to bp 3377720 in BamHI site of pRL838 (21) |
| anc3515 | Cmr Emr; Anabaena sp. chromosomal DNA from bp 1262840 to bp 1283836 in BamHI site of pRL838 (21) |
| pRL2814 | Apr Cmr Emr; an internal fragment of alr1086, PCR amplified from Anabaena sp. with primers 5′- GCGAGTAATGGTGAAGAAGGAA-3′ and 5′- CGGTAAGGCAATTTAAGACCTG-3′, was inserted in pGEM-T Easy (Promega, Madison, WI), and a PstI-Cmr Emr-oriT(RK2)-PstI fragment from pRL2665b (19) was placed in the unique PstI site of the resulting plasmid |
| pRL2833b | Cmr Emr; same as pRL2833a (11), but PglnA oppositely oriented |
| pRL2855a | Cmr Emr; alr0117-containing BsrFI fragment from anc0709 fused with XmaI-BamHI linkers from pIC20H (29) to the BamHI site of pRL2833a (11) |
| pRL2858a | Cmr Emr; all2760-containing MfeI fragment from anc1982 fused with EcoRI-SacI linkers from pIC20H (29) to the SacI site of pRL2833b |
| pRL3055 | Smr Spr (34); all0187-containing PvuI (blunted)-AclI fragment of anc1418 transferred between the StuI and ClaI sites of pRL2831a (19) |
| pRL3120 | Apr; alr1086-containing PCR product, generated with primers 5′-TCTCCCGAATTCATGTTTCAAATTTTGATAATTGATGAT-3′ and 5′-GCTTCTCGAGTTAGTCAAACTTAACTAGTAGGA-3′ and wild-type Anabaena sp. DNA as template, cloned between the EcoRI and XhoI sites of pGilda (BD Biosciences Clontech) |
| pRL3141 | Cmr Emr; all0185- and all0186-containing XbaI-ScaI fragment of anc1418 linked with the short XbaI-PstI and SmaI-EcoICRI fragments of pIC20H (29) to the NsiI and StuI sites of pRL2833a (11) |
| pRL3178 | Cmr Emr; sacB vector based on pRL271 (3), deleted from SpeI to XbaI |
| pRL3183 | Bmr Cmr Emr Kmr Smr; the all2760-bearing SstI fragment from pRL2858a was first inserted into the SstI site of pRL3178, and into the unique XbaI site of that fragment was then added a SacI-luxAB-XbaI-oriV-Kmr Bmr Smr-BglII fragment from pRL1063a by means of BlnI-containing linkers; finally, a supernumerary oriV sequence was deleted by cutting with SgrAI and XbaI, blunting, and religating |
| pRL3190 | Apr; alr1086-containing EcoRI-XhoI fragment of pRL3120 transferred between the same sites of pIC20H (29) |
| pRL3194 | Cmr Emr; alr1086-containing XhoI-PstI fragment of pRL3190 transferred between the XhoI and NsiI sites of pRL2833a (11) |
Ap, ampicillin; Bm, bleomycin; Cm, chloramphenicol; Em, erythromycin; Nm, neomycin; Sm, streptomycin; Sp, spectinomycin.
As discussed below, ORFs all0187, alr1086, and all2760, presumptively encoding, respectively, a transcriptional regulator, a response regulator, and a Ser/Thr kinase, were shown for the first time to be Fox genes (see below and Table 1). We also confirm and extend findings presented by Ning and Xu (32) regarding alr0117, whose presumptive product is a histidine kinase, and by Fan et al. (11) concerning alr5348, one of whose predicted domains shows limited similarity to a Spo0J transcriptional regulator.
Testing of presumptive Fox genes.
Plasmids pRL2855a, pRL2858a, pRL3055, and pRL3194 (Table 2) replicate in Anabaena sp. and contain, intact, only the wild-type gene assessed for complementation. FQ671 and other alr0117 mutants were complemented by plasmid pRL2855a. pRL3141 contains two genes 3′ from all0187 and on the same strand of DNA. The all0187 mutant FQ1062 grew aerobically on N2 when bearing pRL3055 with or without pRL3141 but not when bearing pRL3141 alone (data not shown). PCR analysis (11) of complementation of fully segregated FQ1062 by pRL3055 provided no evidence of recombination of pRL3055 with the genome (data not shown), implying that pRL3055 complemented in trans. As noted for the ORF 3′ from alr0117 (32), the ORFs 3′ from alr1086 and all2760 are encoded on the opposite strand of DNA (21), so no polar effect of transposon mutation accounts for the phenotypes of their mutants. FQ621 and another alr1086 mutant were complemented by pRL3194. In addition, an alr1086 mutant was reconstructed by insertional mutagenesis with pRL2814, and the insertions were determined to be where expected by (i) PCR with primers 5′-AGCAGCCGGGACAAAATTA-3′ and, separately, 5′-TTTTACAAAAACAGGGTTATCAAA-3′ and 5′-TTGTAAAACGACGGCCAGT-3′; and (ii) recovery of the integrated plasmid from genomic DNA by excision with HindIII, religation, and transfer to Escherichia coli, followed by restriction, separately, with PstI and XmnI. FQ1487, FQ1641, and other all2760 mutants were complemented by plasmid pRL2858a. We conclude that ORFs all0187, alr1086, and all2760, like alr0117 (32) and alr5348 (11), are Fox genes.
Many genes that are involved in heterocyst differentiation are transcriptionally activated, often after an extended delay, in response to nitrogen step-down (15). Plasmid pRL3183 bears an all2760::luxAB fusion. Strain SR3183 is derived from single, homologous recombination of pRL3183 with the chromosome of Anabaena sp. strain SR3183, and the transpositions in FQ671 (alr0117), FQ1062 (all0187), and FQ621 (alr1086) position luxAB to measure transcription of the corresponding genes. As quantified by use of a luminometer (41), in none of those mutants was a significant increase in transcription in excess of twofold observed in response to deprivation for fixed nitrogen (data not shown). Neither was such an increase observed by use of microarrays for the same ORFs except, at 8 h after nitrogen step-down, for all2760 (9). Therefore, these may be developmentally active genes that, like hetF (43), are transcribed constitutively. It remains possible that their transcription may increase in heterocysts while diminishing in vegetative cells and that their activity may be regulated posttranscriptionally.
Structural and lipid-related phenotypes of Fox− mutants. (i) all0187.
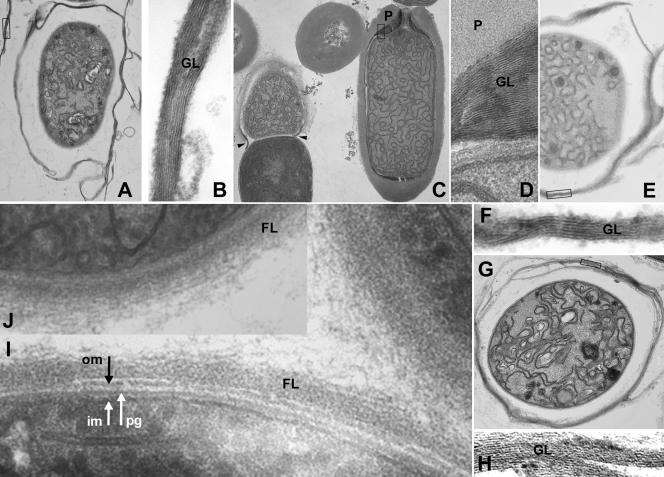
The methods of Black et al. (2) and Nichols and Wood (31) were used for fixation for electron microscopy and thin-layer chromatography of lipid extracts, respectively. Strains were incubated for 7 days (for electron microscopy) or 10 days (for thin layer chromatography) on agar-solidified AA medium. Because the all0187 mutant FQ1062 forms heterocysts with a polysaccharide layer (i.e., is Hep+) and a glycolipid layer (i.e., is Hgl+) (Fig. 1C and D; Fig. 2), its mutation does not appear to affect the biosynthesis, per se, of the components of the heterocyst envelope. Rather, the constriction of the cell and cell wall that normally takes place at the poles of the heterocyst is very often aborted. As a result, the constituents of the heterocyst envelope are deposited aberrantly, leading to the formation of heterocyst envelopes whose ends are widely open (Fig. 1C, arrowheads). Consequently, more oxygen can enter heterocysts and inactivate nitrogenase, presumptively accounting for their inability to fix N2 under a normal atmosphere. Seldom are the heterocysts of FQ1062 as nearly normal at their poles as seen in the right-hand side of Fig. 1C. In an all0187 mutant, the junctions between vegetative cells are also often less clearly constricted than those in the wild-type strain (data not shown). Thus, all0187, like hglK, is a Fox gene whose influence can be seen in the shapes of vegetative cells in series (2). All0187 shows alignment along 87% of the length of a LytR transcriptional regulator domain (COG1316 [28]) that, in other bacteria, is involved in autolysis (6). Bacillus subtilis LytR is described as a transcriptional attenuator of its own gene and of the lytABC operon, where LytC is N-acetylmuramoyl-l-alanine amidase. Although HcwA, also an N-acetylmuramoyl-l-alanine amidase, is a Fox gene product (47), it remains unknown whether All0187 regulates itself and (or) hcwA. FQ1550 (26) and other mutants in our collection show defects in heterocyst envelope formation similar to those seen in FQ1062; the corresponding wild-type genes may possibly be regulated by All0187. Because the phenotype of an all0187 mutant appears to be constriction specific rather than heterocyst specific and All0187 appears to be regulatory, we denote the gene conR.
FIG. 1.
Electron microscopy of heterocyst sections from regulatory Fox− mutants. (A and B) alr0117 mutant FQ671. (C and D) all0187 mutant FQ1062, most of whose heterocysts have terminal pores that are unusually wide (C, arrowheads: the heterocyst on the right is more nearly normal in structure). (E and F) all2760 mutant FQ1641. (G and H) alr5348 mutant FQ1281. (I and J) alr1086 mutant FQ621. Panels B, D, F, and H, magnified from the boxed regions in panels A, C, E, and G, respectively, show glycolipid laminations. im, inner membrane; pg, peptidoglycan; om, outer membrane; GL, envelope glycolipid; P, envelope polysaccharide; FL, fibrous material. Magnifications (103): A, ×11; B, ×142; C, ×9.3; D, ×148; E, ×19; F, ×147; G, ×15; H, ×118; I, ×137; J, ×69.
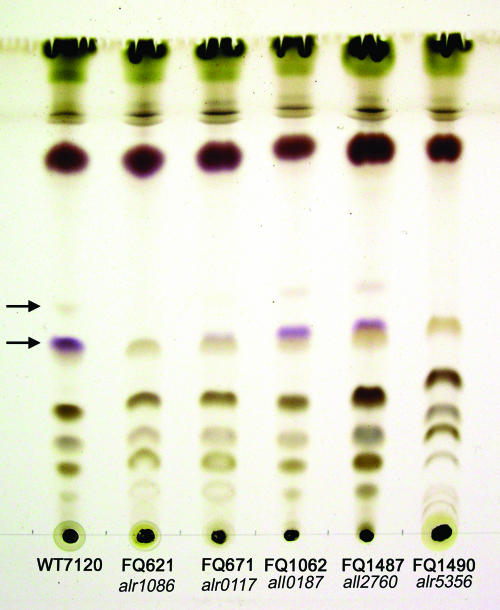
FIG. 2.
Analysis of heterocyst envelope glycolipids in regulatory Fox− mutants, mutant FQ1490 (11; see Table 2), and wild-type Anabaena sp. Heterocyst envelope glycolipids (arrows) are distinguishable from other Anabaena sp. lipids by their violet coloration when incompletely oxidized. The upper glycolipid is less abundant than the lower and may for that reason sometimes not be visualized together with the latter. Concordant with the results in Fig. 1, mutants bearing transposon insertions in ORFs all0187 and all2760 produce abundant heterocyst envelope glycolipids, little of such glycolipid is seen in an alr0117 mutant (see the text), and no more than a trace is seen in an alr1086 mutant, whereas mutant FQ1490 is known to form none (11).
(ii) all2760.
The all2760 mutants tested are Hep− and Hgl+, as assessed by electron microscopy (Fig. 1E and F) and thin-layer chromatography of lipid extracts (Fig. 2) of mutant and wild-type strains, and as corroborated by staining with Alcian Blue (unpublished technique of M. Gantar, J. Elhai, J. Jia, and M. Ow, cited in references 4, 19, and 26—see also references 16 and 32; data not shown). Other mutants were examined similarly. The sole putative regulatory motif in predicted All2760 is characteristic of serine/threonine kinases. Therefore, we denote all2760 hepS. Thirteen ORFs present in Anabaena sp. that have both Ser/Thr and His kinase domains (33, 39) provide presumptive precedents for direct coupling of His and Ser/Thr kinases in signal transduction pathways. What protein(s) All2760 may phosphorylate is unknown.
(iii) alr0117.
Ning and Xu (32) described the predicted product of alr0117 and showed that an alr0117 mutant produces transcripts of two of the seven biosynthetic genes for heterocyst envelope glycolipids that have been documented (11) and lacks staining of heterocyst envelope polysaccharide. Our observations by electron microscopy (Fig. 1A and B) confirm their implication that those heterocysts synthesize heterocyst envelope glycolipids and their interpretation that heterocysts of an alr0117 mutant lack a heterocyst envelope polysaccharide layer. Because alr0117 presumptively encodes a two-component histidine kinase but because a histidine kinase that regulates synthesis of heterocyst envelope polysaccharide has already been named hepK (46, 48), we and X. Xu (personal communication) agree to name alr0117 hepN. Facile loss of the glycolipid layers from Hep− heterocysts can account for the description of those heterocysts as proheterocysts (see the legend to Fig. 1b in reference 32), and breakage of the glycolipid layers into fragments so small that they are not recovered following centrifugal sedimentation can account for the paucity of envelope glycolipid seen in thin-layer chromatograms of lipid extracts of FQ671 (Fig. 2). Concordantly, we were able to visualize the glycolipid layers of the alr0117 mutant FQ671 by transmission electron microscopy only when filaments were scraped from petri dishes.
(iv) alr5348.
In the Fox− mutant FQ1281, the transposon is inserted downstream from the Spo0J-like domain of ORF alr5348. Although present within a cluster of genes required for the synthesis and normal deposition of heterocyst envelope glycolipids (11), fully segregated FQ1281 is Hgl+ (11) and Hep− (Fig. 1G, H). That mutant is complemented by a plasmid whose only intact Anabaena sp. gene is alr5348 (11). Mutations in hepS, hepN, hepK, and devRA, like that of the alr5348 mutation in FQ1281, all result in Hep− Hgl+ ultrastructural mutant phenotypes. Whether the products of any or all of hepS, hepN, and alr5348 form a phosphotransfer cascade with the interacting, two-component regulatory elements HepK and DevRA (46) remains to be determined. alr5348 is the only one of these genes whose product bears what resembles a known DNA-binding domain, spo0J. We are pursuing determination of the role, if any, of the Spo0J-like domain of alr5348 in the Hep− attribute of FQ1281.
(v) alr1086.
Although heterocysts of the alr1086 mutant FQ621 may synthesize traces of envelope glycolipids (Fig. 2), they appear to lack both the heterocyst envelope glycolipid layer present in the wild-type strain (40) and a normal heterocyst envelope polysaccharide layer (Fig. 1). However, those heterocysts produce fibrous extracellular material of variable thickness that appears to be layered parallel to the cell membrane, hinting at lamination (Fig. 1I and J). The fibrous material that lies outside of the plasmalemma of an alr1086 mutant may correspond to the fibrous material identified by Lang and Fay (25) as formed earliest and most peripherally during heterocyst formation in Anabaena cylindrica. Such a mutant phenotype has not previously been reported. Whether or not the material is chemically related to the material that forms the bulk of the polysaccharide layer of the heterocyst envelope remains unknown, but Alcian Blue did not stain it noticeably (data not shown). Because an alr1086 mutation pleiotropically affects regulation of the two principal layers of the heterocyst envelope, we denote that ORF henR.
Whereas sigma factors play important roles in other bacterial differentiation processes (see, e.g., reference 37), no major role of sigma factors has heretofore been recognized in heterocyst differentiation (5, 23). Like the Anabaena sp. developmental proteins DevR (8, 46) and PatA (27), HenR resembles response regulators that lack known DNA-binding motifs. HenR belongs to a group of response regulators in Anabaena sp. that have “a weak but significant similarity to the catalytic domain of Ser/Thr phosphatases of type PP2C” (39). HenR most closely resembles the Bacillus sp. response regulator RsbU, which exhibits phosphatase activity and interacts with RsbV. In turn, RsbV resembles the product of Anabaena sp. ORF all1087, which overlaps the 3′ terminus of henR. RsbV and All1087 show similarity to an anti-sigma factor antagonist. Just beyond all1087 lies all1088, whose predicted product is a two-component histidine kinase. If the effect of HenR should prove to be mediated by All1087, sigma factors may have a more major role than heretofore recognized in heterocyst differentiation. In this regard, it is notable that ORFs alr4800 and all1087, which presumptively encode anti-sigma factor antagonists, show strongly significant increases in transcription at 3 and 8 h, respectively, of nitrogen deprivation (9).
The apparently pleiotropic effect of a henR mutation suggests that HenR is part of a master system that regulates synthesis of, at least, the heterocyst envelope. That system may bifurcate after HenR, one branch controlling deposition of the glycolipid layer and the other regulating deposition of the polysaccharide layer. It remains to be determined how regulation by henR is coordinated with that by the known ORFs (i) abp2, abp3 (24), and devH (35), which encode DNA-binding proteins and whose mutation blocks production of heterocyst envelope glycolipids; and (ii) devRA-hepK (46), hepN (32), hepS, and alr5348, which evidently also regulate synthesis of the polysaccharide layer of the heterocyst envelope.
Acknowledgments
We thank Haixia He, Yi Li, and Elizabeth Wojciuch for technical assistance and Alicia Pastor-Lecha and Marlene Cameron for assistance with electron microscopy and graphics, respectively.
This work was supported by U.S. National Science Foundation grant MCB-0090232 and U.S. Department of Energy grant DOE-FG02-91ER20021.
REFERENCES
- 1.Alex, L. A., and M. I. Simon. 1994. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 10:133-138. [DOI] [PubMed] [Google Scholar]
- 2.Black, K., W. J. Buikema, and R. Haselkorn. 1995. The hglK gene is required for localization of heterocyst-specific glycolipids in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:6440-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 4.Borthakur, P. B., C. C. Orozco, S. S. Young-Robbins, R. Haselkorn, and S. M. Callahan. 2005. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 57:111-123. [DOI] [PubMed] [Google Scholar]
- 5.Brahamsha, B., and R. Haselkorn. 1992. Identification of multiple RNA polymerase sigma factor homologs in the cyanobacterium Anabaena sp. strain PCC 7120: cloning, expression, and inactivation of the sigB and sigC genes. J. Bacteriol. 174:7273-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321-330. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, E. L., K. D. Hagen, M. F. Cohen, M. L. Summers, and J. C. Meeks. 1996. The devR gene product is characteristic of receivers of two-component regulatory systems and is essential for heterocyst development in the filamentous cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 178:2037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehira, S., and M. Ohmori. 2006. NrrA, a novel nitrogen-responsive response regulator, facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 10.Ernst, A., T. Black, Y. Cai, J. M. Panoff, D. N. Tiwari, and C. P. Wolk. 1992. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J. Bacteriol. 174:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, Q., G. Huang, S. Lechno-Yossef, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 58:227-243. [DOI] [PubMed] [Google Scholar]
- 12.Fay, P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56:340-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fay, P., W. D. P. Stewart, A. E. Walsby, and G. E. Fogg. 1968. Is the heterocyst the site of nitrogen fixation in blue-green algae? Nature 220:810-812. [DOI] [PubMed] [Google Scholar]
- 14.Gallon, J. R. 1992. Reconciling the incompatible: N2 fixation and O2. New Phytol. 122:571-609. [Google Scholar]
- 15.Golden, J. W., and H. S. Yoon. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6:557-563. [DOI] [PubMed] [Google Scholar]
- 16.Hebbar, P. B., and S. E. Curtis. 2000. Characterization of devH, a gene encoding a putative DNA binding protein required for heterocyst function in Anabaena sp. strain PCC 7120. J. Bacteriol. 182:3572-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 19.Huang, G., Q. Fan, S. Lechno-Yossef, E. Wojciuch, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for the synthesis of heterocyst envelope polysaccharide in Anabaena sp. strain PCC 7120. J. Bacteriol. 187:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. USA 101:4848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 22.Khudyakov, I., and C. P. Wolk. 1997. hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J. Bacteriol. 179:6971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khudyakov, I. Y., and J. W. Golden. 2001. Identification and inactivation of three group 2 sigma factor genes in Anabaena sp. strain PCC 7120. J. Bacteriol. 183:6667-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koksharova, O. A., and C. P. Wolk. 2002. Novel DNA-binding proteins in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 184:3931-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang, N. J., and P. Fay. 1971. The heterocysts of blue-green algae II. Details of ultrastructure. Proc. R. Soc. Lond. B 178:193-203. [Google Scholar]
- 26.Leganés, F., A. Blanco-Rivero, F. Fernández-Piñas, M. Redondo, E. Fernández-Valiente, Q. Fan, S. Lechno-Yossef, and C. P. Wolk. 2005. Wide variation in the cyanobacterial complement of presumptive penicillin-binding proteins. Arch. Microbiol. 184:234-248. [DOI] [PubMed] [Google Scholar]
- 27.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. USA 89:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 30.Muro-Pastor, A. M., E. Olmedo-Verd, and E. Flores. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 256:171-177. [DOI] [PubMed] [Google Scholar]
- 31.Nichols, B. W., and B. J. B. Wood. 1968. New glycolipid specific to nitrogen-fixing blue-green algae. Nature 217:767-768. [Google Scholar]
- 32.Ning, D., and X. Xu. 2004. alr0117, a two-component histidine kinase gene, is involved in heterocyst development in Anabaena sp. PCC 7120. Microbiology 150:447-453. [DOI] [PubMed] [Google Scholar]
- 33.Ohmori, M., M. Ikeuchi, N. Sato, P. Wolk, T. Kaneko, T. Ogawa, M. Kanehisa, S. Goto, S. Kawashima, S. Okamoto, H. Yoshimura, H. Katoh, T. Fujisawa, S. Ehira, A. Kamei, S. Yoshihara, R. Narikawa, and S. Tabata. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:271-284. [DOI] [PubMed] [Google Scholar]
- 34.Prentki, P., A. Binda, and A. Epstein. 1991. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene 103:17-23. [DOI] [PubMed] [Google Scholar]
- 35.Ramírez, M. E., P. B. Hebbar, R. Zhou, C. P. Wolk, and S. E. Curtis. 2005. Anabaena sp. strain PCC 7120 gene devH is required for synthesis of the heterocyst glycolipid layer. J. Bacteriol. 187:2326-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 37.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 38.Tsygankov, A. A., A. S. Fedorov, S. N. Kosourov, and K. K. Rao. 2002. Hydrogen production by cyanobacteria in an automated outdoor photobioreactor under aerobic conditions. Biotechnol. Bioeng. 80:777-783. [DOI] [PubMed] [Google Scholar]
- 39.Wang, L., Y. P. Sun, W. L. Chen, J. H. Li, and C.-C. Zhang. 2002. Genomic analysis of protein kinases, protein phosphatases and two-component regulatory systems of the cyanobacterium Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 217:155-165. [DOI] [PubMed] [Google Scholar]
- 40.Winkenbach, F., C. P. Wolk, and M. Jost. 1972. Lipids of membranes and of the cell envelope in heterocysts of a blue-green alga. Planta 107:69-80. [DOI] [PubMed] [Google Scholar]
- 41.Wolk, C. P., J. Elhai, T. Kuritz, and D. Holland. 1993. Amplified expression of a transcriptional pattern formed during development of Anabaena. Mol. Microbiol. 7:441-445. [DOI] [PubMed] [Google Scholar]
- 42.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 43.Wong, F. C., and J. C. Meeks. 2001. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J. Bacteriol. 183:2654-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon, H.-S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, C.-C., A. Friry, and L. Peng. 1998. Molecular and genetic analysis of two closely linked genes that encode, respectively, a protein phosphatase 1/2A/2B homolog and a protein kinase homolog in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 180:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, R., and C. P. Wolk. 2003. A two-component system mediates developmental regulation of biosynthesis of a heterocyst polysaccharide. J. Biol. Chem. 278:19939-19946. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, J., K. Jäger, T. Black, K. Zarka, O. Koksharova, and C. P. Wolk. 2001. HcwA, an autolysin, is required for heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 183:6841-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, J., R. Kong, and C. P. Wolk. 1998. Regulation of hepA of Anabaena sp. strain PCC 7120 by elements 5′ from the gene and by hepK. J. Bacteriol. 180:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]