Abstract
Pseudomonas aeruginosa is a microorganism associated with the disease cystic fibrosis. While environmental P. aeruginosa strains are generally nonmucoid and motile, isolates recovered from the cystic fibrosis lung frequently display a mucoid, nonmotile phenotype. This phenotypic conversion is mediated by the alternative sigma factor AlgT. Previous work has shown that repression of fleQ by AlgT accounts for the loss of flagellum biosynthesis in these strains. Here, we elucidate the mechanism involved in the AlgT-mediated control of fleQ. Electrophoretic mobility shift assays using purified AlgT and extracts derived from isogenic AlgT+ and AlgT− strains revealed that AlgT inhibits fleQ indirectly. We observed that the AlgT-dependent transcriptional regulator AmrZ interacts directly with the fleQ promoter. To determine whether AmrZ functions as a repressor of fleQ, we mutated amrZ in the mucoid, nonmotile P. aeruginosa strain FRD1. Unlike the parental strain, the amrZ mutant was nonmucoid and motile. Complementation of the mutant with amrZ restored the mucoid, nonmotile phenotype. Thus, our data show that AlgT inhibits flagellum biosynthesis in mucoid, nonmotile P. aeruginosa cystic fibrosis isolates by promoting expression of AmrZ, which subsequently represses fleQ. Since fleQ directly or indirectly controls the expression of almost all flagellar genes, its repression ultimately leads to the loss of flagellum biosynthesis.
Several gram-negative bacterial species show evidence of a reciprocal regulation of flagellum expression and exopolysaccharide synthesis. It has been suggested that this mechanism enables microbes to optimize their interaction with their prospective hosts or with particular niches in the environment. In Vibrio cholerae, for instance, epsD and epsE are involved in the coordinate control of flagellum and exopolysaccharide expression during the formation of biofilms (31). Disruption of the flagellar regulatory genes flrA and flrC in this microorganism results not only in the loss of flagellum expression but also in the induction of exopolysaccharide synthesis (1, 32). Recently, it has also been suggested that the sodium-driven flagellar motor may play a role in controlling the expression of exopolysaccharide in this microorganism (19). In Escherichia coli, an increase in the biosynthesis of colanic acid exopolysaccharide is evident upon repression of flagellum synthesis (24). In the symbiont Sinorhizobium meliloti, the ExoR protein and the ExoS/ChvI two-component system have been shown to control both succinoglycan and flagellum synthesis (35). The inverse regulation of flagellum biosynthesis and exopolysaccharide expression has also been observed in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis (CF) isolates and is mediated by the alternative sigma factor AlgT (AlgU, σE) (11, 28).
The function of AlgT in the regulation of alginate synthesis has been well documented (12, 27). In nonmucoid P. aeruginosa isolates, the activity of AlgT is negligible due to the suppressive effect of the anti-sigma factor MucA (12, 34). However, in the majority of mucoid CF isolates, mutations in mucA result in a nonfunctional protein, which ultimately leads to a deregulation of AlgT (12, 27). Subsequently, AlgT positively controls several intermediate regulatory genes, including algB, algR, and amrZ (formerly algZ). Their activities result in the expression of algD, which encodes a GDP-mannose dehydrogenase and ultimately commits the bacterium to the production of alginate (27). One of the mentioned AlgT-dependent intermediates, AmrZ, is a DNA-binding protein of the ribbon-helix-helix family (3) that is homologous to the repressors Mnt and Arc of Salmonella enterica serovar Typhimurium bacteriophage 22 (30). In each of these proteins, the amino terminus consists of a β-sheet involved in recognizing and binding to the DNA (18). Mutation of AmrZ residue K18 or R22, which reside within the proposed β-sheet, results in the loss of DNA binding (4, 26). In addition to its function in alginate production, AmrZ has also been shown to play a role in twitching motility and type IV pilus biosynthesis (4).
In contrast to what is known regarding the function of AlgT in alginate regulation, the AlgT-mediated repression of flagellum biosynthesis remains to be further elucidated. In P. aeruginosa, a four-tiered transcriptional hierarchy tightly controls flagellum synthesis. In this cascade, proper expression of genes belonging to each particular tier/class requires the expression of genes of the previous tier (8). Previously published data indicate that AlgT inhibits flagellum synthesis by repressing the class I gene fleQ (28), which encodes an NtrC-like transcriptional activator (2). The FleQ protein has been referred to as the “master switch” of the flagellar regulatory circuit, as it is required for the expression of all other known flagellar genes with the exception of fliA (8).
The goal of this study was to elucidate the mechanism of the AlgT-mediated repression of fleQ in mucoid, nonmotile P. aeruginosa CF isolates. Biochemical approaches using the mucoid, nonmotile reference strain FRD1 (mucA22), as well as clinical mucoid, nonmotile CF isolates carrying mutations in mucA, revealed that AlgT inhibits fleQ by an indirect pathway. AmrZ, an AlgT-dependent regulator required for alginate production (3, 26, 33) and twitching motility/type IV pilus synthesis (4), was identified as the intermediate involved in the repression of fleQ. Electrophoretic mobility shift assays (EMSA) showed that AmrZ specifically binds the fleQ promoter and that this interaction is abolished if critical DNA-binding residues of the protein are mutated, which implied that AmrZ may function as a repressor of fleQ. This hypothesis was supported by results obtained from promoter fusion assays, Western blot analysis, and microscopy, which showed that mutation of amrZ in the mucoid, nonmotile P. aeruginosa CF isolate FRD1 results in increased fleQ promoter activity and restores flagellum expression as well as motility. Our data indicate that AlgT indirectly mediates the negative control of flagellum biosynthesis in mucoid, nonmotile P. aeruginosa CF isolates by increasing the expression of AmrZ. AmrZ subsequently represses the flagellar regulator fleQ, which ultimately results in loss of flagellum production.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides, and DNA manipulations.
Pseudomonas aeruginosa PAO1, FRD1 (mucA22), FRD440 (mucA22 algT::Tn501), FRD831 (mucA22 algB::aacC1), FRD840 (mucA22 algR::aacC1), FRD1200 (mucA22 amrZ::xylE-aacC1), FRD2234 (mucA22 amrZ17; expresses AmrZ K18A) (4), and FRD2238 (mucA22 amrZ19; expresses AmrZ R22A) (26) were used for this study. Other P. aeruginosa strains used included the mucoid, nonmotile CF isolates CF1 and CF2, and their isogenic algT mutants (28), as well as a collection of other CF-derived mucoid strains (5, 33). E. coli strain JM109 (Promega) was utilized for all cloning experiments. Strains SM10 and HB101/pRK2013 (10) were used to transfer plasmids to P. aeruginosa. Oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| fleQ8 | TTTGATCAGCTGCCTTGCATC |
| Q10F | GCCGTTACTTTGGCGCAAGGC |
| CTX1 | CCTCGTTCCCAGTTTGTTCC |
| attB2 | GTCGCCGCCGGCGATGC |
| attB4 | CGCCCTATAGTGAGTCG |
| attB5 | CGCCCCAACCTCGCTGG |
Plasmid pAT2 (28) was used to insert fleQ::lacZ fusions at the neutral attB site in the P. aeruginosa chromosome. Plasmid pFlp2 (13) was utilized to remove unwanted mini-CTX lacZ vector sequences from fusions that were integrated into the P. aeruginosa chromosome. Primers CTX1, attB2, attB4, and attB5 were used to verify proper insertion of the fusions as well as the loss of unwanted vector sequences. Plasmid pDJW585 (5) contains a functional copy of amrZ and was used for complementation analysis. Plasmid pKMG168 (kindly supplied by Kalai Mathee) was used for expression of AlgT. Plasmid pDR2 (26) was used for production of AmrZ-His6. Plasmids pPJ155 (amrZ17) (4), pPJ157 (amrZ19) (4), and pPJ141 (amrZ) (3) were used to generate extracts for EMSA.
P. aeruginosa genomic DNA was purified with Wizard genomic DNA isolation reagents according to the instructions of the manufacturer (Promega). PCR assays were performed with 100 to 150 ng of genomic DNA, as described previously (3, 28). For PCRs requiring labeling, techniques similar to those described previously were used (3, 4, 5, 26).
Media, antibiotics, and enzyme assays.
Luria broth (LB) (10 g tryptone/liter, 5 g yeast extract/liter, 5 g NaCl/liter) and LBNS (LB without NaCl) were used throughout the study. In addition, LB and LBNS agar plates (broth and 15 g agar/liter) were used. Plasmids used in this study were maintained in E. coli by antibiotic selection with 15 μg/ml of tetracycline, 100 μg/ml of ampicillin, and 30 μg/ml of kanamycin. For P. aeruginosa, antibiotics were used at 100 μg/ml of tetracycline and 300 μg/ml of carbenicillin. For counterselection, sucrose (5%) and irgasan (25 μg/ml) were used. For experiments that involved expression of AmrZ, media were supplemented with 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The β-galactosidase assays were performed as previously described (22, 28).
EMSA.
The radiolabeled fleQ promoter fragment was generated by PCR using primers fleQ8 and Q10F (Table 1). Protein extracts were prepared by growing cells to mid-exponential phase, centrifuging 1.0 ml for 3 min at 14,000 rpm, and resuspending the pellet in 100 μl FB (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM MgCl2). The samples were sonicated for 20 s and immediately put on ice. The samples were centrifuged for 11 min at 14,000 rpm. Supernatants were harvested, and 0.5 μl of 0.1 M phenylmethylsulfonyl fluoride (Sigma) was added. Recombinant AmrZ-enriched extracts were prepared from E. coli JM109/pPJ14 (wild type), JM109/pPJ155 (amrZ17), or JM109/pPJ157 (amrZ19), as previously described (3). AlgT-His6 and AmrZ-His6 were purified with QIAGEN Ni-nitrilotriacetic acid resin as described by the manufacturer (QIAGEN). Binding reactions and EMSA were performed as described elsewhere (3, 4, 5, 26) with protein extract (3 μg) or purified AlgT-His6 or AmrZ-His6. Gels were run for 3 h at 200 V (4°C), dried for 50 min, and exposed to a PhosphorImager screen (Molecular Dynamics) for 45 min prior to development with a Typhoon Scanner.
TEM and phase-contrast microscopy.
For transmission electron microscopy (TEM), bacteria were grown in LBNS to an optical density at 600 nm of 0.5. Formvar-coated copper grids were hydrophilized by immersion in 100% ethanol. One drop of bacterial culture was added per grid. After 1 min, excess liquid was wicked off without completely drying the grid to avoid flagellar shearing. Grids were washed twice by floating them on ultrapure water. Subsequently, a drop of 2% uranyl acetate was added and wicked off after 1 min. TEM was performed on a Philips TEM 400 operated at 80 kV. For phase-contrast microscopy, bacteria were grown overnight in LBNS. A drop of the suspension was added to a glass microscopy slide (Fisher) and covered with a glass coverslip (Fisher), and flagellar motility was examined at ×100 with a Nikon Eclipse E400 microscope.
Western blot analysis.
Western blotting was performed with whole-cell lysates. The lysates were prepared from P. aeruginosa grown in LBNS to an optical density at 600 nm of 0.5. A 1.0-ml volume of cells was centrifuged for 3 min at 14,000 rpm, and the pellet was resuspended in 100 μl FB (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM MgCl2). A 10-μl volume of each suspension was examined by Western blotting. Western blotting was performed with rabbit anti-flagellin serotype B and anti-AmrZ antiserum as described previously (11, 33). Blots were developed with Kodak Image Station 2000RT.
RESULTS
AlgT controls expression of fleQ indirectly.
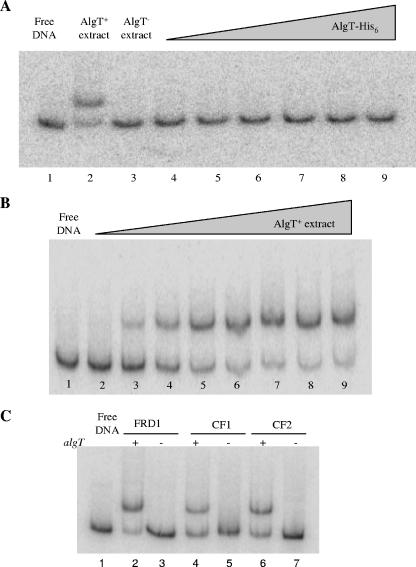
Recent work in our laboratory revealed that AlgT controls P. aeruginosa flagellum synthesis by inhibiting expression of the flagellar master regulator fleQ (28). However, the underlying mechanism of the repression of fleQ remains unknown. To test whether fleQ expression is inhibited directly or indirectly, we first examined the fleQ promoter region for the AlgT promoter recognition sequence, GAACT-16/17 bp-TCTGA (12), but were unable to detect this consensus. Subsequently, we incubated radiolabeled fleQ promoter DNA either with purified AlgT-His6 or with protein extracts derived from the mucoid, nonmotile AlgT+ CF isolate FRD1 or its isogenic algT mutant and analyzed these binding reactions by EMSA. There were no DNA-protein complexes in lanes containing purified AlgT-His6 (Fig. 1A, lanes 4 to 9), which suggested that AlgT does not directly repress fleQ. The lack of binding observed for purified AlgT is not likely due to the loss of activity upon purification, as the same preparation of AlgT-His6 was used in an in vitro transcription assay and found to be active (unpublished data).
FIG. 1.
AlgT represses fleQ indirectly. (A) Autoradiograph of a radiolabeled 250-bp fleQ promoter fragment which was incubated with protein extracts derived from isogenic AlgT+ or AlgT− P. aeruginosa or with purified AlgT-His6 and separated by polyacrylamide gel electrophoresis under nondenaturing conditions. Lane 1, free fleQ promoter DNA; lane 2, AlgT+ extract (3 μg); lane 3, AlgT− extract (3 μg); lanes 4 to 9, increasing amounts of AlgT-His6 (25, 50, 100, 250, 500, and 750 ng, respectively). (B) Binding of extracts from AlgT+ to the fleQ promoter. Lane 1, free fleQ promoter DNA; lanes 2 to 9, increasing amounts of AlgT+ extract (50, 100, 250, 500, 750, 1,500, 3,000, and 5,000 ng, respectively). (C) Binding of protein in extracts from AlgT+ P. aeruginosa to the fleQ promoter is conserved among mucoid, nonmotile CF isolates. Lane 1, free fleQ promoter DNA; lanes 2, 4, and 6, extracts (3 μg) derived from mucoid, nonmotile AlgT+ P. aeruginosa CF isolates; lanes 3, 5, and 7, extracts (3 μg) derived from the corresponding isogenic algT mutants.
However, a protein-DNA complex was evident for the AlgT+-derived protein extract (Fig. 1A, lane 2). A gradual loss of free DNA was observed as increasing quantities of the AlgT+-derived protein extract were added to fleQ (Fig. 1B, lanes 2 to 9). In contrast, the protein-DNA complex was missing in the lane containing extracts derived from the isogenic algT mutant (Fig. 1A, lane 3). Thus, the EMSA results suggest that under the conditions employed here, AlgT does not directly interact with fleQ; rather, a gene product under the control of AlgT binds to the fleQ promoter DNA.
To determine whether the observed AlgT-mediated binding activity is conserved among mucoid, nonflagellated P. aeruginosa cells, we analyzed extracts from two additional clinical mucoid, nonmotile CF isolates and their isogenic algT mutants by EMSA. As with the AlgT+ reference strain FRD1 (Fig. 1C, lane 2), a protein-DNA complex was observed for each extract derived from AlgT+ P. aeruginosa (Fig. 1C, lanes 4 and 6) but was absent when extracts from the isogenic algT mutants were used (Fig. 1C, lanes 3, 5, and 7).
AmrZ interacts directly with the fleQ promoter.
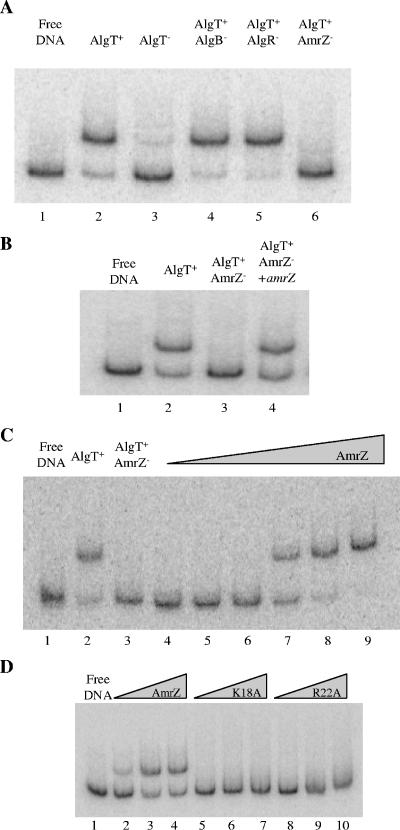
Previously published data show that there is an AlgT-mediated inverse regulation of alginate and flagellum expression in mucoid, nonmotile P. aeruginosa CF isolates (11, 28). In the alginate regulatory pathway, algB, algR, and amrZ are known AlgT-dependent genes, all of which encode regulators of alginate synthesis. We hypothesized that one or more of these gene products might also be involved in the repression of fleQ and, ultimately, flagellum biosynthesis. To test this hypothesis, radiolabeled fleQ promoter DNA was incubated with protein extracts isolated from isogenic FRD1-derived AlgT+ strains carrying null mutations either in algB, algR, or amrZ. The binding reactions were subsequently analyzed by EMSA. DNA-protein complexes, which migrated to a position identical to that seen with the extract derived from the parental strain, were observed in both the algB and the algR mutants (Fig. 2A, lanes 4 and 5). This indicated that neither AlgB nor AlgR directly interacts with fleQ. On the other hand, no DNA-protein complex was observed for the amrZ mutant (Fig. 2A, lane 6). To be certain that the observed loss of binding to the fleQ promoter DNA was indeed due to amrZ, another EMSA was performed with protein extract derived from an amrZ mutant which had been complemented with a functional copy of amrZ (Fig. 2B). While no DNA-protein complex was evident for the amrZ mutant (Fig. 2B, lane 3), a DNA-protein complex was observed when extract of the complemented amrZ mutant was used (Fig. 2B, lane 4). This suggested that either AmrZ or an AmrZ-dependent gene product binds to the fleQ promoter.
FIG. 2.
AmrZ binds to the fleQ promoter. (A) Mutation of amrZ in AlgT+ P. aeruginosa abolishes fleQ binding activity. Lane 1, free fleQ promoter DNA; lane 2, AlgT+ extract (3 μg); lane 3, AlgT− extract (3 μg); lane 4, AlgT+ AlgB− extract (3 μg); lane 5, AlgT+ AlgR− extract (3 μg); lane 6, AlgT+ AmrZ− extract (3 μg). (B) Complementation of the amrZ mutant restores binding to fleQ promoter DNA. Lane 1, free fleQ promoter DNA; lane 2, AlgT+ extract (3 μg); lane 3, AlgT+ AmrZ− extract (3 μg); lane 4, extract derived from an AlgT+ AmrZ− strain complemented with amrZ (3 μg). (C) AmrZ binds to the fleQ promoter. Lane 1, free fleQ promoter DNA; lane 2, AlgT+ extract (3 μg); lane 3, AlgT+ AmrZ− extract (3 μg); lanes 4 to 9, increasing amounts of recombinant AmrZ (1, 5, 10, 25, 50, and 100 ng, respectively). (D) Mutation of critical residues abolishes the ability of AmrZ to bind to fleQ. Lane 1, free fleQ promoter DNA; lanes 2 to 4, increasing amounts of recombinant AmrZ (20, 40, and 60 ng, respectively); lanes 5 to 7, increasing amounts of recombinant AmrZ K18A (20, 40, and 60 ng, respectively); lanes 8 to 10, increasing amounts of recombinant AmrZ R22A (20, 40, and 60 ng, respectively).
Baynham et al. (5) showed that recombinant AmrZ derived from overexpression in E. coli specifically binds at algD. Thus, to examine whether AmrZ directly interacts with the fleQ promoter, radiolabeled fleQ promoter DNA was incubated with increasing amounts of recombinant AmrZ (Fig. 2C). A gradual loss of free DNA was observed as increasing quantities of AmrZ were added to fleQ (Fig. 2C, lanes 4 to 9). The position of the AmrZ-fleQ complexes was identical to that observed for the extract derived from the AlgT+ strain (Fig. 2C; compare lane 2 with lanes 7 to 9).
Previous work showed that alanine substitutions of AmrZ residues K18 and R22, which reside in the proposed β-sheet DNA-binding domain, resulted in a loss of DNA binding at algD and amrZ (4, 26). Therefore, we wanted to determine whether these residues are also essential for DNA binding at fleQ. Radiolabeled fleQ promoter DNA was incubated with increasing amounts of recombinant wild-type, K18A, or R22A mutant AmrZ proteins (Fig. 2D). Mutation of either K18 or R22 resulted in a loss of interaction with fleQ DNA (Fig. 2D, lanes 5 to 7 and 8 to 10, respectively). Thus, AmrZ recognizes fleQ DNA in a fashion similar to that seen at algD and amrZ. Together, these data led to the hypothesis that AmrZ is the AlgT-dependent repressor responsible for the inhibition of fleQ expression and, ultimately, flagellum synthesis in the mucoid, nonmotile P. aeruginosa CF isolate FRD1.
AmrZ controls flagellum biosynthesis.
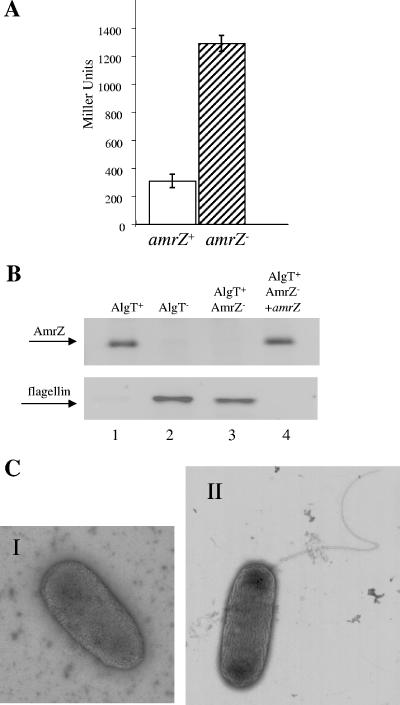
To explore whether AmrZ represses fleQ, the fleQ promoter was fused with a promoterless lacZ and the fusion was integrated into the chromosome of the mucoid, nonflagellated AlgT+ P. aeruginosa CF isolate FRD1 and its isogenic amrZ mutant. The data revealed that fleQ promoter activity was approximately fourfold higher in the amrZ mutant than in the parental AlgT+ strain (Fig. 3A). To examine whether the binding of AmrZ to the fleQ promoter (Fig. 2) and the observed reduction in fleQ promoter activity translate into a flagellum phenotype, we analyzed the mucoid, nonmotile AlgT+ CF isolate FRD1 and its isogenic amrZ mutant for flagellin and AmrZ expression. The results showed that the AlgT+ strain expressed AmrZ and lacked flagellin (Fig. 3B, lane 1). In contrast, the amrZ mutant lacked AmrZ but expressed flagellin (Fig. 3B, lane 3). Upon complementation with amrZ, flagellin expression was inhibited and AmrZ expression was restored (Fig. 3B, lane 4), which was accompanied by the synthesis of alginate (data not shown). Moreover, TEM revealed that ∼95% of the amrZ mutant cells expressed a characteristic single polar flagellum (Fig. 3C, panel II), which was absent in the parental strain (Fig. 3C, panel I). Examination of swimming behavior by phase-contrast microscopy showed that the mucoid, nonmotile AlgT+ strain was nonmotile whereas its isogenic amrZ mutant was motile. Upon complementation of the mutant with amrZ, motility was lost (data not shown). These data provide further evidence that the AlgT-dependent regulatory protein AmrZ plays a role in the direct repression of fleQ in mucoid, nonmotile P. aeruginosa CF isolates and thus in flagellum biosynthesis.
FIG. 3.
AmrZ inhibits flagellum biosynthesis. (A) fleQ::lacZ fusions were integrated into the chromosomes of mucoid, nonmotile P. aeruginosa CF isolate FRD1 (open bar) and its isogenic amrZ mutant (hatched bar) at the neutral attB site. Promoter activity was measured by β-galactosidase assays with ONPG (o-nitrophenyl-β-d-galactopyranoside) used as a substrate and is expressed as the amount of ONPG hydrolyzed per minute as a function of cell density. Shown are the averages of four independent experiments and standard deviations. (B) Whole-cell lysates of the indicated strains were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and examined for AmrZ and flagellin expression by Western blotting with AmrZ (top) and flagellin B (bottom) antiserum. Lane 1, AlgT+ P. aeruginosa; lane 2, AlgT− P. aeruginosa; lane 3, AlgT+ AmrZ− P. aeruginosa; lane 4, AlgT+ AmrZ− P. aeruginosa complemented with amrZ. As a loading control, a second gel containing comparable amounts of total protein was simultaneously prepared and processed by Coomassie staining. (C) TEM of nonflagellated AlgT+ P. aeruginosa (I) and its isogenic, flagellated amrZ mutant (II). Magnification, ×15,600.
DISCUSSION
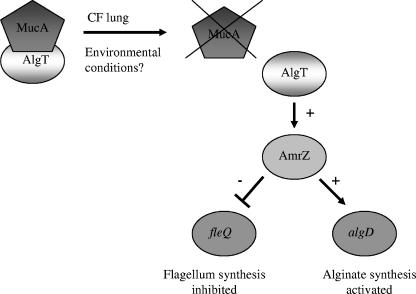
P. aeruginosa has a host of virulence factors at its disposal to successfully establish and maintain infections; these factors include endotoxin, elastase, alkaline protease, exotoxin A, cytotoxin, hemolysin, type III secretion proteins, type IV pili, flagella, and the exopolysaccharide alginate (12, 29). Previous work in our laboratory revealed that in mucoid, nonmotile P. aeruginosa CF isolates, alginate expression and flagellum biosynthesis are inversely controlled by the alternative sigma factor AlgT (11, 28). AlgT modulates the increased expression of several intermediates, including algB, algR, and amrZ, which result in the expression of the algD operon and thus in alginate production (27). Our lab discovered that in mucoid, nonmotile P. aeruginosa strains, inactivation of algT restores flagellum expression and motility (11, 28). Recent work suggested that AlgT mediates the loss of flagellum synthesis in mucoid, nonmotile CF isolates by repressing the flagellar regulator fleQ (28), which is often referred to as the “master switch” of the flagellar regulon, as fleQ is vital for the expression of almost all other known flagellar genes (2, 8, 17). In this study, we propose a model (Fig. 4) in which AlgT represses fleQ by an indirect mechanism. Under physiological conditions, AlgT activity is controlled by the anti-sigma factor MucA. In the CF lung, mutations in mucA result in a nonfunctional MucA and, subsequently, the deregulation of AlgT. AlgT then promotes expression of amrZ, which functions as both a repressor of fleQ and an activator of algD, thus resulting in the loss of flagellum biosynthesis and alginate production, respectively.
FIG. 4.
Proposed model for AlgT-mediated inverse regulation of flagellum synthesis and alginate production in mucoid, nonmotile P. aeruginosa CF isolates. Under most physiological conditions, the activity of the alternative sigma factor AlgT is inhibited by the anti-sigma factor MucA. Unique environmental conditions in CF airways result in mutations of mucA and, subsequently, deregulation of AlgT. AlgT is now free to up-regulate expression of the ribbon-helix-helix-protein AmrZ, which has dual functions as a repressor of fleQ and an activator of algD.
Baynham et al. (5) first discovered AmrZ as a DNA-binding activity upstream of the algD promoter. Here, AmrZ binds an A/T-rich sequence centered 282 bp upstream of the transcriptional start site. Mutation of this site abolishes AmrZ binding in vitro and results in an almost complete loss of algD promoter activity (5). Expression of AmrZ is conserved among mucoid P. aeruginosa CF isolates and is absolutely required for both activation of algD transcription and alginate expression (3). Ramsey et al. (26) discovered that AmrZ binds two sites at the amrZ promoter and functions as an autorepressor. While ribbon-helix-helix proteins usually function as repressors, AmrZ plays a role in both activation and repression of particular target genes. More recent data revealed that AmrZ is also required for twitching motility (4). However, while the data convincingly showed that the DNA-binding activity of AmrZ is required in this process, the AmrZ-dependent genes involved in twitching motility remain to be identified. In the present work, AmrZ is ascribed yet another novel function as a repressor of flagellum synthesis in mucoid, nonmotile P. aeruginosa CF isolates.
Each of the diverse functions of the AmrZ protein described above requires its ability to bind DNA. AmrZ residues K18 and R22, which reside within the proposed β sheet, are required for DNA binding to sites at algD, amrZ (4, 26), and fleQ (Fig. 2D). Replacement of either of these residues with an alanine results in a complete loss of DNA binding in each case, suggesting that the requirement for particular AmrZ residues in DNA binding is conserved independently of the target DNA. When AmrZ binds to sites at algD (3, 5) or amrZ (26), several protein-DNA complexes are evident, which are likely due to AmrZ oligomerization. Interestingly, only a single protein-DNA complex is present when AmrZ binds to fleQ.
Comparison of the three known AmrZ-binding sites at algD and amrZ resulted in a proposed consensus motif, 5′-gGCCAttACCaggcc-3′, where uppercase letters indicate nucleotides conserved among all three known AmrZ-binding sites and lowercase letters represent nucleotides that are found at only two binding sites (26). We searched the 250-bp fleQ promoter fragment utilized in the DNA binding assays for evidence of the proposed AmrZ consensus motif but found only a minimal match. Therefore, footprinting assays will be necessary to identify the specific AmrZ-binding site(s) at fleQ. It should also be pointed out that currently only three genes are known to be under the direct control of AmrZ. Therefore, identification of additional AmrZ-dependent genes and corresponding binding sites is vital to eventually derive a more concise AmrZ consensus motif.
There are many other gram-negative bacterial species that coordinately regulate flagellum synthesis and expression of exopolysaccharides. It has been proposed that this mechanism enables microbes to optimize their interaction with prospective hosts or with particular niches in the environment. For example, Cano et al. (6) reported that in Salmonella enterica, the igaA gene encodes a pleiotropic regulator that positively controls the flagellar master operon flhDC and inhibits expression of the colanic acid gene cluster wca. Mutation of igaA activates the two-component system RscB-RscC, which in turn results in the repression of flhDC and a derepression of the wca genes, leading to nonmotile, mucoid S. enterica variants. This is functionally similar to the mechanism involved in the reciprocal control of flagellum synthesis and alginate expression in mucoid, nonmotile P. aeruginosa, as described in this study. Here, AlgT controls expression of AmrZ, which in turn represses the flagellar master regulator fleQ and promotes production of alginate. Therefore, AlgT is the functional equivalent of IgaA, and AmrZ plays a role functionally similar to that of the two-component system RscB-RscC in S. enterica.
It has been suggested that both the mucoid phenotype and the lack of flagella provide P. aeruginosa with a selective advantage in the CF lung (7, 21). The copious amounts of alginate form a barrier that shields the bacteria from some antimicrobials. For instance, Learn et al. (20) showed that alginate is able to scavenge hypochlorite produced by phagocytic cells, and Pedersen et al. (23) reported that alginate reduces the chemotaxis of polymorphonuclear leukocytes into the CF lung and inhibits activation of the complement system. Moreover, Cobb et al. (7) presented evidence that infection of Calu-3 cells with mucoid P. aeruginosa results in increased expression of genes with antiapoptotic effects in the infected cells. Thus, the presence of alginate not only attenuates host responses but also aids in bacterial circumvention of host defenses. In contrast, flagellin, the major structural subunit of the bacterial flagellum, induces a potent proinflammatory response (7, 9, 15, 16, 25). In fact, flagellin appears to be the major proinflammatory signal of P. aeruginosa (9, 16). Therefore, the ability to shut off flagellum expression may provide P. aeruginosa with yet another way to successfully evade host immune defenses and facilitate its persistence in the CF lung.
While the AlgT-mediated inverse control of flagellum expression and alginate production seems particularly beneficial to the bacterium in the CF lung, it also provides an interesting target for future therapeutic strategies aimed at controlling chronic P. aeruginosa infections. Today, early aggressive antibiotic treatment is used to delay the onset of chronic P. aeruginosa infection and the appearance of mucoid, nonmotile variants (14). However, once these variants arise, they are generally a poor prognostic indicator for CF patients, as it is impossible to eradicate them (12). Thus, the possibility of being able to reverse the mucoid, nonmotile phenotype to a nonmucoid, motile one would be an appealing therapeutic strategy for successfully managing chronic P. aeruginosa infections in the CF lung. Together with traditional therapeutic approaches, this strategy may therefore yield an improved prognosis for patients suffering from CF.
Acknowledgments
We thank the Electron Microscopy Laboratory for assistance with TEM and Kalai Mathee for providing plasmid pKMG168.
This work was supported by grant HL58334 (to D.J.W.) and American Heart Association grant 0515325U (to A.H.T.).
REFERENCES
- 1.Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzger, T. D. Connell, J. G. Morris, and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baynham, P., A. L. Brown, L. L. Hall, and D. J. Wozniak. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33:1069-1080. [DOI] [PubMed] [Google Scholar]
- 4.Baynham, P. J., D. M. Ramsey, B. V. Gvozdyev, E. M. Cordonnier, and D. J. Wozniak. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baynham, P. J., and D. J. Wozniak. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22:97-108. [DOI] [PubMed] [Google Scholar]
- 6.Cano, D. A., G. Dominguez-Bernal, A. Tierrez, F. G. Portillo, and J. Casadesus. 2002. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobb, L. M., J. C. Mychalecky, D. J. Wozniak, and Y. S. Lopez-Boado. 2004. Pseudomonas aeruginosa flagellin and alginate elicit very distinct gene expression patterns in airway epithelial cells: implications for cystic fibrosis disease. J. Immunol. 173:5659-5670. [DOI] [PubMed] [Google Scholar]
- 8.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, M., R. Bryan, S. Rajan, L. Scheffler, S. Brunnert, H. Tang, and A. Prince. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 66:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett, E., and D. J. Wozniak. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 181:7401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: applications for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 14.Hoiby, N., B. Frederiksen, and T. Pressler. 2005. Eradication of early Pseudomonas aeruginosa infection. J. Cystic Fibrosis 4:49-54. [DOI] [PubMed] [Google Scholar]
- 15.Honko, A. N., and S. B. Mizel. 2004. Mucosal administration of flagellin induces innate immunity in the mouse lung. Infect. Immun. 72:6676-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hybiske, K., J. K. Ichikawa, V. Huang, S. J. Lory, and T. E. Machen. 2004. Cystic fibrosis airway epithelial cell polarity and bacterial flagellin determine host response to Pseudomonas aeruginosa. Cell. Microbiol. 6:49-63. [DOI] [PubMed] [Google Scholar]
- 17.Jyot, J., N. Dasgupta, and R. Ramphal. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 184:5251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight, K. L., and R. T. Sauer. 1992. Biochemical and genetic analysis of operator contacts made by residues within the beta-sheet DNA binding motif of Mnt repressor. EMBO J. 11:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 186:4864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Learn, D. B., E. P. Brestel, and S. Seetharama. 1987. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect. Immun. 55:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., M. E. Campbell, and D. P. Speert. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 62:596-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria, p. 493-495. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 23.Pedersen, S. S., A. Kharazmi, F. Espersen, and N. Høiby. 1990. Pseudomonas aeruginosa alginate in cystic fibrosis sputum and the inflammatory response. Infect. Immun. 58:3363-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prigent-Combaret, C., C. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos, H. C., M. Rumbo, and J. C. Sirard. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12:509-517. [DOI] [PubMed] [Google Scholar]
- 26.Ramsey, D. M., P. Baynham, and D. J. Wozniak. 2005. Binding of Pseudomonas aeruginosa AlgZ to sites upstream of the algZ promoter leads to repression of transcription. J. Bacteriol. 187:4430-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309-322. [DOI] [PubMed] [Google Scholar]
- 28.Tart, A. H., M. C. Wolfgang, and D. J. Wozniak. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibition expression of fleQ. J. Bacteriol. 187:7955-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasil, M. L. 1986. Pseudomonas aeruginosa: biology, mechanisms of virulence, epidemiology. J. Pediatr. 108:800-805. [DOI] [PubMed] [Google Scholar]
- 30.Waldburger, C. D., and R. T. Sauer. 1995. Domains of the Mnt repressor: roles in tetramer formation, protein stability, and operator DNA binding. Biochemistry 34:13109-13116. [DOI] [PubMed] [Google Scholar]
- 31.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wozniak, D. J., A. B. Sprinkle, and P. J. Baynham. 2003. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J. Bacteriol. 185:7297-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie, Z. D., C. D. Hershberger, S. Shankar, R. W. Ye, and A. M. Chakrabarty. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J. Bacteriol. 178:4990-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao, S.-Y., L. Luo, K. J. Har, A. Becker, S. Rueberg, G. Yu, J. Zhu, and H. Cheng. 2004. Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J. Bacteriol. 186:6042-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]