Abstract
ATP-citrate lyase (ACL) is an essential enzyme of the reductive tricarboxylic acid (RTCA) pathway of CO2 assimilation. The RTCA pathway occurs in several groups of autotrophic prokaryotes, including the green sulfur bacteria. ACL catalyzes the coenzyme A (CoA)-dependent and MgATP-dependent cleavage of citrate into oxaloacetate and acetyl-CoA, representing a key step in the RTCA pathway. To characterize this enzyme from the green sulfur bacterium Chlorobium tepidum and determine the role of its two distinct polypeptide chains, recombinant holo-ACL as well as its two individual subunit polypeptides were synthesized in Escherichia coli. The recombinant holoenzyme, prepared from coexpressed large and small ACL genes, and the individual large and small subunit polypeptides, prepared from singly expressed genes, were all purified to homogeneity to high yield. Purified recombinant holo-ACL was isolated at high specific activity, and its kcat was comparable to that of previously prepared native C. tepidum ACL. Moreover, the purified recombinant large and small subunit polypeptides were able to reconstitute the holo-ACL in vitro, with activity levels approaching that of recombinant holo-ACL prepared from coexpressed genes. Stoichiometric amounts of each subunit protein were required to maximize the activity and form the most stable structure of reconstituted holo-ACL. These results suggested that this reconstitution system could be used to discern the catalytic role of specific amino acid residues on each subunit. Reconstitution and mutagenesis studies together indicated that residues of each subunit contributed to different aspects of the catalytic mechanism, suggesting that both subunit proteins contribute to the active site of C. tepidum ACL.
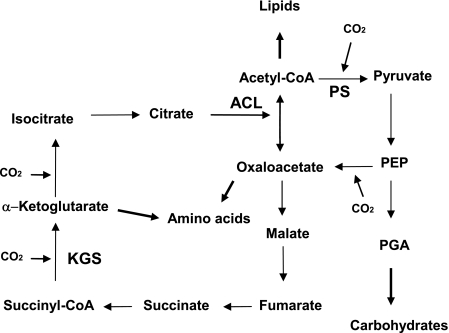
The reductive tricarboxylic acid (RTCA) cycle is employed as a carbon dioxide fixation pathway in a variety of autotrophic bacteria and archaea (1, 2, 6, 9, 11, 12, 26-28). In the RTCA cycle, oxaloacetate is synthesized upon the incorporation of four molecules of CO2, and it is replenished from acetyl coenzyme A (CoA). ATP-citrate lyase (ACL) is an essential enzyme in the cycle, since it catalyzes the formation of oxaloacetate and acetyl-CoA from the cleavage of citrate (Fig. 1). ACL is dependent on CoA and MgATP (13), and studies with the eukaryotic enzyme indicate that catalysis is initiated by autophosphorylation of a histidine residue, generating an unstable citryl-phosphate intermediate. The unstable intermediate is then converted into oxaloacetate and acetyl-CoA by nucleophilic attack of CoA (33, 34), thus completing the reaction.
FIG. 1.
The RTCA cycle. KGS, α-ketoglutarate synthase; PS, pyruvate synthase.
Recent genomic sequencing of the green sulfur photosynthetic bacterium Chlorobium tepidum (3) and biochemical studies with the enzyme from the related organism C. limicola (16, 17) indicated that ACL is comprised of two subunits which are encoded by separate adjacent genes. At this point it is not clear whether each subunit contributes to form the active site of the enzyme or if there are distinct roles of the large and small subunits in catalysis. In addition, it is not clear whether the overall catalytic mechanism of this structurally unique bacterial ACL may differ from the ACL found in eukaryotes, which is a homotetramer (α4) of a large single polypeptide (18, 29) whose synthesis and activity are regulated by transcriptional control and allosteric interactions (4, 21-23, 25). Analysis of deduced amino acid sequences of the large and small subunits of C. tepidum ACL indicated that there were regions identified from both subunit proteins (CLS and CLL) that showed similarities to previously described motifs from other proteins that interacted with common substrates and products, such as ATP, acetyl-CoA, and oxaloacetate (10, 20). Sequence comparisons of such motifs suggested that both subunits of C. tepidum ACL might be important for catalysis, with residues from both subunits potentially comprising the active site. Of particular note was succinyl-CoA synthetase (SCS) from Escherichia coli, another two-subunit protein, where comparisons of motifs important for binding and interacting with common substrates and products appeared to indicate that motifs and residues important for ATP binding might be localized on separate subunits of ACL. In this study, individual recombinant subunits of the C. tepidum protein were prepared and used for reconstitution assays to investigate the necessity and potential role of both subunits for catalytic activity. Particular emphasis was placed on amino acid residues of a putative ATP-grasp motif, which was found on both subunits.
MATERIALS AND METHODS
Construction of overexpression vectors for individual subunit proteins.
The overexpression vectors for C. tepidum ACL subunit genes were constructed by cloning the ACL genes into pET11a (Novagen, La Jolla, CA). For amplification of aclA (encoding the large subunit, CLL) and aclB (encoding the small subunit, CLS), genomic DNA of C. tepidum and paclB containing aclB were used as templates, respectively. Cycling parameters for the amplifications were 95°C for 5 min and 25 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2.5 min. The PCR samples contain 0.25 mM deoxynucleoside triphosphates (dNTPs), 0.2 μM primers, 10× polymerase buffer, 2.5 mM MgCl2, and 10× PCRx enhancer solution (Invitrogen, Carlsbad, CA). For PCR of aclB, 5 U of Pfu polymerase (Stratagene, La Jolla, CA) was used, while aclA was amplified with 5 U of Pfu polymerase (Stratagene) plus 0.5 U of Taq polymerase (Invitrogen). For amplification of aclA, aclaF (5′-CCCCCCATATGAGCATTCTCGCAAATAAAGATACCCGG-3′) and aclaR (5′-AAAAAAGATCTTTACTTCTTGTCGGGAACCGGGC-3′) were used as primers (underlined portions of the two primers indicate restriction sites of NdeI and BglII, respectively). For amplification of aclB, aclbF (5′-AAAAACATATGGCTAAAATTCTTGAAGGGCCTG-3′) and aclbR (5′-AAAAAGGATCCTCAAGACTTCAGAGCCAGGTCGACAATA-3′) were used as primers (underlined portions of the two primers indicate restriction sites of NdeI and BamHI, respectively). After PCR, the products from the amplifications were inserted into pCRII-TOPO (Invitrogen). The aclA and aclB genes were recovered from agarose gels (QIAGEN gel extraction kit; QIAGEN, Hilden, Germany) after digestion with NdeI plus BglII and NdeI plus BamHI, respectively. The genes were cloned into pET11a (Novagen) separately; the resulting vectors were then chemically transformed into E. coli (24) DH5α competent cells. Plasmids were purified from the transformants, and DNA sequencing was performed in the OSU Plant-Microbe Genomics Facility to determine whether point mutations might have been generated as a result of the PCRs for the genes in pET11a. A point mutation found in aclA was corrected by replacing a region of the gene containing the mutation with wild-type sequence. Resulting vectors containing wild-type aclA or aclB were chemically transformed into E. coli BL21(DE3)(pLysS) competent cells (Stratagene) for overexpression.
Construction of an overexpression vector for the holoenzyme containing both subunits.
The vector used to overexpress the C. tepidum holo-ACL was constructed by cloning the aclA and aclB genes into the overexpression vector pCDF-Duet1 (Novagen) as follows. Each of the genes was first amplified with 5 units of Platinum Pfx polymerase, 10× reaction buffer, 1 mM MgSO4, 10× PCRx enhancer solution (Invitrogen), 0.3 mM dNTPs, and 0.3 μM primers. Oligonucleotides for amplification of aclB were clsF (5′-AAAAACCATGGCTAAAATTCTTGAAGGG-3′; underline indicates NcoI restriction site) and aclbR. PCR conditions for aclB were 94°C for 2 min and 35 repeated cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 2 min. Two primers, aclaF and cllR (5′-AAAAACTCGAGTTACTTCTTGTCGGGAA; underline indicates XhoI restriction site), were used for amplification of aclA. PCR cycling parameters for aclA were identical to those for aclB except for the extension time, 2.5 min. Amplified aclB was inserted into the pCR-BluntII-TOPO vector (Invitrogen) and chemically transformed into E. coli DH5α. Plasmid DNA was purified from the transformants, and sequencing verified that aclB contained no point mutations. aclB in the TOPO vector was then digested with NcoI plus BamHI, purified from agarose gels using a QIAGEN gel extraction kit, and cloned into pCDF-Duet1. The resulting plasmid was named pCLS-1. PCR for aclA initially produced a faint 1.8-kb DNA band; the sample was reamplified under the same PCR conditions, and reamplified aclA was inserted into the ZeroBlunt TOPO vector, followed by transformation into E. coli DH5α. Plasmid DNA was purified from the transformants and sequenced to verify that no mutations had been generated. This aclA-containing TOPO plasmid was restricted with NdeI plus XhoI, gel purified, and inserted into the NdeI-XhoI site of pCLS-1, completing construction of plasmid pCLLCLS. Plasmid pCLLCLS, containing both the aclA and aclB genes, was chemically transformed into E. coli BL21(DE3)(pLysS) competent cells for overexpression.
Overexpression of the ACL proteins.
Overnight cultures of E. coli BL21(DE3)(pLysS) strains, harboring overexpression vectors for the C. tepidum ACL proteins, were incubated in 1 liter or 0.5 liter LB broth (containing 100 μg/ml ampicillin for expression of CLL and CLS and 50 μg/ml spectinomycin for expression of the holoenzyme, CLLCLS) at 37°C. Expression was induced by adding 0.15 to 0.30 mM isopropyl thio-β-d-galactoside (31) into cultures that reached an A600 of approximately 0.4. Induction proceeded at room temperature for 12 h. Expression levels of each of the proteins were estimated after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Enzyme assays.
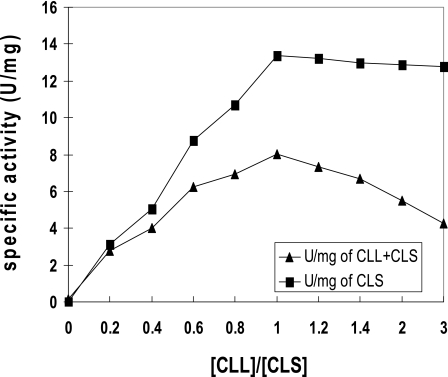
ACL activity was measured using a malate dehydrogenase-coupled assay (30). The assay was performed at room temperature in 1-ml volumes containing 100 mM Tris-HCl (pH 8.4), 20 mM sodium citrate, 10 mM dithiothreitol, 10 mM MgCl2, 0.25 mM NADH, 3.3 U of malate dehydrogenase, 0.44 mM CoA, 2.5 mM ATP, and ACL proteins. For assays of reconstituted ACL, subunit preparations were mixed and preincubated at 42°C for 5 min prior to initiating the reaction. One unit of activity was defined as 1 μmol of NADH oxidized per min in the reaction. To determine the optimum amounts (molar ratios) of subunit polypeptides required for maximum activity and holoenzyme formation during reconstitution, different amounts of CLS were added to a fixed amount of CLL (6.1 μg). The activities for each molar ratio mixture of CLS to CLL were measured at room temperature; these mixtures were also examined by nondenaturing PAGE. Both activity measurements and electrophoretic examination helped to establish the stoichiometry of CLL and CLS required for optimal reconstitution. Specific activities from the reconstitution experiments were expressed as units/mg (CLL) and as units/mg (CLS plus CLL).
Molecular weight and nondenaturing PAGE analysis of recombinant proteins.
Nondenaturing stacking gels were prepared with 4% polyacrylamide, and resolving (separating) gels contained 6% polyacrylamide in the absence of SDS and β-mercaptoethanol. After the standards and samples were loaded, gel electrophoresis was conducted at 50 V in a cold room for 4 to 5 h; the gels were then stained with Coomassie blue solution.
Protein purification.
A fast protein liquid chromatography system (Bio-Rad, Hercules, CA) was employed for conventional purification of the ACL proteins. For purification of CLS, 1-liter E. coli cultures containing overexpressed CLS were harvested by centrifugation (4,000 × g, 10 min) and resuspended in 30 ml lysis buffer (100 mM potassium phosphate buffer, pH 7.2, containing 10 mM β-mercaptoethanol [β-ME], a droplet of DNase, and 2 mM phenylmethylsulfonyl fluoride). The cell suspension was disrupted by passing the suspension twice through a French pressure cell at high pressure (15,400 lb/in2). The disrupted cell suspension was then centrifuged at low speed (17,000 × g for 20 min), and the resulting supernatant was centrifuged at high speed (100,000 × g for 1 h). To confirm solubility of CLS, supernatant and pellet fractions were examined by SDS-PAGE. The soluble CLS-containing cell extract was then filtered using 0.45-μm filters (Fisher Scientific, Pittsburgh, PA) and loaded onto a Q-Sepharose fast flow anion exchange column (9.5 by 2 cm). After loading, the column was first washed with 70 ml of buffer A (20 mM phosphate pH 7.2, 10 mM β-ME) containing 75 mM KCl; CLS was eluted after application of a 300-ml linear gradient of 75 to 400 mM KCl at a flow rate of 1 ml/min. Fractions containing CLS were collected and concentrated using an Amicon ultracentrifugal filter device (Millipore, Billerica, MA) and then loaded onto a hydroxyapatite column (11 by 2 cm). The column was washed with 75 ml of buffer A (10 mM phosphate pH 7.2, 10 mM β-ME) followed by application of a 350-ml gradient of 10 to 150 mM phosphate at a flow rate of 0.95 ml/min. CLS-containing fractions were collected, and purity was confirmed by SDS-PAGE.
For purification of CLL, a 1-liter E. coli culture containing overexpressed CLL was harvested (4,000 × g for 10 min), and the resulting cell pellet was resuspended in 30 ml of lysis buffer (100 mM potassium phosphate, pH 6.5, containing 10 mM β-ME, 1 mM PMSF, a droplet of DNase, and 25% sucrose). The CLL-containing E. coli cell extract was lysed as above, and soluble CLL (verified by SDS-PAGE) was filter sterilized and loaded onto an SP-Sepharose cation exchange column (9.5 by 1 cm) equilibrated with buffer A (20 mM potassium phosphate buffer, pH 6.45, containing 10 mM β-ME, 20% sucrose). The column was washed with 60 ml of buffer A containing 150 mM KCl, and CLL was eluted with 200 ml of a linear gradient of 150 to 250 mM KCl at a flow rate of 0.85 ml/min.
The recombinant holoenzyme was overexpressed in a 0.5-liter E. coli culture. The cell extract was prepared as described above, and the holoenzyme was first loaded onto a Q-Sepharose fast flow column. Holoenzyme was eluted with 250 ml of a 0 to 500 mM KCl gradient at a flow rate of 1.0 ml/min. Active fractions were concentrated as above and then loaded onto a Superose 6 gel filtration column (Pharmacia Biotech, Piscataway, NJ). The gel exclusion column was run at a flow rate of 0.2 ml/min with buffer A containing 0.1 M KCl. Half of the active material from the gel filtration column was then loaded onto the hydroxyapatite column described above and eluted with a 200-ml gradient of 0 to 500 mM phosphate at a flow rate of 0.9 ml/min. Homogeneity of the eluted proteins was determined by SDS-PAGE. All mutant CLS and CLL proteins were prepared similar to the wild-type proteins. Protein concentrations were estimated using commercial Bradford assay reagents (Bio-Rad).
Site-directed mutagenesis.
Point mutations were introduced into the aclA and aclB genes using the QuikChange site-directed mutagenesis kit (Stratagene) or by standard PCR, which was performed with 2.5 units of Pfu Ultra High-Fidelity DNA polymerase or PfuTurbo DNA polymerase, 5 μl 10× reaction buffer (Stratagene), 0.125 μg of two primers, 5 μl of 10× PCRx enhancer solution (Invitrogen), 0.2 mM dNTPs, and 25 to 50 ng of pCLS or pCLL in a total volume of 50 μl. Cycling parameters for PCR were 95°C for 30 s and 18 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 12 to 14 min. After PCR, 10 units of DpnI (Stratagene) was added to the PCR samples and incubated for 1 to 2 h at 37°C to restrict parental DNA strands. Then, the restricted samples were chemically transformed into E. coli DH5α or XL-1 BlueMRF′ competent cells (Stratagene). Transformants from the transformation mixture were incubated in LB medium, and plasmids were purified using a mini-prep kit (QIAGEN). The aclA and aclB genes in pET11a were sequenced, and plasmids containing the point mutations were chemically transformed into E. coli BL21(DE3)(pLysS). The oligonucleotide primers for the PCR were as follows: CLSK50N-F, 5′-CAAAACTTGTCGTCAACGCTCACGAAGCGATCG-3′; CLSK50N-R, 5′-CGATCGCTTCGTGAGCGTTGACGACAAGTTTTG-3′; CLSG56D-F, 5′-GGCTCACGAAGCGATCGACGGCCGGTTCAAGCTTG-3′; CLSG56D-R, 5′-CAAGCTTGAACCGGCCGTCGATCGCTTCGTGAGCC-3′; CLSG57D-F, 5′-CGAAGCGATCGGCGACCGGTTCAAGCTTGG-3′; CLSG57D-R, 5′-CCAAGCTTGAACCGGTCGCCGATCGCTTCG-3′; CLSR58L-F, 5′-CGAAGCGATCGGCGGCCTGTTCAAGCTTGG-3′; CLSR58L-R, 5′-CCAAGCTTGAACCGGACGCCGATCGCTTCG-3′; CLSE97A-F, 5′-GGTCATCGTGGCAGCAATGCTCGATCATGACG-3′; CLSE97A-R, 5′-CGTCATGATCGAGCATTGCTGCCACGATGACC-3′; CLSE104A-F, 5′-GCTCGATCATGACGCAGCATTCTATGTCTCC-3′; CLSE104A-R, 5′-GGAGACATAGAATGCTGCGTCATGATCGAGC-3′; CLSE188Q-F, 5′-GGACGCACAGTCTATCCAAATCAACCCGCTGGTC-3′; CLSE188Q-R, 5′-GACCAGCGGGTTGATTTGGATAGACTGTGCGTCC-3′; CLSN190K-F, 5′-CAGTCTATCGAAATCAAGCCGCTGGTCATCCGC-3′; CLSN190K-R, 5′-GCGGATGACCAGCGGCTTGATTTCGATAGACTG-3′; CLSD205V-F, 5′-CGCTGCGCTCGTCGCCGTGATGAACG-3′; CLSD205V-R, 5′-CGTTCATCACGGCGACGAGCGCAGCG-3′; CLLS176A-F, 5′-GGTGTTCTAACCAAGGCCGGCGGTCTGTCCAAC-3′; CLLS176A-R, 5′-GTTGGACAGACCGCCGGCCTTGGTTAGAACACC-3′; CLLG177F-F, 5′-GTTCTAACCAAGTCCTTCGGTCTGTCCAACGAG-3′; CLLG177F-R, 5′-CTCGTTGGACAGACCGAAGGACTTGGTTAGAAC-3′; CLLG178F-F, 5′-CTAACCAAGTCCGGCTTTCTGTCCAACGAGGCG-3′; CLLG178F-R, 5′-CGCCTCGTTGGACAGAAAGCCGGACTTGGTTAG-3′; CLLE231L-F, 5′-GTCGTCATGATCGGCTTGGTTGGCGGTAATCTC-3′; CLLE231L-R, 5′-GAGATTACCGCCAACCAAGCCGATCATGACGAC-3′. (The bold letters indicate nucleotides changed after site-directed mutagenesis; underlining indicates the specific codon.)
Kinetic studies with wild-type and mutant proteins.
Km(ATP) values for the ACL proteins were determined using at least six different ATP concentrations. The measurements were repeated three times, and Km values for each sample were determined using the Enzyme Kinetics program from Sigma Plot 8.0, which estimates Km values according to the Michaelis-Menten equation.
RESULTS
Analysis of C. tepidum ACL sequences.
Amino acid alignments between C. tepidum CLS and E. coli SucC revealed 33% overall amino acid identity; most important was the potential presence of several residues of a highly conserved ATP grasp region on CLS (Fig. 2). Despite the fact that amino acid sequences from C. tepidum CLL and E. coli SucD shared relatively low similarity, based on comparisons with SucD there also appeared to be potential conserved residues for ATP binding and/or hydrolysis on the CLL subunit of ACL (Fig. 2). The C terminus of CLL contained a domain that is not homologous to E. coli SCS and, in particular, the modified Rossman fold motif previously identified from the N terminus of E. coli SucD (5, 7) was completely absent from either CLS or CLL of C. tepidum ACL.
FIG. 2.
Alignment of amino acid sequences from C. tepidum ACL and E. coli SCS. The amino acid residues mutated in this research, as well as those previously shown to be involved with ATP binding of E. coli SCS, and the histidine residue for phosphorylation are shown in shaded backgrounds.
Further sequence comparisons indicated that residues from the C terminus of C. tepidum CLL possessed 28% identity to E. coli CS citrate synthase. Most interestingly, conserved residues potentially involved with the binding of acetyl-CoA and oxaloacetate were identified on C. tepidum CLL (Fig. 3) (20).
FIG. 3.
Alignment of amino acid sequences from C. tepidum CLL and E. coli citrate synthase (CS). The deduced amino acid residues for acetyl-CoA binding are shown with a black background, and the deduced residues for oxaloacetate binding are shown in gray boxes.
Synthesis of recombinant ACL subunit proteins.
Initially, attempts to produce significant amounts of recombinant CLL and CLS were unsuccessful when the requisite genes were induced in E. coli cells cultured at 37°C for 4 h. However, after induction of the aclA and aclB genes using E. coli-containing pET vectors at room temperature and after increasing the induction time to 12 h, significant amounts of recombinant CLL and CLS were obtained. Both CLS and holo-recombinant ACL produced from tandemly expressed aclA aclB genes were highly soluble and thus localized in the supernatant fraction after centrifugation of cell extracts. However, recombinant CLL was prone to aggregate, and most of the CLL protein was found in the particulate (insoluble) fraction of centrifuged cell lysates. It was subsequently found that harvesting aclA-induced E. coli cultures to late stationary phase (A600 of 1.8 to 2.0) provided large amounts of soluble CLL.
Purification of recombinant ACL proteins.
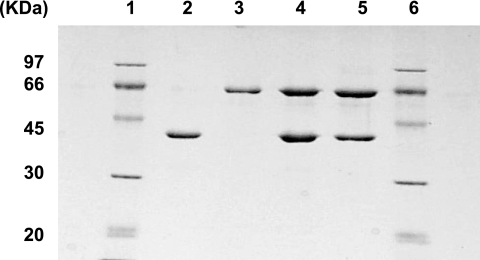
Recombinant ACL proteins were purified to homogeneity by conventional column chromatography from constructs containing the aclA and aclB genes expressed in E. coli using pET11a vectors. Attempts to produce recombinant proteins with His-tag-containing pET vectors resulted in preparations that yielded very little enzymatic activity. It was further found that the inclusion of 20 to 25% sucrose helped to prevent aggregation of CLL; thus, sucrose was included in all buffers during purification to stabilize recombinant CLL. A previous report had indicated that sucrose enhanced the stability of native holo-ACL from C. tepidum (32). After purification (Fig. 4), the expected molecular sizes of the CLL (65 kDa) and CLS (42 kDa) proteins were obtained. Recombinant mutant ACL subunit proteins were also successfully purified by these methods; however, for the K50T mutant of CLS (CLSK50T), recovery yields were substantially lower (data not shown).
FIG. 4.
Purification of ACL proteins (SDS-PAGE). Lanes 1 and 6, standard markers; lane 2, purified recombinant CLS (approximately 0.9 μg); lane 3, purified recombinant CLL (approximately 0.8 μg); lane 4, reconstituted CLS and CLL (approximately 2.7 μg); lane 5, purified recombinant holo-ACL from coexpressed aclA and aclB on a single plasmid (approximately 2 μg).
Reconstitution of ACL activity from individual purified recombinant subunit preparations.
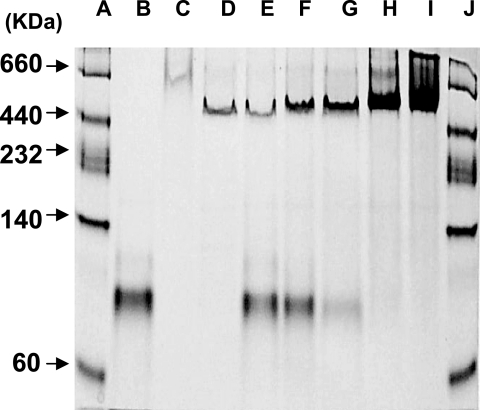
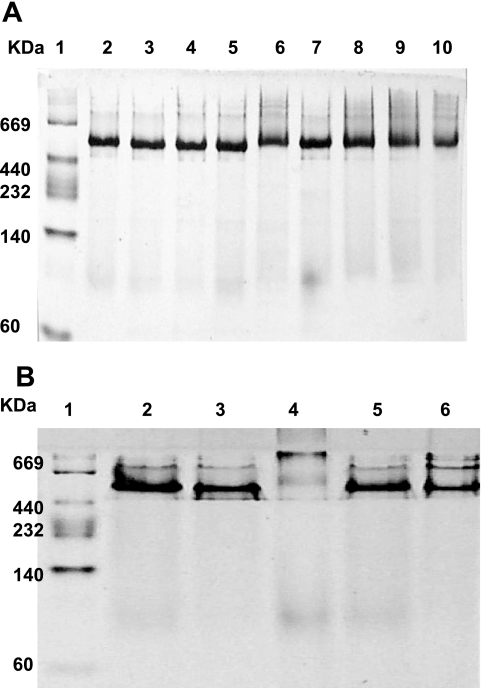
Neither purified recombinant subunit preparation alone, CLL nor CLS, possessed any enzymatic activity. However, at optimum levels of each subunit preparation, significant enzymatic activity was obtained (Fig. 5; Table 1). The CLL/CLS complex contained 72% of the activity of the recombinant holo-ACL enzyme complex prepared from tandemly expressed aclA aclB genes (Table 1). Furthermore, mixing experiments with different molar ratios of CLL and CLS indicated that maximum activity was attained at CLL/CLS molar ratios of 1 or greater (Fig. 5). This result was confirmed by nondenaturing gel electrophoresis in which CLS/CLS molar ratios of 1 or greater yielded the most stable protein preparation (Fig. 6). Unequal molar ratios of the subunits did not result in a stable single band. In addition, the apparent molecular weights of each individual CLL and CLS preparation appeared to be considerably larger than monomers, indicating that both CLL and CLS formed high-molecular-weight aggregates in the absence of the companion subunit polypeptide. From the results of these reconstitution experiments, the stable reconstituted single band on these nondenaturing gels exhibited a molecular weight of approximately 550,000, very close to the molecular weight of the recombinant holoenzyme complex (Fig. 6) and the holoenzyme isolated from C. tepidum itself (32). By contrast, complexes of certain mutant preparations, CLSK50T/CLL (data not shown) and CLS/CLLG177F complexes, failed to form a stable single band in nondenaturing gels (Fig. 7).
FIG. 5.
Titration of CLL and CLS and reconstitution of ACL activity. Maximum activity was achieved when the molar ratio of the subunits was 1:1.
TABLE 1.
Kinetic studies with purified mutant reconstituted recombinant proteins
| Protein(s) | Sp act ± SD (μmol/min/mg) | Km(ATP)a ± SD (μM) | kcat(ATP)a ± SD (s−1) |
|---|---|---|---|
| Holo-ACL | 7.88 ± 1.65 | 417 ± 59 | 17.5 ± 3.45 |
| CLL + CLS | 5.67 ± 0.23 | 382 ± 52 | 12.2 ± 0.45 |
| CLL + CLSK50T | NAb | NDc | ND |
| CLL + CLSG56D | 0.03 ± 0.005 | ND | 0.07 ± 0.01 |
| CLL + CLSG57D | 0.66 ± 0.06 | 5050 ± 570 | 1.50 ± 0.15 |
| CLL + CLSR58L | 0.07 ± 0.015 | ND | 0.13 ± 0.015 |
| CLL + CLSE97A | 4.94 ± 1.12 | 590 ± 140 | 11.3 ± 2.60 |
| CLL + CLSE104A | 1.68 ± 0.05 | 420 ± 17 | 3.9 ± 0.01 |
| CLL + CLSE188Q | 0.02 ± 0.006 | ND | 0.04 ± 0.01 |
| CLL + CLSN190K | NA | ND | ND |
| CLL + CLSD205V | 0.15 ± 0.01 | 454 ± 38 | 0.34 ± 0.02 |
| CLLS176A + CLS | 3.29 ± 0.14 | 173 ± 27 | 7.58 ± 0.37 |
| CLLG177F + CLS | 0.02 ± 0.001 | ND | 0.07 ± 0.003 |
| CLLG178F + CLS | NA | ND | ND |
| CLLE231L + CLS | NA | ND | ND |
Kinetic constants for ATP are “apparent” Km and Vmax values. The other substrates were held at fixed concentrations for the kinetic determinations.
NA, no activity.
ND, not determined.
FIG. 6.
Nondenaturing gel analysis after titration of the subunits. Lanes: A and J, standard molecular mass markers; B, CLS; C, CLL; D, holo-ACL; E, [CLL]/[CLS] of 0.125; F, [CLL]/[CLS] of 0.25; G, [CLL]/[CLS] of 0.5; H, [CLL]/[CLS] of 1.0; I, [CLL]/[CLS] of 2.0. Approximately 3 μg of CLS and different amounts of CLL were reconstituted and loaded onto the gel. Approximately 3.5 μg of the recombinant holo-ACL was loaded into lane D.
FIG. 7.
Nondenaturing PAGE analysis for enzyme reconstituted from mutant subunits. A. Lanes: 1, standard molecular mass markers; 2, CLL plus CLS; 3, CLL plus CLSG56D; 4, CLL plus CLSG57D; 5, CLL plus CLSR58L; 6, CLL plus CLSE97A; 7, CLL plus CLSE104A; 8, CLL plus CLSE188Q; 9, CLL plus CLSN190K; 10, CLL plus D205V. B. Lanes: 1, standard molecular mass markers; 2, CLL plus CLS; 3, CLLS176A plus CLS; 4, CLLG177F plus CLS; 5, CLLG178F plus CLS; 6, CLLE231L plus CLS. Approximately 10 μg of the reconstituted proteins was loaded onto the gel.
Properties of mutant forms of CLS and CLL.
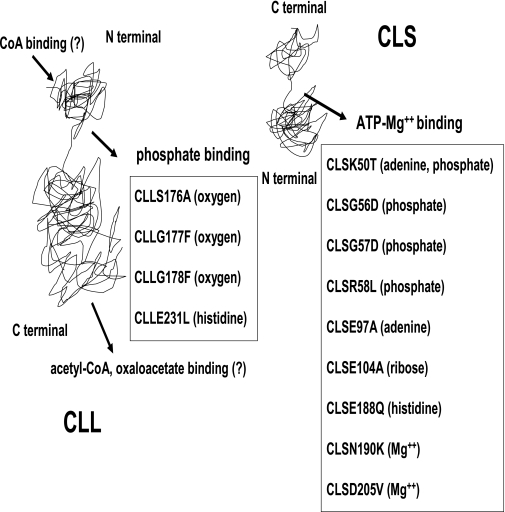
Having established a valid reconstitution system with individual recombinant CLL and CLS polypeptides, it became feasible to determine the functional roles of each subunit in generating the active site of ACL. Based on sequence homologies with known motifs important for ATP binding and other functions, as well as previous X-ray crystallography studies with E. coli SCS (15, 35), various residues on each subunit of ACL were chosen to be modified by site-directed mutagenesis. Based on these comparisons, Lys-50 and Glu-97 on CLS (equivalent to E. coli SCS46β and E. coli SCSE99β on the small subunit of succinyl-CoA synthetase, respectively) would be expected to bind to the adenine ring of ATP. In addition, it was expected that Glu-104 on CLS (equivalent to E. coli SCSE107β) would bind to the ribose ring of ATP (15). Furthermore, similar comparisons of residues on CLS suggested that the γ-phosphate moiety of ATP may bind to Lys-50 as well as Gly-56 (equivalent to E. coli SCSG52β), Gly-57 (equivalent to E. coli SCSG53β), and Arg-58 (equivalent to E. coli SCSG54β). With respect to the possible coordination between magnesium ions, which are essential for ACL catalysis (13), similar comparative analyses suggest the importance of Asn-190 (equivalent to E. coli SCSN199β) and Asp-205 (equivalent to E. coli SCSD213β) in C. tepidum CLS.
With respect to the catalytically significant histidine residue (His-273), the presumed site for phosphorylation (17), this residue is located on CLL of C. tepidum ACL. This residue is equivalent to E. coli SCS231α. The resulting phosphohistidine structure must be stabilized prior to formation of the protein-bound citryl-phosphate intermediate. Among potential deduced amino acids involved in stabilizing this intermediate are residues from both subunits, Glu-188 of CLS (equivalent to E. coli SCS197β) and Glu-231 of CLL (equivalent to E. coli SCS209α) (8), which are presumed to interact with the imidazole ring of His-273 on CLL (equivalent to E. coli SCS231α). With respect to binding to the oxygen atoms of the phosphate moiety of the phosphohistidine, residues Ser-176 of CLL (equivalent to E. coli SCS154α), Gly-177 of CLL (equivalent to E. coli SCS155α), and Gly-178 of CLL (equivalent to E. coli SCS156α) are predicted to be important (35).
Each of the residues predicted to be significant as discussed above were changed by site-directed mutagenesis, and the mutant recombinant proteins were purified and subsequently reconstituted with purified wild-type polypeptides of the opposite subunit. Kinetic parameters, including specific activity, Km(ATP), and kcat, were calculated from the activities of the reconstituted ACL complex (Table 1). The functional properties for the reconstituted CLSE97A/CLL complex were similar to the wild-type CLS/CLL complex, with the possible exception of a slightly higher Km(ATP). The CLSE104A/CLL complex yielded 30% of the activity of the ACL activity of the wild-type complex, with a slightly higher difference in the Km(ATP) value of the complex compared to that of wild-type ACL. A much more drastic effect was noted for the CLSG57D mutation, as this protein when combined with wild-type CLL decreased the activity to 12% of the wild-type CLS/CLL complex. Moreover, this drastic effect on activity also resulted in a 13-fold increase in the Km(ATP). The CLSG56D, CLSR58L, CLSE188Q, and CLSD205V mutant proteins all yielded extremely low reconstituted activities, from 1 to 3% of the activity of the wild-type complex. These low activities precluded making accurate Km(ATP) measurements. The CLSK50T/CLL and CLSN190K/CLL complexes did not yield any ACL activity whatsoever. With respect to mutations on CLL, the CLS/CLLG177F, CLS/CLLE231L, and CLS/CLLG178F complexes also failed to yield significant ACL activity, while the CLLS176A/CLS enzyme yielded 58% of the wild-type complex activity but nearly a twofold decrease in the Km(ATP) (Table 1).
DISCUSSION
In this investigation an in vitro reconstitution system, using individual recombinant subunit preparations, was successfully developed for the analysis of C. tepidum ACL. This reconstitution system allowed facile study of the roles of each subunit during catalysis. Indeed, no activity whatsoever was obtained using CLL or CLS alone, and significant ACL activity required both CLS and CLL such that these individual polypeptides formed a stoichiometric CLS/CLL complex. Collectively, these results strongly suggested that the two subunits of C. tepidum ACL associate to form the active site of this enzyme. In all cases, reconstituted complexes that contained site-directed mutations in either CLS or CLL were examined using nondenaturing gels to determine if any of the mutations grossly modified the holo-ACL structure. With the exception of the reconstituted complexes containing CLSK50T (data not shown) and CLLG177F, these analyses demonstrated no observable alteration of the structure, such that the resultant holoenzyme complexes migrated on nondenaturing gels similar to the wild-type ACL complex. Residues Lys-50 and Gly-177 might thus be responsible for subunit interactions important for the stabilization of the CLS/CLL complex; thus, the effects of these residues need to be considered apart from any specific role in catalysis.
Since ACL activity resulted from the obligatory interaction of the two subunits, site-directed mutagenesis experiments were initiated to confirm the role of likely residues on each subunit, in particular amino acids involved in ATP binding and phosphohistidine stabilization. For these studies, advantage was taken of homologous residues implicated for similar functions on the two subunits of E. coli SCS, with subsequent kinetic studies with mutant ACL proteins confirming the roles of specific residues involved in ATP binding. For example, by analogy to E. coli SCS (14), Gly-57 of CLS and Arg-58 of CLS were assumed to be involved in transferring the γ-phosphate group of ATP to the key histidine residue (His-273) in the active site of ACL. The drastically lowered ACL activities from the CLSG57D and CLSR58L mutations, as well as the greatly increased Km(ATP) for the CLSG57D protein, after reconstitution with wild-type CLL, strongly suggested that some key function such as phospho-transfer or ATP binding was definitely impaired in reconstituted mutant CLS/wild-type CLL complexes. Further site-directed mutations of the neighboring residue, Gly-56, to yield the CLSG56D mutant protein also severely reduced ACL activity although its role in catalysis is not apparent, since its resultant reconstituted activity with CLL, much like for the CLSR58L protein, was too low for accurate kinetic studies. The substantial loss of activity exhibited by the CLSN190K and CLSD205V proteins, again by analogy to E. coli SCS (Asn-199β and Asp-213β, respectively), implies that Asn-190 and Asp-205 of CLS are most likely involved with the coordination of magnesium ions.
In summary, those mutations of amino acids that stabilized the phosphohistidine in the active site also had significant effects on kinetic parameters. The substantially lower kcat values for the specific mutant CLS proteins suggest that residues on CLS also stabilize the phosphohistidine structure of the CLS/CLL complex and that this function is indispensable for effective ACL catalysis.
The CLSE104A, CLSD205V, and CLLS176A mutants, when complexed with their respective wild-type subunit polypeptide, all exhibited significantly reduced kcat values; however, none of these proteins possessed greatly changed Km(ATP) values. At this point, the role of these residues in catalysis is not understood. As for a specific role for Glu-97 of CLS, this residue was considered to be important for ACL catalysis since the equivalent residue in E. coli SCS not only binds to the nucleotide but also is considered to be a conserved residue for nucleotide specification (7, 14). However, after preparing the CLSE97A/CLL complex, there was no significant reduction in kcat, indicating that Glu-97 of CLS might not be a critical residue for ATP grasp and ACL activity.
Mutations in CLL also affected reconstitution activity, i.e., the CLLG177F, CLLG178F, and CLLE231L mutants all drastically affected the kcat of the resultant complex with wild-type CLS. Of particular interest is the CLLS176A mutant, where the kcat was reduced about 40%; however, the Km(ATP) was improved over twofold compared to the wild-type CLS/CLL complex. Again, these studies reiterate the involvement of residues from both subunits to reconstitute a functional active site. Based on the current studies, and via consideration of E. coli SCS, a model for the ability of both the CLS and CLL proteins of the complex to reconstitute a full ACL active site might be envisioned (Fig. 8). Certainly, further structural work will be necessary to fully understand the stoichiometry and nature of CLS and CLL interactions, other than the fact that our experimental results clearly indicate a 1:1 ratio of CLS and CLL for maximum reconstitution activity and structural stability.
FIG. 8.
Schematic representation of catalytically important residues from each separate subunit of C. tepidum ATP-citrate lyase.
Finally, as studies continue with this complicated enzyme, which is involved in multiple partial reactions, it will be important to examine the role of potential sites that interact with other substrates, cofactors, and products, including coenzyme A, acetyl-CoA, and oxaloacetate. Putative binding motifs for these ligands may also be deduced by first comparing known motifs specific to these metabolites to sequences found on CLS and CLL. From such bioinformatics analyses, the CoA binding mechanism for C. tepidum ACL is likely to be much different from E. coli SCS. In E. coli SCS, the modified Rossman motif for CoA binding is located at the N terminus of SucD; however, this motif is missing in the N terminus of C. tepidum CLL, perhaps substantiating the recent indication that diverse structures for CoA binding motifs exist in various enzymes (5). However, conserved binding motifs for acetyl-CoA and oxaloacetate suggest that C. tepidum ACL activity might be regulated by its two final products. A previous study with the Chlorobium limicola enzyme indicated that catalysis proceeds only in the direction toward synthesis of acetyl-CoA and oxaloacetate from citrate and CoA. There was no reversibility exhibited for this enzyme (16), unlike other prokaryotic ACL enzymes (2, 27). If this were also true for C. tepidum ACL, then it would appear that the indicated binding motifs are not involved with the biosynthesis of citrate and CoA. Since earlier reports with both eukaryotic and prokaryotic ACL indicated that the enzyme could be inhibited by oxaloacetate, this metabolic product might play a significant role in ACL activity regulation (16, 19) as well as control carbon flux through the RTCA cycle.
Acknowledgments
This work was supported by DOE grant DE-FG02-91-ER20033.
REFERENCES
- 1.Atomi, H. 2002. Microbial enzymes involved in carbon dioxide fixation. J. Biosci. Bioeng. 94:497-505. [DOI] [PubMed] [Google Scholar]
- 2.Beh, M., G. Strauss, R. Huber, K. O. Stetter, and G. Fuchs. 1993. Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 160:306-311. [Google Scholar]
- 3.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elshourbagy, N. A., J. C. Near, P. J. Kmetz, G. M. Sathe, C. Southan, J. E. Strickler, M. Gross, J. F. Young, T. N. C. Wells, and P. H. E. Groot. 1990. Rat ATP citrate-lyase. Molecular cloning and sequence analysis of a full-length cDNA and mRNA abundance as a function of diet, organ, and age. J. Biol. Chem. 265:1430-1435. [PubMed] [Google Scholar]
- 5.Engel, C., and R. Wierenga. 1996. The diverse world of coenzyme A binding proteins. Curr. Opin. Struct. Biol. 6:790-797. [DOI] [PubMed] [Google Scholar]
- 6.Evans, M. C. W., B. B. Buchanan, and D. I. Arnon. 1966. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 55:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser, M. E., M. N. G. James, W. A. Bridger, and W. T. Wolodko. 1999. A detailed structural description of Escherichia coli succinyl-CoA synthetase. J. Mol. Biol. 285:1633-1653. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, M. E., M. A. Joyce, D. G. Ryan, and W. T. Wolodko. 2002. Two glutamate residues, Glu 208α and Glu 197β, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase. Biochemistry 41:537-546. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs, G., E. Stupperich, and G. Eden. 1980. Autotrophic CO2 fixation in Chlorobium limicola. Evidence for the operation of a reductive tricarboxylic acid cycle in growing cells. Arch. Microbiol. 128:64-71. [Google Scholar]
- 10.Galperin, M. Y., and E. V. Koonin. 1997. A diverse superfamily of enzymes with ATP dependent carboxylate-amine/thiol ligase activity. Protein Sci. 6:2639-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hügler, M., H. Huber, K. O. Stetter, and G. Fuchs. 2003. Autotrophic CO2 fixation pathways in archaea (Crenarchaeota). Arch. Microbiol. 179:160-173. [DOI] [PubMed] [Google Scholar]
- 12.Hügler, M., C. O. Wirsen, G. Fuchs, C. D. Taylor, and S. M. Sievert. 2005. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ɛ subdivision of proteobacteria. J. Bacteriol. 187:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanovsky, R. N., N. V. Sintsov, and E. N. Konsratieva. 1980. ATP-linked citrate lyase activity in the green sulfur bacterium Chlorobium limicola forma thiosulfatophilum. Arch. Microbiol. 128:239-241. [Google Scholar]
- 14.Joyce, M. A., M. E. Fraser, E. R. Brownie, M. N. G. James, W. A. Bridger, and W. T. Wolodko. 1999. Probing the nucleotide-binding site of Escherichia coli succinyl-CoA synthetase. Biochemistry 38:7273-7283. [DOI] [PubMed] [Google Scholar]
- 15.Joyce, M. A., M. E. Fraser, M. N. G. James, W. A. Bridger, and W. T. Wolodko. 2000. ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by X-ray crystallography. Biochemistry 39:17-25. [DOI] [PubMed] [Google Scholar]
- 16.Kanao, T., T. Fukui, H. Atomi, and T. Imanaka. 2001. ATP-citrate lyase from the green sulfur bacterium Chlorobium limicola is a heteromeric enzyme composed of two distinct gene products. Eur. J. Biochem. 268:1670-1678. [PubMed] [Google Scholar]
- 17.Kanao, T., T. Fukui, H. Atomi, and T. Imanaka. 2002. Kinetic and biochemical analyses on the reaction mechanism of a bacterial ATP-citrate lyase. Eur. J. Biochem. 269:3409-3416. [DOI] [PubMed] [Google Scholar]
- 18.Linn, T. C., and P. A. Srere. 1979. Identification of ATP citrate lyase as a phosphoprotein. J. Biol. Chem. 254:1691-1698. [PubMed] [Google Scholar]
- 19.Pentyala, S. N., and W. B. Benjamin. 1995. Effect of oxaloacetate and phosphorylation on ATP-citrate lyase activity. Biochemistry 34:10961-10969. [DOI] [PubMed] [Google Scholar]
- 20.Pereira, D. S., L. J. Donald, D. J. Hosfield, and H. W. Duckworth. 1994. Active site mutants of Escherichia coli citrate synthase. Effects of mutations on catalytic and allosteric properties. J. Biol. Chem. 269:412-417. [PubMed] [Google Scholar]
- 21.Pierce, M. W., J. L. Palmer, H. T. Keutmann, and J. Avruch. 1981. ATP-citrate lyase. Structure of a tryptic peptide containing the phosphorylation site directed by glucagon and the cAMP-dependent protein kinase. J. Biol. Chem. 256:8867-8870. [PubMed] [Google Scholar]
- 22.Potapova, I. A., M. R. El-Maghrabi, S. V. Doronin, and W. B. Benjamin. 2000. Phosphorylation of recombinant human ATP: citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry 39:1169-1179. [DOI] [PubMed] [Google Scholar]
- 23.Ramakrishna, S., G. D'Angleo, and W. B. Benjamin. 1990. Sequence of sites on ATP-citrate lyase and phosphatase inhibitor 2 phosphorylated by multifunctional protein kinase (a glycogen synthase kinase 3 like kinase). Biochemistry 29:7617-7624. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2003. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sato, R., A. Okamoto, J. Inoue, W. Miyamoto, Y. Sakai, N. Emoto, H. Shimano, and M. Maeda. 2000. Transcriptional regulation of the ATP citrate-lyase gene by sterol regulatory element-binding proteins. J. Biol. Chem. 275:12497-12502. [DOI] [PubMed] [Google Scholar]
- 26.Schäfer, S., C. Barkowski, and G. Fuchs. 1986. Carbon assimilation by the autotrophic thermophilic archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 146:301-308. [Google Scholar]
- 27.Shauder, R., F. Widdel, and G. Fuchs. 1987. Carbon assimilation pathways in sulfate-reducing bacteria. II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter thermophilus TK-6. Arch. Microbiol. 148:218-225. [Google Scholar]
- 28.Shiba, H., T. Kawasumi, Y. Igarashi, T. Kodama, and Y. Minoda. 1985. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141:198-203. [Google Scholar]
- 29.Singh, M., E. G. Richards, A. Mukherjee, and P. A. Srere. 1976. Structure of ATP citrate lyase from rat liver. Physicochemical studies and proteolytic modification. J. Biol. Chem. 251:5242-5250. [PubMed] [Google Scholar]
- 30.Srere, P. A. 1959. The citrate cleavage enzyme. I. Distribution and purification. J. Biol. Chem. 234:2544-2547. [PubMed] [Google Scholar]
- 31.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 32.Wahlund, T. M., and F. R. Tabita. 1997. The reductive tricarboxylic acid cycle of carbon dioxide assimilation: initial studies and purification of ATP-citrate lyase form the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 179:4859-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh, C. T. 1979. Enzymatic reaction mechanisms. W.H. Freeman & Co., New York, N.Y.
- 34.Wells, T. N. C. 1991. ATP-citrate lyase from rat liver. Characterization of the citryl-enzyme complexes. Eur. J. Biochem. 199:163-168. [DOI] [PubMed] [Google Scholar]
- 35.Wolodko, W. T., M. E. Fraser, M. N. G. James, and W. A. Bridger. 1994. The crystal structure of succinyl-CoA synthetase from Escherichia coli at 2.5-Å resolution. J. Biol. Chem. 269:10883-10890. [DOI] [PubMed] [Google Scholar]