Abstract
Vibrio cholerae has multiple iron acquisition systems, including TonB-dependent transport of heme and of the catechol siderophore vibriobactin. Strains defective in both of these systems grow well in laboratory media and in the infant mouse intestine, indicating the presence of additional iron acquisition systems. Previously uncharacterized potential iron transport systems, including a homologue of the ferrous transporter Feo and a periplasmic binding protein-dependent ATP binding cassette (ABC) transport system, termed Fbp, were identified in the V. cholerae genome sequence. Clones encoding either the Feo or the Fbp system exhibited characteristics of iron transporters: both repressed the expression of lacZ cloned under the control of a Fur-regulated promoter in Escherichia coli and also conferred growth on a Shigella flexneri mutant that has a severe defect in iron transport. Two other ABC transporters were also evaluated but were negative by these assays. Transport of radioactive iron by the Feo system into the S. flexneri iron transport mutant was stimulated by the reducing agent ascorbate, consistent with Feo functioning as a ferrous transporter. Conversely, ascorbate inhibited transport by the Fbp system, suggesting that it transports ferric iron. The growth of V. cholerae strains carrying mutations in one or more of the potential iron transport genes indicated that both Feo and Fbp contribute to iron acquisition. However, a mutant defective in the vibriobactin, Fbp, and Feo systems was not attenuated in a suckling mouse model, suggesting that at least one other iron transport system can be used in vivo.
Iron is essential for nearly all life, and gram-negative bacterial pathogens have evolved numerous systems for the acquisition of this element. Many of these systems involve the synthesis of outer membrane receptors that bind a specific ligand and transport it across the outer membrane. Some of these receptors transport ferrisiderophores, which are small, iron-binding molecules synthesized by microorganisms. Other receptors recognize specific iron-containing complexes synthesized by their hosts, such as lactoferrin, transferrin, heme, or hemoglobin. Heme receptors transport the intact heme moiety into the periplasm, whereas receptors for host iron proteins remove the iron or heme from the protein at the cell surface and only the iron or heme is transported into the cell (9, 47, 55). Energy for transport across the outer membrane requires an active TonB-ExbBD complex (40). Following transport into the periplasm, the iron ligand is bound by a specific periplasmic binding protein, which delivers its ligand to an ATP binding cassette (ABC) transport system located in the inner membrane. This system usually consists of two integral membrane proteins, which form the channel, and two hydrophilic proteins with ATPase activity. This ATP hydrolysis provides the energy for transport of the ligand across the inner membrane into the cytoplasm (10).
Although iron is essential, high levels of iron are toxic. For this reason, the expression of iron acquisition genes is tightly regulated. In gram-negative bacteria, iron homeostasis is maintained primarily through the action of the regulatory protein Fur. Under iron-replete conditions, ferri-Fur binds to the promoter of iron-regulated genes and prevents their expression. Under low-iron conditions, apo-Fur is released from the DNA, and genes of the Fur regulon are expressed (4, 19).
Vibrio cholerae, the causative agent of the severe diarrheal disease cholera, requires iron and must obtain this element in each of the environments it inhabits. A number of iron acquisition systems have been characterized for V. cholerae, including those for the synthesis and transport of the catechol siderophore vibriobactin (7, 15, 64, 65), the transport and utilization of heme (22, 33, 37, 63), and the transport of several siderophores made by other microorganisms, including ferrichrome (15, 44) and enterobactin (35, 65). In addition, two sets of tonB exbBD genes have been identified (37), and these systems have partially redundant functions (49). V. cholerae encodes a Fur protein, which regulates the genes for iron acquisition (29, 34).
In this report we identified uncharacterized potential iron uptake systems in the published V. cholerae genome sequence (21). We show that two of these systems, an ABC transport system termed Fbp and an apparent Feo system, transport ferric and ferrous iron, respectively. Mutational analysis of V. cholerae suggests that additional, uncharacterized iron transport systems are present in its genome.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and iron utilization assays.
The strains and plasmids used in this study are listed in Table 1. The iron chelator ethylenediamine di(ortho-hydroxyphenylacetic acid) (EDDA) was deferrated by the method of Rogers (43). The antibiotic concentrations used were 250 μg/ml carbenicillin, 50 μg/ml kanamycin, and 50 μg/ml (for Escherichia coli) or 5 μg/ml (for V. cholerae) chloramphenicol. The bioassay for siderophore utilization was performed as previously described (64). For the growth of strain SM193w, the medium was supplemented with sterile culture supernatant of Shigella flexneri strain SA101 as a source of the siderophore aerobactin as previously described (46).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference(s) or source |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| O395 | Classical biotype | 31 |
| ALV101 | O395 ΔvibB | This study |
| ALV102 | O395 ΔvibB feo::tmp | This study |
| ARM591 | O395 ΔvibB fbp::cam | This study |
| CFV1 | O395 ΔvibB fbp::cam feo::tmp | This study |
| EWV125 | O395 ΔvibB fbp::cam feo::tmp VCA0685-0686::kan | This study |
| CA401 | Classical biotype | 14 |
| Lou15 | El Tor biotype | 52 |
| E. coli | ||
| DH5α | Cloning strain | 16 |
| H1771 | MC4100 aroB feoB7 fhuF::λplac Mu | 18, 24 |
| ARM100 | W3110 tonB::kan entF::cam | 32 |
| S. flexneri | ||
| SM193 | SM100 iuc::Tn5 feo::tmp sitA::cam | 46 |
| SM193w | Avirulent derivative of SM193 | This study |
| SA101 | Avirulent derivative of SA100 | 28 |
| Plasmids | ||
| pGEM-T Easy | High-copy-number cloning vector | Stratagene |
| pWKS30 | Low-copy-number cloning vector | 61 |
| pMTLcam | cam resistance gene | 62 |
| pUC4K | kan resistance gene | Pharmacia |
| pHM5 | pGP704 carrying sacB | 45 |
| pCVD442 | pGP704 carrying sacB | 11 |
| pCVD442N | pCVD442 with a NotI adaptor in the SacI site | This study |
| pUH18E | E. coli feo genes in pT7-5 | 24 |
| pFbp1 | pWKS30 carrying fbpABC | This study |
| pFeo101 | pWKS30 carrying feoABC | This study |
| pVfu4 | pWKS30 carrying VCA0685-0687 | This study |
| pVtu100 | pWKS30 carrying VCA0601-0603 | This study |
Colony size assays were performed by diluting overnight cultures 10-fold into LB broth. Cultures were grown until late exponential phase and then diluted and plated at low density on LB agar or LB agar containing added iron or EDDA as indicated. Colony diameters were measured after incubation at 37°C for 24 h. Data are the averages of results for 10 colonies.
Sequence analysis and gene nomenclature.
DNA sequencing was performed by the University of Texas Institute for Cellular and Molecular Biology DNA Core Facility using an Applied Biosystems Prism 377 DNA sequencer (Perkin-Elmer Corp.). Amino acid sequence alignments were performed using the ClustalW feature of the MacVector package (38). The BLAST program (2) was used to search the National Center for Biotechnology Information (NCBI) protein database. Some DNA sequence was obtained from The Institute for Genomic Research (TIGR) website (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?database=gvc) (21).
Plasmid construction.
To construct pFeo101, the feoABC genes of V. cholerae strain Lou15 were amplified with Pfx DNA polymerase (Invitrogen) using primers Feo102 (5′ CAAGAAAACCTTACCCTGCGTATG) and Feo103 (5′ GCGGCACCTTCAAGTTGGAAG). The resulting PCR product was gel purified and cloned into the SmaI site of pWKS30. pFbp1 was constructed by amplification of the fbpABC genes from Lou15 with Pfx polymerase using the primers Fbp100 (5′ GAAGAAGAGATGGAAGGGATGGAC) and Fbp101 (5′ ATGAAGATGGGGCGAAAGTCTAC). The PCR product was cloned into the SmaI site of pWKS30. pVfu4 was constructed by amplification of the VCA0685-0687 genes of Lou15 using the primers VCV12 (5′ CTACCCCATGTCACTGGCTATTG) and VCV13 (5′AGCTTAAAAGTAACGCCACCTCAG). The PCR fragment was cloned into the SmaI site of pWKS30. pVtu100 is the PCR fragment generated by amplification of Lou15 DNA using Pfx polymerase and the primers vtu1 (5′ CGGTGTCAGCAGTAGATAAGCATTC) and vtu2 (5′ TTGAGTGGGGTCATAGCGTAAAG) cloned into the SmaI site of pWKS30. pCVD442N was constructed by self-annealing the adaptor oligonucleotide 5′ TAGCGGCCGCTAAGCT and inserting it into the SacI site of pCVD442.
Mutant construction.
SM193w is a spontaneous mutant of S. flexneri strain SM193 (46) that fails to bind Congo red and is noninvasive. Characterization of this mutant indicated that it has the same iron transport defects found in its parental strain. All site-directed mutants were constructed by cloning the genes as described below into a suicide vector containing the sacB gene. These plasmids were then mated into the desired V. cholerae strain, and allelic exchange mutations were obtained as previously described (33). To construct mutations in vibB, the region upstream of the V. cholerae strain O395 vibB was amplified using Taq DNA polymerase with the primers Liz 256 (5′ GCTTTGGTGGAAGGGGATTATG) and Liz 257 (5′ TTGATATCCAGCCAACTGAGC) and the region downstream of vibB was amplified using Liz 258 (5′ GGCTGGATATCAACCTTGCTCTGCGTGAT) and Liz 259 (5′ GTCGACGCTATTTTTGTCAGCACAGCC). The two resulting PCR products were gel purified and used as template in a PCR using the primers Liz 256 and Liz 259. The product of this reaction was cloned into pGEM-T Easy (Promega). The SalI fragment carrying the insert was subcloned into the SalI site of pHM5 to create the plasmid pSvibBΔ. To construct strains containing feoB mutations, the feo region from V. cholerae strain CA401 was PCR amplified using the primers feofor (5′ GCATAGGCGGATTTGTGTGATTAG) and feorev (5′ CACCAGGATAGAGGGTCTTTTTCAG). The product was cloned into pGEM-T Easy, and a trimethoprim cassette was inserted into the BclI site. The insert was excised as a NotI fragment, made blunt with Klenow, and inserted into the EcoRV site of pHM5 to create pSfeoB::tmp. Mutations in the VCA0685-0687 genes were constructed by PCR amplification of the VCA0685-0687 region of strain Lou15 with Taq polymerase and the primers VCV10 (5′ GCTTTGGCTCTGTTATCGCATC) and VCV11 (5′ GAGGTAGTTTTTGAAGGTCAGGGTG). The PCR fragment was cloned into pGEM-T Easy. In the resulting plasmid, the region between the BstZ17I and PshAI sites, which contains most of VCA0685 and a portion of VCA0686, was replaced with a HincII fragment containing the kanamycin gene from pUC4K. The NotI fragment containing VCA0685-0687::kan was subcloned into the NotI site of pCVD442N to create pVfu3. The FbpA mutation was constructed by PCR amplification of V. cholerae O395 DNA using Taq polymerase and the primers FbpA.1 (5′ TCGATAACCCGGGAGGTGGC) and FbpA.2 (5′ GAATGCGGTCTAGAAGATCGC). The fragment was cloned into pGEM-T Easy. The cam cassette from pMTLcam was inserted into the NruI site within fbpA. The fbpA::cam gene was then inserted as an XbaI/SmaI fragment into the XbaI/EcoRV sites of pHM5 to create pSfbpA::cam.
Iron transport assays.
S. flexneri strain SM193w containing the indicated plasmid was grown overnight in LB broth supplemented with 1/10 volume of sterile supernatant from S. flexneri strain SA101 as a source of aerobactin. Overnight cultures were diluted 1/25 into MM9 minimal medium supplemented with SA101 culture supernatant and grown to late exponential phase with aeration at 37°C. Cells were pelleted, washed one time in transport buffer, and resuspended in transport buffer at a concentration of 1.5 (optical density at 650 nm) per ml of buffer. The transport buffer contained the following per liter: 6 g sodium acetate, 3.15 g dibasic sodium phosphate, 2.05 g monobasic sodium phosphate, 0.6 g ammonium sulfate, 0.44 g monobasic potassium phosphate, and 0.232 g dibasic potassium phosphate (12). The buffer was prepared as a 10× stock, treated with Chelex 100, and stored frozen. It was supplemented with MgCl2 (final concentration, 0.39 μM) and KOH (final concentration, 10 mM) immediately prior to use. 55Fe (Perkin-Elmer Life and Analytical Sciences, Boston, MA) was diluted into 1× transport buffer and filtered through a Microcon filter (Millipore, Bedford, MA) to remove iron that precipitated during storage. The transport reaction was initiated by the addition of approximately 3.4 μCi of 55Fe per ml of resuspended cells, and 100-μl samples were removed at the indicated times. Cells from each sample were collected by filtration through GN-6 Metricel membrane filters (Pall Life Sciences, Ann Arbor, MI) and washed with 15 ml of citrate wash buffer (100 mM sodium citrate, 1 mM MgCl2, 0.25 mM CaCl2) (39). The filters were dried, and the amount of radioactive iron was determined by scintillation counting. Transport assays were performed in triplicate at room temperature and on ice. The values obtained for the assays performed on ice were subtracted from the values obtained with the same strain at room temperature to correct for iron nonspecifically bound to the cell surface.
In vivo competition assays.
In vivo competition assays were performed by the method of Taylor et al. (58). Strains were grown to mid-log phase, and approximately 5 × 105 CFU in 50 μl of saline solution containing 0.5% (wt/vol) sucrose and 0.02% (wt/vol) Evans blue dye was inoculated intragastrically into prestarved 5-day-old BALB/c mice. After 24 h, the small intestines were removed and homogenized in sterile phosphate-buffered saline. The titer of each of the competing strains was determined, and the competitive index (CI) was calculated using the formula CI = (mutant output/wild-type output)/(mutant input/wild-type input). For each strain pair, we report the average and standard deviation calculated for an experiment that included at least five mice.
RESULTS
Identification of potential iron transport systems.
The V. cholerae genome (21) was scanned for uncharacterized genes that potentially encode iron transporters. One set of these genes appeared to encode a member of the Feo family of ferrous iron transporter systems. The Feo system consists of at least two genes, feoA and feoB, which encode, respectively, a small protein of unknown function and a GTPase essential for transport activity (18, 24). In some organisms, an additional small open reading frame (ORF) referred to as yhgG, or feoC, is present immediately downstream of feoB (17). The significance of this ORF is unknown. V. cholerae FeoA (VC2078) has a molecular weight of approximately 8,300 and a predicted pI of 9.4. It has 40% amino acid identity with E. coli FeoA (24), as well as significant similarity with FeoA proteins of other organisms. FeoB (VC2077) has a molecular weight of about 83,000 and a pI of 6.1. It has 40% identify with E. coli FeoB, including conservation of all of the G protein signature motifs (24, 30). A small ORF (VC2276), initiating with a GTG that overlaps the feoB stop codon, is also present. We refer to this ORF as feoC, although the predicted protein has limited similarity (11% amino acid identity and 30% similarity) with E. coli FeoC. Upstream of feoA is an apparent Fur box, and transcription of feoABC is regulated by Fur and iron (34).
The V. cholerae genome contains numerous uncharacterized periplasmic binding protein-dependent ABC transport system genes. One of these systems, called Fbp, consists of three genes. fbpA (VC0608) encodes the apparent periplasmic binding protein, while fbpB (VC0609) and fbpC (VC0610) appear to encode the inner membrane permease and ATPase components, respectively. Upstream of fbpA is a potential Fur box, and regulation of fbpA mRNA by Fur and iron was observed by microarray analysis (34). fbpB and fbpC appear to be transcribed separately from fbpA. The promoter region for the fbpBC transcript does not contain a Fur box, and the fbpBC genes are not significantly regulated by Fur or iron (34). The fbpABC genes have high amino acid similarity with many genes annotated as iron transporters. The most closely related system for which the function has been experimentally determined is the Fbp system of Mannheimia haemolytica (previously called Pasteurella haemolytica) (26, 50, 51). V. cholerae FbpA, FbpB, and FbpC have 60%, 52%, and 48% amino acid identity with FbpA, FbpB, and FbpC of M. haemolytica, respectively. The M. haemolytica fbpABC genes promoted growth of an E. coli mutant deficient in siderophore production under iron-depleted conditions (26), and the crystal structure of FbpA bound to ferric iron has been obtained (50, 51).
Two other ABC transport systems that are annotated as ferric iron transporters were evaluated in some assays. One of these, encoded by VCA0685-0687 (21), is similar to the AfuABC system of Actinobacillus pleuropneumoniae, which stimulated the growth of an E. coli siderophore mutant in the presence of an iron chelator (8). VCA0685, VCA0686, and VCA0687 have 69%, 65%, and 60% amino acid identity with AfuA, AfuB, and AfuC, respectively. No Fur box is apparent in the promoter regions, and none of the VCA0685-0687 genes was regulated by Fur or iron (32). A similar system is present in E. coli O157:H7 and other organisms, but the function of these genes in these organisms has not been characterized. The distribution of genes similar to the VCA0685-0687 system does not correlate well with the phylogenetic relationships, suggesting that horizontal transfer of these genes has occurred.
Another ABC transport system investigated in this study is encoded by the VCA0601-0603 genes (21), which have 27%, 33%, and 28% amino acid identity with VCA0686, VCA0687, and VCA0685, respectively. As observed with the VCA0685-0687 genes, no Fur box is apparent in the promoter regions, and none of these genes was regulated by iron or Fur (34). The VCA0601-0603 genes are highly conserved among the Vibrionaceae.
Functional complementation of iron transport defects.
The potential iron transport systems were first evaluated for their abilities to supply iron to E. coli strain H1771, in which the intracellular level of iron can be monitored via a chromosomal fhuF-lacZ operon fusion. The fhuF promoter is controlled by Fur, and thus the expression of lacZ is iron and Fur regulated in this strain. Since H1771 carries mutations in both feoB and siderophore biosynthesis genes, it is starved for iron, and the level of β-galactosidase is high, even in iron-replete medium (24). Introduction of a functional iron transport system on a plasmid reduces expression of lacZ from the fhuF promoter due to increased intracellular iron (24, 30). We observed that H1771 carrying only the plasmid vector pWKS30 expressed high levels of lacZ under high-iron conditions (Table 2) , but lacZ expression was repressed in H1771 carrying the E. coli feoABC genes, as has been previously shown (24, 30). When the V. cholerae feo genes were introduced, the level of β-galactosidase activity was low in iron-replete medium (Table 2), indicating that the amount of iron transported into the cell was sufficient to repress expression of the lacZ gene. Similarly, introduction of the V. cholerae fbp genes also reduced lacZ expression. In contrast, lacZ expression in strains carrying the VCA0685-0687 or VCA0601-0603 genes was similar to that of the strain carrying the vector alone (data not shown), indicating that, under the conditions used, any potential transporters encoded by these systems did not raise intracellular iron to a level sufficient to repress lacZ expression. As expected, β-galactosidase activity was high in each strain when grown under iron-restricted conditions (Table 2).
TABLE 2.
β-Galactosidase activities of H1771 strains carrying clones encoding potential iron transport systems
| Plasmid | Vector or genes | β-Galactosidase activitya
|
|
|---|---|---|---|
| High iron | Low iron | ||
| pWKS30 | Vector | 1,299 ± 192 | 2,691 ± 129 |
| pUH18E | E. coli feoABC | 38 ± 6.3 | 2,117 ± 178 |
| pFeo101 | V. cholerae feoABC | 12 ± 1.1 | 2,497 ± 217 |
| pFbp1 | V. cholerae fbpABC | 15 ± 2.6 | 2,652 ± 503 |
β-Galactosidase activities were calculated in Miller units (36). All data are the averages ± standard deviations from three experiments. High- and low-iron conditions were obtained by use of LB broth containing 40 μM FeSO4 or 5 μg EDDA per ml, respectively.
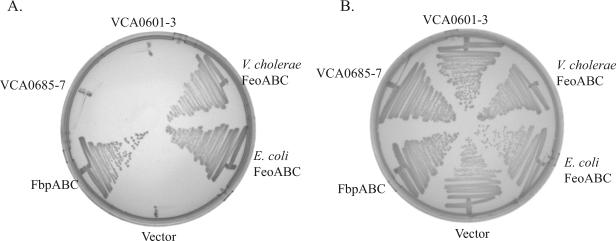
Clones carrying the putative iron transport systems were also tested for their abilities to supply iron to an S. flexneri iron acquisition mutant. This strain, SM193w, is defective in the synthesis of the siderophore aerobactin, as well as in the Feo and Sit systems (46), and is able to grow only when the medium is supplemented with a usable siderophore, such as aerobactin or ferrichrome. As seen in Fig. 1A, SM193w carrying the plasmid vector pWKS30 did not grow on LB agar. However, strains carrying plasmids with E. coli feoABC, V. cholerae feoABC, or V. cholerae fbpABC grew well on the same plate. Neither the VCA0685-0687 nor the VCA0601-0603 system genes stimulated the growth of SM193w in this assay (Fig. 1A). All strains grew well when the medium was supplemented with aerobactin (Fig. 1B). These experiments provide strong evidence that the Fbp and Feo systems transported iron into the cells whereas the VCA0685-0687 and VCA0601-0603 systems did not promote iron transport in SM193w.
FIG. 1.
Growth of S. flexneri strain SM193w. SM193w carrying genes for the indicated iron transport systems was streaked on either (A) LB agar or (B) LB agar supplemented with an aerobactin-containing culture supernatant. Genes were carried on the following plasmids: pWKS30 (vector), pFbp1 (fbpABC), pVfu4 (VCA0685-0687), pVtu100 (VCA0601-0603), pFeo101 (V. cholerae feoABC), and pUH18E (E. coli feoABC).
Iron transport assays.
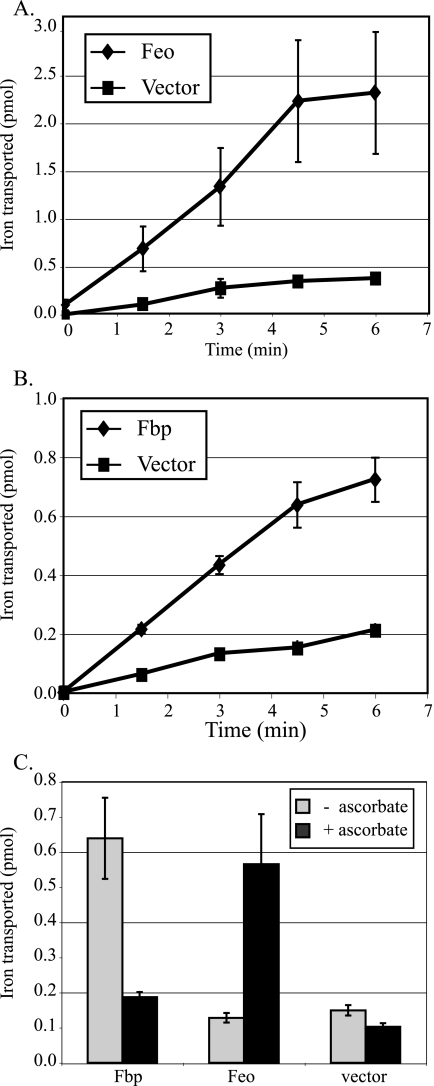
To directly determine whether Fbp or Feo stimulates the transport of iron into the cell, the uptake of radioactive iron into S. flexneri strain SM193w containing one of these systems was monitored. The presence of V. cholerae feoABC stimulated the uptake of iron approximately sixfold over the strain carrying the vector plasmid alone (Fig. 2A). Similarly, iron uptake was stimulated more than threefold by the presence of the fbp genes (Fig. 2B). To determine whether ferric or ferrous iron was transported by these systems, the assays were performed in the presence or absence of the reducing agent ascorbate to promote the conversion of ferric iron to the reduced ferrous form. Iron transport by the Fbp system was inhibited by ascorbate, indicating that it is likely a ferric iron transporter (Fig. 2C). Conversely, transport by Feo was stimulated by ascorbate, indicating that it transports ferrous iron (Fig. 2C). This is consistent with previously reported data indicating that the E. coli Feo system transports ferrous iron (18, 24).
FIG. 2.
Iron transport assays. SM193w carrying a plasmid encoding the indicated iron transport system was grown to late exponential phase and resuspended in the transport buffer. The assay was begun with the addition of 55Fe, and samples were removed at the times shown (A and B) or after 5 min (C) and filtered to determine the amounts of cell-associated radioactivity. SM193w carrying plasmids with (A) feoABC (pFeo101) or (B) fbpABC (pFbp1) was compared to strains carrying the vector (pWKS30). (B) The transport reaction mixture contained 5 μM sodium ascorbate. (C) Uptake of iron by SM193w containing the indicated system genes was carried out in buffer with (+) or without (−) supplementation with 5 μM sodium ascorbate. The data are the averages from three experiments. The error bars represent 1 standard deviation.
TonB independence of Fbp system.
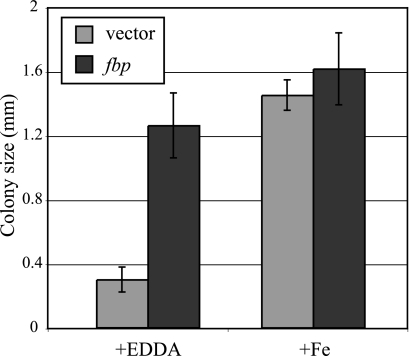
It is unclear how ferric iron reaches the periplasm for transport by Fbp. Under the oxic, neutral-pH conditions used for the growth and transport assays reported here, it is expected that ferric iron would be present primarily in insoluble complexes that might not pass freely through the outer membrane porins. All known systems for the transport of iron complexes across the outer membrane require ligand-specific outer membrane receptors that are dependent upon TonB for their activity. To test whether any TonB-dependent transport system is required for Fbp function, the sizes of colonies formed by an E. coli tonB mutant containing either the vector or V. cholerae fbpABC were measured on agar containing either the iron chelator EDDA or supplemental iron. As shown in Fig. 3, the growth of the tonB mutant under iron-limited conditions was strongly stimulated by the presence of the fbp genes, indicating that TonB activity is not required for iron transport by the Fbp system. As expected, both strains grew well under high-iron conditions.
FIG. 3.
Growth of an E. coli tonB mutant containing the V. cholerae fbpABC genes. E. coli tonB entF mutant strain ARM100 carrying either the vector plasmid pWKS30 or the fbp genes on the plasmid pFbp1 was spread at low density on LB agar containing either 1 μg EDDA per ml or 20 μM FeSO4. Colony diameters were measured after 24 h at 37°C. The data are the mean diameters of 10 colonies from one representative experiment. The error bars indicate 1 standard deviation.
Analysis of iron transport mutants in V. cholerae.
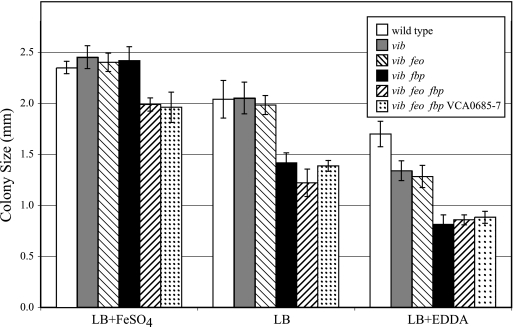
A series of strains carrying mutations in one or more iron transport systems was constructed from the classical V. cholerae strain O395. To determine the relative abilities of these strains to grow under high- or low-iron conditions, colony size assays were performed with cells spread on LB agar or on LB agar containing either additional iron to create high-iron conditions or the iron chelator EDDA to reduce available iron (Fig. 4). The wild-type strain O395 formed large colonies on LB containing supplemental iron. Although the sizes of the colonies were progressively smaller as the amount of available iron was reduced, healthy colonies were formed even in the presence of the iron chelator. The vibB mutant formed colonies the same size as those of its wild-type parent, except at the lowest level of available iron, where the colonies were smaller than those formed by the wild-type strain (P < 0.001). All of the strains carrying mutations in the fbp, feo, or VCA0685-0687 genes included a mutation in the vibriobactin biosynthetic gene vibB in order to prevent the activity of this efficient iron acquisition system from masking the effects of mutations in other potential iron transport genes. The feo mutant grew the same as the parental vibB strain at all iron levels tested in this experiment. In contrast, the fbp mutant formed colonies that were smaller than those of the parent, except when the medium was supplemented with iron (P < 0.001) (Fig. 4). This indicates that Fbp, but not Feo, contributes to the amount of iron transported under both intermediate- and low-iron conditions. Consistent with these observations, the feo fbp mutant formed colonies that were the same size as those formed by the fbp mutant on LB and on LB containing EDDA, indicating that the Feo system does not contribute to iron transport under these conditions. However, under high-iron conditions, the feo fbp mutant formed smaller colonies than either the feo or the fbp mutant, suggesting that under high-iron conditions the Fbp and the Feo systems are functionally redundant and that the presence of either system is sufficient for optimal growth. The presence of a mutation in the VCA0685-0687 system genes in the vib feo fbp genetic background had no effect on colony size under any of the conditions of iron availability tested in this experiment. This is consistent with the observation that the cloned VCA0685-0687 genes did not promote iron transport in E. coli or S. flexneri (Fig. 1).
FIG. 4.
Growth of V. cholerae iron transport mutants. Bacteria were spread on LB agar or on LB agar containing either 40 μM FeSO4 or 5 μg EDDA per ml. The sizes of the colonies formed were measured after incubation at 37°C for 24 h. The strains used in this assay were O395 (wild type), ALV101 (vib), ALV102 (vib feo), ARM591 (vib fbp), CFV1 (vib fbp feo), and EWV125 (vib feo fbp VCA0685-0687). The data are the mean diameters of 10 colonies from one representative experiment. The error bars indicate 1 standard deviation.
Virulence assays of iron transport mutants.
An infant mouse model (58) was used to determine whether the strain EWV125 (vib feo fbp VCA0685-0687) was at a competitive disadvantage in vivo relative to the wild-type parental strain O395. In these experiments, an equal number of cells from the mutant and the parental strain were inoculated intragastrically into infant mice, and the ratio of viable mutant cells relative to wild-type cells within the mouse intestine was determined at 24 h. The competitive index (the output ratio normalized to the input ratio) was not reduced in the iron transport mutant strain (CI = 1.4 ± 0.6). Since the growth of EWV125 might have been stimulated by siderophore produced by the wild-type strain, the ability of EWV125 to compete with the vibB mutant constructed in the O395 genetic background was also tested. In this experiment, EWV125 was not significantly attenuated relative to the vibB mutant (CI = 0.8 ± 0.5). This indicates that EWV125 was able to acquire sufficient iron for growth in vivo and that additional functional iron transport systems must be present in this mutant.
DISCUSSION
In this work we show that V. cholerae has two systems for the transport of free iron: the Feo system, which transports ferrous iron, and the Fbp system, which transports ferric iron. The mechanism of iron transport by the Feo system is unknown. FeoA, FeoB, and FeoC lack sequence similarity with other classes of metal transporter proteins. E. coli FeoB, which has eight transmembrane domains, is a GTPase, and at least one of the signature G protein motifs is required for iron transport (30). However, it is unclear whether FeoB is itself the iron transport protein, with the GTPase activity supplying the energy for iron transport, or whether it stimulates the activity of an unidentified ATPase that is the actual transporter (17, 20, 30). The presence of an unidentified protein with activity stimulated by FeoB is unlikely, since such a protein has not been isolated in genetic screens in E. coli (20). Further, our observation that a clone containing only the V. cholerae feoABC genes promotes iron transport in both E. coli and S. flexneri indicates that any proteins requiring the FeoB GTPase activity must be either contained on the feoABC clone or functionally conserved between V. cholerae and E. coli.
A feo mutation had only a small effect in colony size assays, and this was observed only under iron-replete conditions. These assays were performed under aerobic conditions in the absence of reducing agents, suggesting that, even under high-iron conditions, only a minimal amount of ferrous iron is available for transport by the Feo system. In virulence assays, our data do not indicate that a V. cholerae feo mutation reduces colonization in an infant mouse model. This is in contrast with data that E. coli (54), Salmonella enterica serovar Typhimurium (59), and Helicobacter pylori (60) feo mutants have reduced gastric or intestinal colonization, while a Legionella pneumophila feo mutant shows reduced intracellular growth (42). The reason for this difference is not clear, but V. cholerae may possess iron transport systems, not found in the other species, that are functionally redundant with Feo.
The Fbp system is a periplasmic binding protein-dependent ABC transporter. ABC transporters constitute a large family of proteins that couple ATP hydrolysis to the transport of ligands across cell membranes (10, 27). ABC transporters that facilitate the uptake of iron in bacteria can be divided into several groups, each of which has only minimal sequence identity with iron transporters in the other groups. One of these groups transports ligands, such as ferrisiderophores, heme, and B12, that enter the periplasm through the activity of TonB-dependent receptors (27). Five members of this ABC transporter group are present in the V. cholerae genome: three for siderophores (35, 44, 65), one for heme (37), and an apparent B12 transporter (21). The second group is ABC transporters of the iron metal type (27). This large family of proteins includes the SitABCD system of Salmonella (25, 66) and the YfeABC system of Yersinia pestis (5). There is considerable variation in metal specificity shown by members of this group, with many members binding ions such as manganese or zinc in preference to iron. The only proteins in V. cholerae related to this group are the putative zinc transport system proteins ZnuABC (VC2081-2083).
FbpABC, VCA0685-0687, and VCA0601-0603 are classified as members of a third group, which consists of ferric iron-type ABC transporters (27). This class of iron transporters was first identified in Serratia marcescens (3) and was later found to occur in Neisseria spp. (48) and various other organisms. Several periplasmic binding proteins from this family have been crystallized; their three-dimensional structure is similar to that of eukaryotic transferrins, and like the transferrins, they bind an exogenous anion that serves as an iron ligand (6, 50, 67). The ferric binding proteins and transferrins bind the anion at similar positions within the protein structure, and that position is similar to the anion-binding site in periplasmic sulfate and phosphate binding proteins (41). In contrast, the locations of the amino acids that form ligands with the iron are not well-conserved between ferric binding proteins and eukaryotic transferrins, and thus it has been proposed that these proteins are derived from a common ancestral anion-binding protein and evolved the capacity to bind iron independently from each other (6). This descent from non-iron-binding proteins may explain why ferric transport proteins are assigned to clusters of orthologous groups (COG) (57) that contain more than one ligand. The periplasmic binding proteins FbpA, VCA0685, and VCA0603 are all assigned to COG1840, “ABC-type iron/thiamine transport systems, periplasmic component” (http://www.ncbi.nlm.nih.gov/COG/old/palox.cgi?COG1840), and similarly, FbpB, VCA0686, and VCA0601 are all in COG1178, “ABC-type iron/thiamine transport systems, permease components” (http://www.ncbi.nlm.nih.gov/COG/old/palox.cgi?COG1178). Perhaps surprisingly, FbpC, VCA0687, and VCA0602 are assigned to COG3842, “ABC-type spermidine/putrescine transport systems, ATPase components” (http://www.ncbi.nlm.nih.gov/COG/old/palox.cgi?COG3842). A large number of proteins have been assigned to these COGs, but functional data are available on only a small fraction of these. In the absence of functional data on these proteins, it is difficult to ascertain the reliability of ligand assignments based solely on bioinformatics.
The characterized ABC transport system with the greatest similarity to V. cholerae Fbp is the Fbp system of M. haemolytica. Similarly to our observations with V. cholerae Fbp, the M. haemolytica Fbp genes stimulate growth of an E. coli siderophore mutant under iron-restricted growth conditions (26). The crystal structure of the M. haemolytica FbpA protein confirms that it is a member of the transferrin structural superfamily that binds both iron and a synergistic carbonate anion (50, 51). The three tyrosines that coordinate the ferric iron, as well as the amino acids that bind the carbonate anion, are conserved between the M. haemolytica and V. cholerae Fbp proteins. Thus, it is likely that the two proteins are both structurally and functionally conserved. M. haemolytica Fbp has not been directly demonstrated to be a ferric iron transporter; however, its high sequence similarity with V. cholerae Fbp is further evidence that it is also an iron transporter.
It is not known how the ferric iron transported by V. cholerae Fbp reaches the periplasm. Ferric iron forms insoluble particles under the oxic conditions at neutral pH used in the growth experiments in this report, making it unlikely that significant amounts of ferric iron would pass freely through the outer membrane porins. In some species, ferric iron is delivered to the periplasmic binding protein by a TonB-dependent outer membrane receptor, either following removal of ferric iron from a host protein, such as transferrin, or from transport of iron from a low-molecular-weight complex, such as ferric citrate. However, none of the characterized TonB-dependent receptors present in either E. coli or V. cholerae is predicted to deliver free, ferric iron to the periplasm under the conditions used in these experiments. Introduction of the V. cholerae fbpABC genes into an E. coli tonB mutant stimulated its growth under low-iron conditions, while they had minimal effect under high-iron conditions (Fig. 3). These data show that TonB function is not required for iron transport by V. cholerae FbpABC in E. coli. We suggest three mechanisms for the TonB-independent transport of ferric iron across the outer membrane. Ferric complexes could be disrupted, perhaps by a mechanism that includes extracellular reductases, so that iron could pass through the outer membrane porins. Alternatively, an unidentified iron transport system that does not require TonB could transport iron across the outer membrane. Finally, there is some evidence that the Neisseria sp. FbpA protein, which has been shown to bind metal complexes (1, 67) and which has homology to V. cholerae FbpA, may be exposed on the cell surface (13). This leads to the intriguing possibility that the FbpA protein may itself facilitate iron transport across the outer membrane.
Our experiments do not support a role for either VCA0685-0687 or VCA0601-0603 in iron transport. Neither system stimulated the growth of SM193w, an S. flexneri strain defective in iron transport, nor did clones of these genes allow for the transport of the iron needed to promote Fur-dependent repression of the fhuF-lacZ gene. Sequence analysis of these clones ruled out defects caused by PCR errors (data not shown), although it is possible that expression was poor from these plasmid clones under the conditions tested. Further, a V. cholerae mutant defective in vib, feo, and fbp formed colonies of the same size as those of an isogenic strain which also carries a mutation in VCA0685-0687 (Fig. 4). Microarray analysis indicated that the VCA0685-0687 and VCA0601-0603 genes are expressed but not regulated by iron or Fur (reference 34 and data not shown), and the periplasmic binding protein component from neither of these systems possesses the vicinal tyrosines characteristic of the periplasmic binding protein of ferric iron transporters (67). The identification of these systems as ferric iron transporters is based only on bioinformatic analysis of the genome. Further investigation is needed to determine the function of these proteins.
The iron source used by V. cholerae during colonization of its host is not known. Previous work has shown that strains defective in the synthesis and transport of vibriobactin or in the utilization of heme do not have a significant colonization defect in animal models (23, 33, 53, 56). Evidence that at least one high-affinity iron transport system plays a role in vivo was provided by the observation that TonB function is required for optimal colonization of the infant mouse intestine (49). Further work is required to determine which iron transport system or combination of systems is used to acquire iron during colonization of the intestine.
It was known previously that V. cholerae can use siderophores or hemin to fulfill its requirement for iron. In this report we show that V. cholerae also carries genes for the direct transport of both ferric and ferrous iron. Since V. cholerae mutants defective in all known systems for the acquisition of ferric and ferrous iron formed colonies in the absence of heme or exogenous siderophores (Fig. 4), it is apparent that V. cholerae has at least one additional system for iron acquisition.
Acknowledgments
We thank Laura Runyen-Janecky and Charles Earhart for helpful advice and Thomas Marlovits, Nicola Davies, Valerie Virta, and Bonnie Reus for strains and plasmids. Cynthia Nau Cornelissen gave invaluable advice with the iron transport assays.
This work was supported by National Institutes of Health grant AI50669.
REFERENCES
- 1.Alexeev, D., H. Zhu, M. Guo, W. Zhong, D. J. Hunter, W. Yang, D. J. Campopiano, and P. J. Sadler. 2003. A novel protein-mineral interface. Nat. Struct. Biol. 10:297-302. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Angerer, A., S. Gaisser, and V. Braun. 1990. Nucleotide sequences of the sfuA, sfuB, and sfuC genes of Serratia marcescens suggest a periplasmic-binding-protein-dependent iron transport mechanism. J. Bacteriol. 172:572-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 5.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 6.Bruns, C. M., A. J. Nowalk, A. S. Arvai, M. A. McTigue, K. G. Vaughan, T. A. Mietzner, and D. E. McRee. 1997. Structure of Haemophilus influenzae Fe+3-binding protein reveals convergent evolution within a superfamily. Nat. Struct. Biol. 4:919-924. [DOI] [PubMed] [Google Scholar]
- 7.Butterton, J. R., J. A. Stoebner, S. M. Payne, and S. B. Calderwood. 1992. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J. Bacteriol. 174:3729-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin, N., J. Frey, C.-F. Chang, and Y.-F. Chang. 1996. Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 143:1-6. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen, C. N. 2003. Transferrin-iron uptake by gram-negative bacteria. Front. Biosci. 8:d836-d847. [DOI] [PubMed] [Google Scholar]
- 10.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. [DOI] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection vector. Infect. Immunol. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, S. L., J. E. Arceneaux, B. R. Byers, M. E. Martin, and H. Aranha. 1986. Ferrous iron transport in Streptococcus mutans. J. Bacteriol. 168:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreiros, C., M. T. Criado, and J. A. Gomez. 1999. The neisserial 37 kDa ferric binding protein (FbpA). Comp. Biochem. Physiol. B 123:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, E. W., S. T. Lyles, and C. E. Langkford. 1964. A comparison of virulence in Vibrio cholerae strains for embryonated egg. J. Infect. Dis. 114:412. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths, G. L., S. P. Sigel, S. M. Payne, and J. B. Neilands. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259:383-385. [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Hantke, K. 2004. Ferrous iron transport, p. 178-184. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. American Society for Microbiology, Washington, D.C.
- 18.Hantke, K. 1987. Ferrous iron transport mutant in Escherichia coli K12. FEMS Microbiol. Lett. 44:53-57. [Google Scholar]
- 19.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 20.Hantke, K. 2003. Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol. 11:192-195. [DOI] [PubMed] [Google Scholar]
- 21.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson, D. P., and S. M. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson, D. P., and S. M. Payne. 1994. Vibrio cholerae iron transport systems: role of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62:5120-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirby, S. D., F. A. Lainson, W. Donachie, A. Okabe, M. Tokuda, O. Hatase, and A. B. Schryvers. 1998. The Pasteurella haemolytica 35 kDa iron-regulated protein is an FbpA homologue. Microbiology 144:3425-3436. [DOI] [PubMed] [Google Scholar]
- 27.Koster, W. 2005. Cytoplasmic membrane iron permease systems in the bacterial cell envelope. Front. Biosci. 10:462-477. [DOI] [PubMed] [Google Scholar]
- 28.Lawlor, K. M., P. A. Daskaleros, R. E. Robinson, and S. M. Payne. 1987. Virulence of iron transport mutants of Shigella flexneri and utilization of host iron compounds. Infect. Immun. 55:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litwin, C. M., S. A. Boyko, and S. B. Calderwood. 1992. Cloning, sequencing, and transcriptional regulation of the Vibrio cholerae fur gene. J. Bacteriol. 174:1897-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marlovits, T. C., W. Haase, C. Herrmann, S. G. Aller, and V. M. Unger. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. USA 99:16243-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mekalanos, J. J., D. J. Swartz, G. D. Pearson, N. Harford, F. Groyne, and M. de Wilde. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551-557. [DOI] [PubMed] [Google Scholar]
- 32.Mey, A. R., and S. M. Payne. 2003. Analysis of residues determining specificity of Vibrio cholerae TonB1 for its receptors. J. Bacteriol. 185:1195-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 34.Mey, A. R., E. E. Wyckoff, V. Kanukurthy, C. R. Fisher, and S. M. Payne. 2005. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect. Immun. 73:8167-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mey, A. R., E. E. Wyckoff, A. Oglesby, E. Rab, R. K. Taylor, and S. M. Payne. 2002. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect. Immun. 70:3419-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 38.Olson, S. A. 1994. MacVector: an integrated sequence analysis program for the Macintosh. Methods Mol. Biol. 25:195-201. [DOI] [PubMed] [Google Scholar]
- 39.Paik, S., A. Brown, C. L. Munro, C. N. Cornelissen, and T. Kitten. 2003. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J. Bacteriol. 185:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postle, K., and R. A. Larsen. 2004. The TonB, ExbB and ExbD proteins, p. 96-112. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, D.C.
- 41.Quiocho, F. A., and P. S. Ledvina. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 20:17-25. [DOI] [PubMed] [Google Scholar]
- 42.Robey, M., and N. P. Cianciotto. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers, H. J. 1973. Iron-binding catechols and virulence in Escherichia coli. Infect. Immun. 7:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers, M. B., J. A. Sexton, G. J. DeCastro, and S. B. Calderwood. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182:2350-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Runyen-Janecky, L. J., M. Hong, and S. M. Payne. 1999. Virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect. Immun. 67:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Runyen-Janecky, L. J., S. A. Reeves, E. G. Gonzales, and S. M. Payne. 2002. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to growth in the intracellular environment of the host. Infect. Immun. 71:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schryvers, A. B., R. Bonnah, R. H. Yu, H. Wong, and M. Retzer. 1998. Bacterial lactoferrin receptors. Adv. Exp. Med. Biol. 443:123-133. [DOI] [PubMed] [Google Scholar]
- 48.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 49.Seliger, S. S., A. R. Mey, A.-M. Valle, and S. M. Payne. 2001. The two TonB systems in Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801-812. [DOI] [PubMed] [Google Scholar]
- 50.Shouldice, S. R., D. R. Dougan, P. A. Williams, R. J. Skene, G. Snell, D. Scheibe, S. Kirby, D. J. Hosfield, D. E. McRee, A. B. Schryvers, and L. W. Tari. 2003. Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins. J. Biol. Chem. 278:41093-41098. [DOI] [PubMed] [Google Scholar]
- 51.Shouldice, S. R., R. J. Skene, D. R. Dougan, G. Snell, D. E. McRee, A. B. Schryvers, and L. W. Tari. 2004. Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens. J. Bacteriol. 186:3903-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sigel, S. P., and S. M. Payne. 1982. Effect of iron limitation on growth, siderophore production, and expression of outer membrane proteins of Vibrio cholerae. J. Bacteriol. 150:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sigel, S. P., J. A. Stoebner, and S. M. Payne. 1985. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect. Immun. 47:360-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stojiljkovic, I., M. Cobeljic, and K. Hantke. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108:111-115. [DOI] [PubMed] [Google Scholar]
- 55.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 56.Tashima, K. T., P. A. Carroll, M. B. Rogers, and S. B. Calderwood. 1996. Relative importance of three iron-regulated outer membrane proteins for in vivo growth of Vibrio cholerae. Infect. Immun. 64:1756-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsolis, R. M., A. J. Baumler, F. Heffron, and I. Stojiljkovic. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64:4549-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 61.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 62.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]
- 63.Wyckoff, E. E., M. Schmitt, A. Wilks, and S. M. Payne. 2004. HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wyckoff, E. E., A.-M. Valle, S. L. Smith, and S. M. Payne. 1999. A multifunctional ABC transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J. Bacteriol. 181:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, D., W.-D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centrisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, H., D. Alexeev, D. J. Hunter, D. J. Campopiano, and P. J. Sadler. 2003. Oxo-iron clusters in a bacterial iron-trafficking protein: new roles for a conserved motif. Biochem. J. 376:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]