Abstract
The possibility of the Rv3782 protein of Mycobacterium tuberculosis being a putative galactosyl transferase (GalTr) implicated in galactan synthesis arose from its similarity to the known GalTr Rv3808c, its classification as a nucleotide sugar-requiring inverting glycosyltransferase (GT-2 family), and its location within the “possible arabinogalactan biosynthetic gene cluster” of M. tuberculosis. In order to study the function of the enzyme, active membrane and cell wall fractions from Mycobacterium smegmatis containing the overexpressed Rv3782 protein were incubated with endogenous decaprenyldiphosphoryl-N-acetylglucosaminyl-rhamnose (C50-P-P-GlcNAc-Rha) as the primary substrate for galactan synthesis and UDP-[14C]galactopyranose as the immediate precursor of UDP-[14C]galactofuranose, the ultimate source of all of the galactofuranose (Galf) units of galactan. Obvious increased and selective synthesis of C50-P-P-GlcNAc-Rha-Galf-Galf, the earliest product in the pathway leading to the fully polymerized galactan, was observed, suggesting that Rv3782 encodes a GalTr involved in the first stages of galactan synthesis. Time course experiments pointed to a possible bifunctional enzyme responsible for the initial synthesis of C50-P-P-GlcNAc-Rha-Galf, followed by immediate conversion to C50-P-P-GlcNAc-Rha-Galf-Galf. Thus, Rv3782 appears to be the initiator of galactan synthesis, while Rv3808c continues with the subsequent polymerization events.
Members of the genus Mycobacterium are responsible for major health problems in humans. Tuberculosis, leprosy, and opportunistic mycobacterioses in immunocompromised individuals are characterized by disease persistence in the host, resistance to common drugs, and overall difficulties associated with treatment. One of the factors that significantly contributes to the success of mycobacteria as pathogens is their extremely hydrophobic and largely impermeable cell wall (11).
The backbone of the mycobacterial cell wall consists of a complex of covalently linked mycolic acids, arabinogalactan, and peptidoglycan. Within this, the d-galactofuran, composed of about 30 alternating 5- and 6-linked β-d-galactofuranose (Galf) units (6), is attached to the peptidoglycan by a dedicated linker unit, α-l-rhamnopyranosyl-(1→3)-N-acetyl-α-d-glucosaminyl-phosphate [Rha-(1→3)-GlcNAc-P] (16). Two to three arabinan chains containing approximately 23 d-arabinofuranose (Araf) units each, composed mostly of 5-linked α-d-Araf with branching introduced by 3,5-α-d-Araf, are bound to the galactan (6). Long (C70 to C90) branched α-alkyl β-hydroxy fatty acids are esterified to the terminal and penultimate Araf units of the arabinan (17).
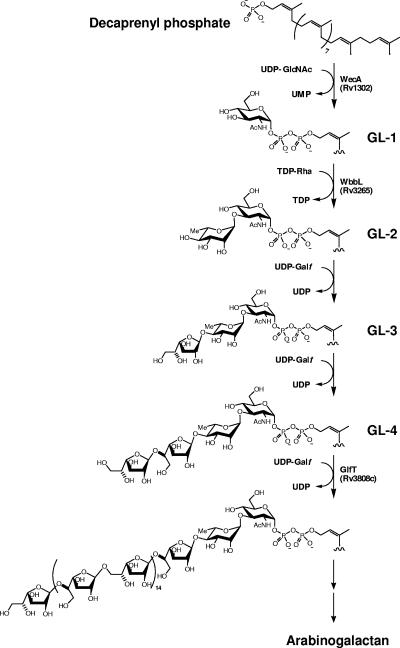
We have addressed the question of the biosynthetic origins of the arabinogalactan (AG) using cell extracts of Mycobacterium smegmatis (20, 21). In one of our initial studies of the subject, we showed that the synthesis of AG begins with the transfer of GlcNAc-1-P from UDP-GlcNAc to the polyprenyl phosphate, decaprenyl-P (C50-P), followed by the addition of Rha from dTDP-Rha, thus forming the linker region and initiating AG biogenesis (20). The polymerization of the galactan and arabinan then takes place on this C50-P-P-GlcNAc-Rha unit by single sugar additions (21) (Fig. 1) provided by UDP-Galf and C50-P-Araf, respectively. Subsequently, the complete sequence of the Mycobcterium tuberculosis H37Rv genome (4) enabled the identification of several enzymes involved in this process. Rv1302 from M. tuberculosis H37Rv was suggested to be the gene responsible for catalysis of the initial step, i.e., the transfer of GlcNAc-1-P to C50-P (15), based on similarity to wecA, which catalyzes a related reaction in Escherichia coli (18, 19). The subsequent addition of Rha is catalyzed by the essential rhamnosyltransferase WbbL (Rv3265c) (22). Based on the assortment of glycosyl linkages within the galactan, there are likely two or three galactosyl transferases (GalTr) involved in the subsequent galactan polymerization process. However, thus far, only a single candidate GalTr gene (Rv3808c) has been implicated (12, 21). Evidence was presented that this bifunctional enzyme catalyzes the synthesis of the alternating 5- and 6-linked, linear, “bulk” galactan in a processive manner. In this report, we present evidence that Rv3782 is an additional GalTr responsible for the initial transfer of Galf residues from their donor, UDP-Galf, onto the C50-P-P-GlcNAc-Rha unit, initiating the subsequent polymerization events catalyzed by Rv3808c, a bifunctional GalTr.
FIG. 1.
Proposed pathway for the biosynthesis of mycobacterial arabinogalactan. AG synthesis is initiated by a transfer of GlcNAc-1-phosphate onto a decaprenyl phosphate and continues with the sequential addition of glycosyl residues to this lipid carrier. Enzymes that have been suggested to be involved in this process are presented.
MATERIALS AND METHODS
PCR amplification, cloning, and expression of Rv3782 from the M. tuberculosis H37Rv genome.
On the basis of the nucleotide sequence of Rv3782 (4), the gene was amplified using the oligonucleotide primers 5′-CGTCCACATATGACTGAATCGGTCTTCGCC-3′ and 5′-AGTAAGCTTTGCAGATCCTCCAGGCTTGCC-3′, containing a HindIII and an NdeI restriction site (underscored). PCR using a Perkin-Elmer GeneAmp 2400 and rTth polymerase (PE Biosystems, Foster City, CA) resulted in the amplification of a single DNA fragment of the expected size (0.92 kb). The PCR fragment was purified after agarose gel electrophoresis, digested with NdeI and HindIII, and ligated into the pVV2 vector (7), which had been similarly digested. Plasmids were propagated in E. coli DH5α cells (Invitrogen, Carlsbad, CA) and were isolated using QIAGEN plasmid purification kits (QIAGEN, Valencia, CA). General molecular biology manipulations were conducted as described previously (26). The resulting pVV2-Rv3782 construct was electroporated into competent M. smegmatis mc2155 at 2.5 V, 800 Ω, and 25 microfarads. The cells were allowed to recover in Luria-Bertani (LB) broth for 90 min and were plated on LB agar containing kanamycin (50 μg/ml). A single colony was chosen to start a liquid culture in LB broth containing kanamycin (50 μg/ml), which was grown to mid-log phase. This genetic construct expressed the M. tuberculosis open reading frame Rv3782 with an N-terminal six-histidine fusion tag, which was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12%) and Western blotting. Transfer of the proteins to the nitrocellulose membrane was accomplished at a constant voltage of 0.6 V/cm2 for 1 h. For visualization of the His-tagged protein, a mouse monoclonal anti-His antibody (Sigma) diluted 1:2,000 and a secondary antibody, goat anti-mouse immunoglobulin G conjugated with alkaline phosphatase (Sigma) diluted 1:8,000, were used; color development was performed with a nitroblue tetrazolium chloride 5-bromo-4-chloro-3-indolylphosphate system (BCIP/NBT tablets; Sigma).
Purification of the recombinant His-tagged Rv3782 protein.
For purification of the recombinant His-tagged Rv3782 protein, M. smegmatis mc2155/pVV2-Rv3782 (approximately 4 g [wet weight]) was washed twice with purification buffer I (30 mM MOPS [morpholinepropanesulfonic acid], pH 7.9, 500 mM NaCl, 1 mM MgCl2, and 0.4 mM β-mercaptoethanol) and resuspended in 10 ml of the same buffer prior to probe sonication (Soniprep 150; Sanyo, MSE Ltd., Sussex, United Kingdom) for 10 min at 4°C (10 60-s pulses with 90-s cooling intervals between pulses). CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, glycerol, and imidazol were then added to the sonicate to achieve final concentrations of 0.5% (wt/vol), 10% (vol/vol), and 5 mM, respectively, and the mixtures were incubated for 2 h at 4°C. The unbroken cells and bacterial debris were removed by centrifugation of the sonicate at 10,000 × g for 20 min. Recombinant His-tagged Rv3782 protein was purified from the supernatant from this centrifugation using the BD TALON Resin (BD Biosciences, Clontech) according to the supplier's recommendations. The clarified sonicate was incubated with prewashed resin (1.5 ml) for 40 min at 4°C while being gently shaken. The mixture was centrifuged at 4°C and 3,000 × g, and the resin was washed twice with 5 ml of 5 mM imidazole in purification buffer II (purification buffer I with 0.5% [wt/vol] CHAPS and 10% [vol/vol] glycerol) and transferred to the column. Unbound proteins were removed by washing the resin with 5 ml of the same buffer. Proteins bound to the resin were then eluted stepwise with 2.5-ml aliquots of purification buffer II containing 10, 50, 100, and 200 mM imidazole. The individual fractions obtained by this procedure were analyzed by SDS-PAGE (12%) and Western blotting as described above. Recombinant Rv3782 protein was detected in the fraction eluted with 100 and 200 mM imidazole at greater than 90% purity as estimated by Coomassie staining. The protein was desalted by exchanging it into 30 mM MOPS (pH 7.9), 1 mM MgCl2, 0.5% (wt/vol) CHAPS, and 10% (vol/vol) glycerol on a PD-10 column (Amersham). The resulting protein was concentrated on an Amicon Ultra-15 centrifugal filter unit (10,000 molecular weight cutoff; Millipore) to a final volume of 250 to 350 μl, aliquoted, and stored at −20°C.
Preparation of enzymatically active membranes and cell envelope.
M. smegmatis mc2155/pVV2 and M. smegmatis mc2155/pVV2-Rv3782 were grown in LB broth containing 50 μg/ml kanamycin. Enzymatically active membranes and cell envelopes (wall and membrane) were prepared by suspending cells (10 g [wet weight]) in 40 ml of 50 mM MOPS buffer (pH 7.9) containing 5 mM 2-mercaptoethanol and 10 mM MgCl2 (buffer A), subjected to probe sonication as described above, and centrifuged at 23,000 × g for 20 min at 4°C. The pellet was resuspended in buffer A, and Percoll (Amersham Pharmacia Biotech) was added to achieve a 60% suspension, which was centrifuged at 23,000 × g for 60 min at 4°C. The white upper band was isolated, and the Percoll was removed by repeated suspension in buffer A and centrifugation. The fraction (cell envelope) was resuspended in buffer A to a protein concentration of 8 to 10 mg/ml for use. Membranes were obtained by centrifugation of the 23,000 × g supernatant at 100,000 × g for 2 h at 4°C and suspended in buffer A to give a protein concentration of 15 to 20 mg/ml.
Preparation of UDP-Galp mutase.
Escherichia coli BL21(DE3) (Stratagene) cells were transformed with plasmid pORF6 containing glf as described previously (23). UDP-Galp (galactopyranose) mutase was prepared as described previously (13) and stored at −20°C until it was used.
Preparation of dTDP-Rha.
The synthesis of dTDP-Rha relied on the presence of the full array of the Rha synthetic enzymes and endogenous cofactors in M. smegmatis (14). dTDP-glucose (sodium salt; 12 nmol; Sigma) was incubated with 25 μl of the 100,000 × g supernatant of disrupted M. smegmatis (70 μg of cytosolic protein) at 37°C for 1 h. The reaction was stopped by the addition of 100 μl of cold ethanol. After 20 min on ice, the sample was centrifuged at 14,000 × g, and the supernatant was removed and evaporated under a stream of N2. The dried material was dissolved in 25 μl of deionized water and used as a source of dTDP-Rha.
Reaction mixtures and fractionation of reaction products.
To demonstrate the GalTr activity of the recombinant Rv3782, reaction mixtures were prepared in which the membrane and/or cell envelope (1.5 mg of protein) from control and overproducing strains served as enzyme sources. In these reactions, cold sugar nucleotides, UDP-GlcNAc, dTDP-Rha, and UDP-Gal, were added at 20 μM. UDP-[U-14C]GlcNAc (NEN; 288 mCi/mmol; 1 μCi) and UDP-[U-14C]Galp (NEN; 278 mCi/mmol; 1 μCi) replaced their unlabeled counterparts in order to allow the examination of incorporation of radioactive GlcNAc or Gal into lipid-linked intermediates. The volumes of reaction mixtures were adjusted to 320 μl with buffer A. After the reaction mixtures were incubated for 1 h at 37°C, CHCl3-CH3OH (2:1; 6 ml) was added, and the mixture was rocked at room temperature for 10 min and then centrifuged (3,000 × g). The CHCl3-CH3OH phase was removed from the pellet and treated as described below. To remove residual radiolabel from the pellet, 50% CH3OH in H2O containing 0.9% NaCl (1 ml) was added, and the mixture was bath sonicated for 1 min and centrifuged at 3,000 × g. The supernatant was discarded, and the pellet was further extracted with 50% CH3OH in H2O (1 ml) and 100% CH3OH (1 ml), which were also discarded. The washed pellet was extracted with 1 ml CHCl3-CH3OH-H2O (10:10:3) (25) to remove polar-lipid-linked polymers and finally with 1 ml of “E-soak” (water-ethanol-diethyl ether-pyridine-concentrated ammonium hydroxide [15:15:5:1:0.017]) (1) to obtain [14C]Gal-labeled lipid-linked products of even greater polarity. The insoluble pellet was suspended and stored in 1 ml of E-soak. To the CHCl3-CH3OH (2:1) extract, 680 μl of deionized water was added to achieve a biphasic mixture. The upper aqueous phase was removed and discarded, and the bottom phase was backwashed with CHCl3-CH3OH-H2O (3:47:48) (8). The backwashed bottom phase was dried under a stream of N2 at room temperature and redissolved in 200 μl of CHCl3-CH3OH-H2O-NH4OH (65:25:3.6:0.5). Thin-layer chromatography (TLC) of the CHCl3-CH3OH (2:1) extract was performed on silica gel plates (Merck) in CHCl3-CH3OH-NH4OH-1 M ammonium acetate-H2O (180:140:9:9:23), and the radiolabeled lipid bands were visualized by autoradiography.
In order to examine the possible role of glycolipid 2 (GL-2) (C50-P-P- [U-14C]GlcNAc-Rha) as the acceptor for Gal transfer catalyzed by Rv3782, the glycolipid was generated in vitro and purified by preparative TLC. The reaction mixture for GL-2 production contained 4.2 mg of membrane protein from M. smegmatis mc2155, 2 μCi of UDP-[U-14C]GlcNAc, 20 μM TDP-Rha, and buffer A in a final volume of 640 μl. After 1 h of incubation at 37°C, [14C]GlcNAc-radiolabeled glycolipids were extracted as described above and separated by preparative TLC on silica gel plates in CHCl3-CH3OH-H2O-NH4OH (65:25:3.6:0.5). Following autoradiography overnight at −70°C, the band corresponding to GL-2 was scraped off and extracted with CHCl3-CH3OH-H2O-NH4OH (65:25:3.6:0.5). In vitro reactions using radioactive GL-2 as the galactose acceptor were performed as follows. In each of two glass tubes, 40,000 dpm of GL-2 preparation was dried under N2. This was followed by the addition of preincubated UDP-Galp (20 nmol) and mutase (20 μg) in 10 μl of buffer A (in order to generate UDP-Galf), membranes from control or overproducing strains (0.5 mg), and buffer A to a final volume of 80 μl. The whole mixture was subsequently bath sonicated. Incubation was carried out for 2 h at 37°C. Extraction of glycolipids was performed as described above, and the radiolabeled products were analyzed by TLC in CHCl3-CH3OH-NH4OH-1 M ammonium acetate-H2O (180:140:9:9:23).
For optimization of the buffer for purification of Rv3782, five detergents were tested under the following conditions. Reaction mixtures (final volume, 55 μl) containing cell wall protein (0.5 mg) from the control strain, 6 nmol of UDP-GlcNAc, 6 nmol of TDP-Rha, and buffer A were preincubated for 1 h at 37°C before being added to tubes containing dried UDP-[U-14C]Galp (NEN; 278 mCi/mmol; 0.25 μCi). Subsequently, 25 μl of cell envelope protein from an overproducing strain (0.5 mg) in buffer A that had been preincubated with detergents (on ice) in a final concentration of 0.5% were added to the tubes, and incubation at 37°C was carried out for a further 1 h. The reaction products were extracted and analyzed as described above. The detergents tested in this experiment were CHAPS, Triton X-100, Brij, deoxycholic acid, and N-octyl-glucopyranoside.
For testing of the enzymatic activity of the purified recombinant protein Rv3782, two reaction mixtures containing cell wall protein (0.5 mg) from the control strain, UDP-GlcNAc (6 nmol), and TDP-Rha (6 nmol) in a final volume of 55 μl in buffer A were preincubated for 1 h at 37°C before their addition to the tubes with dried UDP-[U-14C]Galp (NEN; 278 mCi/mmol; 0.25 μCi). Purified Rv3782 (25 μl) or purification buffer A was then added to these reaction mixtures, which were further incubated for 1 h at 37°C. The reaction products were extracted and analyzed as described above.
RESULTS
The possibility of Rv3782 being a putative GalTr implicated in galactan synthesis arose from a number of observations. First, there was amino acid sequence similarity to portions of the known GalTr Rv3808c. Secondly, it was a glycosyltransferase (GT) of the nucleotide sugar-requiring, inverting GT-2 family of the GT-A superfamily, according to the Carbohydrate-Active enZymes classification (http://afmb.cnrs-mrs.fr/CAZY/). Thirdly, Rv3782 was located in what has been described as a “possible AG biosynthetic gene cluster in M. tuberculosis” (2).
The gene Rv3782 was cloned into the mycobacterial expression vector pVV2, which allows expression of the target protein with an N-terminal His tag. This construct was transformed into M. smegmatis mc2155, and the cells were grown in liquid medium. Expression of the cloned gene was confirmed by Western blotting, and the presence of the target protein in membranes and the cell wall/envelope fraction was also established (Fig. 2).
FIG. 2.
Localization of recombinant protein in M. smegmatis mc2155/pVV2-Rv3782. (A) Cytosol, membranes, and cell envelope were obtained by differential centrifugation of the cell homogenates from M. smegmatis mc2155/pVV2 and M. smegmatis mc2155/pVV2-Rv3782, and 100-μg aliquots from each were loaded on the gel (12% SDS-PAGE), which was stained with Coomassie brilliant blue R-250. (B) Detection of recombinant protein Rv3782 via immunoblotting was performed with anti-His antibodies. The arrows point to the recombinant protein, Rv3782.
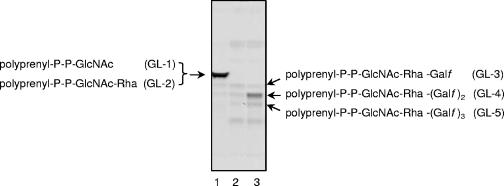
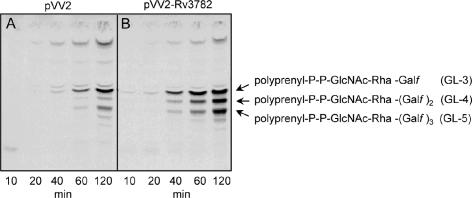
Since Rv3782 conformed in all respects to a GT-2 family member and showed similarities to the known GalTr Rv3808c, the initial screen for activity employed precursors of mycobacterial AG, such as UDP-[14C]GlcNAc, as a source of the GlcNAc-P of the linker region, or UDP-[14C]Galp as a precursor of UDP-[14C]Galf and hence of the Galf units of galactan. Enzymatically active membranes and envelope fractions containing the overexpressed protein Rv3782 were included in these cell-free assays. Products of the reaction, radiolabeled GLs and lipid-linked polymers, were extracted with organic solvents of increasing polarity. TLC of the [14C]GL population in the CHCl3-CH3OH extracts revealed, in the overproducing strain, an obvious increase in the amount C50-P-P-GlcNAc-Rha-Galf-Galf (GL-4) (Fig. 3), presumably one of the earliest products in events leading to the fully polymerized galactan. The results were similar regardless of whether UDP-[14C]Galp or UDP-[14C]GlcNAc was the source of radioactivity. The obvious conclusion was that Rv3782 encodes a GalTr involved in the initial stages of galactan synthesis. Depending on the time of incubation or the enzyme source, i.e., whether membrane or cell envelope derived, or both, GL-3 and/or GL-5 could also be overproduced by the Rv3782 overexpression strain. For example, in a time course experiment in which cell envelope fractions from control or overproducing strains served as sources of enzyme, GL-3 formation in the Rv3782 recombinant strain was much more rapid than in the control strain. In addition, overall synthesis of GL-3, GL-4, and GL-5 was higher in the overexpressing strain (Fig. 4).
FIG. 3.
Galactosyl transferase activity of Rv3782. Reaction mixtures contained 1 μCi of UDP-[14C]Galp, 20 μM UDP-GlcNAc, 20 μM TDP-Rha, membranes (1.5 mg), and cell envelope (1.5 mg) of control (M. smegmatis mc2155/pVV2) and overproducing (M. smegmatis mc2155/pVV2-Rv3782) strains and buffer A in a final volume of 320 μl. Following incubation, lipids were extracted and 10% aliquots were loaded onto a TLC plate and separated in CHCl3-CH3OH-NH4OH-1 M ammonium acetate-H2O (180:140:9:9:23). The TLC plate was exposed to the autoradiography film for 5 days at −70°C. Lanes: 1, glycolipids 1 to 5 radiolabeled with UDP-[14C]GlcNAc (600 dpm); 2, [14C]Gal-labeled lipids from the control (300 dpm); 3, [14C]Gal-labeled lipids from the overproducer (600 dpm).
FIG. 4.
Time course of [14C]Gal-labeled GL-3 to -5 production in control (M. smegmatis mc2155/pVV2) and overproducing (M. smegmatis mc2155/pVV2-Rv3782) strains. Reaction mixtures contained 0.25 μCi of UDP-[14C]Galp, 20 μM UDP-GlcNAc, 20 μM TDP-Rha, 1 mg of protein from the cell envelope fraction of the control (A) or overproducer (B) strain, and buffer A in a final volume of 80 μl. Samples were incubated for the indicated times at 37°C, and subsequently, the radiolabeled lipids were extracted and aliquots were applied to silica gel TLC plates and developed in CHCl3-CH3OH-NH4OH-1 M ammonium acetate-H2O (180:140:9:9:23). The developed TLC plates were exposed to the autoradiography film for 5 days at −70°C.
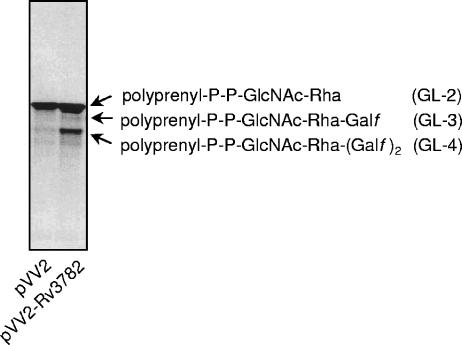
Thus, it appears that GL-2 is the primary acceptor of Galf donated by UDP-[14C]Galf in the reaction catalyzed by Rv3782. To examine this possibility more directly, GL-2 was generated in vitro, purified by preparative TLC, and used as the glycolipid substrate in reactions containing nonradioactive UDP-Gal and membranes from control cells or the overproducer Rv3782 strain. The results showed that the GL-2 was indeed an effective acceptor for Gal transfer and that membranes from the overproducing strain were significantly more effective in the reaction (Fig. 5). However, the primary product of the reaction was not GL-3, containing only one additional Gal residue, but GL-4. Therefore, Rv3782 may be responsible for the initial conversion of GL-2 to GL-3, and in the reaction mixtures as constructed, there may be an immediate conversion of GL-3 to GL-4 catalyzed by a second endogenous GalTr. Alternatively, Rv3782 may be a bifunctional enzyme catalyzing both transfers, namely, synthesis of the 5-linked and the 6-linked Galf residues of GL-4.
FIG. 5.
Identification of GL-2 as the substrate for Rv3782. For testing of GL-2 as a putative acceptor for galactose transfer, the reaction mixtures contained 40,000 dpm of dried GL-2; preincubated UDP-Galp (20 nmol) and mutase (20 μg) in 10 μl of buffer A; membranes from control or overproducing strains (0.5 mg), respectively; and buffer A in a final volume of 80 μl. Incubation was carried out for 2 h at 37°C. Extraction of glycolipids was performed as described in Materials and Methods, and the products were analyzed by TLC in CHCl3-CH3OH-NH4OH-1 M ammonium acetate-H2O (180:140:9:9:23).
In an attempt to resolve these alternatives, through use of the pure enzyme and pure glycolipid acceptor, the recombinant Rv3782 protein was generated in M. smegmatis, since efforts to obtain soluble, active protein by heterologous expression in E. coli were not successful. Purification of the His-tagged overexpressed protein by metal affinity chromatography was preceded by selection of a detergent within the purification buffer capable of solubilizing the active enzyme from membranous/cell wall structures. Of five different ionic and nonionic detergents tested, CHAPS proved to be the most suitable for this purpose, providing a nearly homogeneous product, whereas N-octyl-glucopyranoside and Triton X-100 completely inhibited enzyme activity. Incorporation of the pure Rv3782 into a cell-free assay containing the cell envelope fraction as the source of endogenous acceptor resulted in increased formation of GL-4 (Fig. 6); a similar result was obtained when C50-P-P-[U-14C]GlcNAc-Rha was used as the substrate (data not shown). Thus, the evidence indicates that Rv3782 is the initial GalTr in the series of Gal polymerization steps leading ultimately to cell wall galactan. Apparently, it is a bifunctional enzyme capable of generating the initial 5- and 6-linked Galf residues but not capable of extending the chain beyond this level.
FIG. 6.
Activity of the purified Rv3782. Two tubes containing the cell envelope fraction of the control strain M. smegmatis mc2155/pVV2 (0.5 mg), 6 μmol of UDP-GlcNAc, 6 μmol of TDP-Rha, and buffer A in a final volume of 55 μl were preincubated for 1 h at 37°C to allow formation of GL-2. Then, the reaction mixtures were transferred to tubes with the dried 0.25 μCi of UDP-[14C]Galp and 25 μl of purification buffer III (lane 1) or purified protein Rv3782 in buffer II (lane 2). After 1 h of incubation at 37°C, the reactions were stopped by the addition of CHCl3-CH3OH (2:1). The lipid extract was analyzed by TLC on silica gel plates developed in CHCl3-CH3OH-NH4OH-1 M ammonium acetate-H2O (180:140:9:9:23). The plates were exposed to autoradiography film for 2 days at −70°C.
DISCUSSION
In the course of studies of mycobacterial AG biosynthesis, we have identified several intermediates, including GL-1 (C50-P-P-GlcNAc) through GL-5 (C50-P-P-GlcNAc-Rha-Galf-Galf-Galf) and several incompletely defined polyprenyl-linked polymerized galactan and arabinogalactan precursors (20, 21). These observations, combined with the earlier structural studies on the heteropolysaccharide arabinogalactan, pointed to a large variety of linkage patterns (e.g., 3-, 5-, and 3,5-linked Araf and 5- and 6-linked Galf), suggesting the involvement of an array of GTs acting in a presumably processive manner leading to final assembly of AG and the cell wall core itself. In the case of galactan synthesis, discovery of the bifunctional mycobacterial GalTr Rv3808c and the capability of this one enzyme to synthesize the bulk of the galactan polymer, whether 5- or 6-linked, simplified the search and demonstrated, for the first time, the role of bifunctional enzymes in the context of mycobacterial polysaccharide elongation. The primary remaining goal was definition of the initial galactosylation events, namely, the enzyme(s) responsible for attachment of the initial Galf units to GL-2.
In searching for genes that participate in mycobacterial cell wall biosynthesis, we have focused on the AG biosynthetic gene cluster of M. tuberculosis H37Rv, stretching from Rv3779 to Rv3809c (2), since the genes involved in the biosynthesis of surface polysaccharides in other bacteria are also clustered (2, 10, 28, 29). Moreover, almost identical gene clusters are conserved in other mycobacterial species (M. tuberculosis CDC 1551, Mycobacterium bovis, Mycobacterium leprae, and M. smegmatis) suggesting the essentiality of this region for the physiology of members of the genus Mycobacterium. The Rv3782 gene within this cluster had been initially annotated as encoding a rhamnosyltransferase. However, BLAST analysis revealed similarity to the GalTr Rv3808c, and despite the difference in molecular mass (Rv3808c, 71.506 Da; Rv3782, 33.863 Da), the two proteins showed considerable sequence similarity when analyzed using MultAlin (5) (http://prodes.toulouse.inra.fr/multalin/multalin.html). Both of these proteins belong to the GT-2 family (http://afmb.cnrs-mrs.fr/CAZY), and members of this family typically use an inverting mechanism of catalysis leading to a glycosidic bond in the β configuration. The characteristic amino acid motif DXD found in GTs from this family and identified as Asp256-258 in Rv3808c was also present in Rv3782 as Asp93-95, located in a loop between two β-pleated sheets (Fig. 7).
FIG. 7.
Comparison of predicted secondary structures of Rv3808c and Rv3782, the two known mycobacterial galactosyl transferases. Rv3808c is comprised of 637 amino acids, whereas Rv3782 has 304. The DXD motif in each is shown in boldface and is underlined. a. a., amino acids.
It has been shown that Rv3808c catalyzes the formation of both β-5-linked and β-6-linked galactofuranoses (12) and is responsible for the synthesis of the “bulk” galactan portion of AG (21). In the Rv3808c-overexpressing strain of M. smegmatis, no increase in the production of GL-3, GL-4, or GL-5 was observed compared to the control strain (21). On the contrary, when a similar experiment was performed with the Rv3782-overproducing strain of M. smegmatis, it was particularly in the organic fractions, containing GL-3 to GL-5, that an increase in the incorporation of radioactive label from UDP-[U-14C]Gal was observed, while accumulation of the lipid-linked polymer was detected only occasionally. We thus suggested that Rv3782 is a GalTr involved in catalysis of the initial steps of galactan formation, an initial speculation borne out by the results reported here.
An in silico search for orthologs of Rv3782 revealed that within the genus Mycobacterium, the protein is highly conserved, showing 100% amino acid sequence identity with its counterparts in M. tuberculosis CDC 1551 and Mycobacterium bovis subsp. bovis AF2122/97 and 92.4%, 85.6%, and 88.4% similarities with the proteins from Mycobacterium avium subsp. paratuberculosis, M. smegmatis mc2 155, and M. leprae TN, respectively. In each of these strains, the gene encoding the protein of interest is located in the middle of a predicted operon comprised of three genes, where the first gene encodes a nucleotide binding component and the last one encodes a membrane-spanning protein of the ABC transport system. Also, in close relatives of mycobacteria, Nocardia and Corynebacterium spp., orthologs of Rv3782 are characterized by a rather high degree of similarity of amino acid sequence, ranging from 81.5% in Nocardia farcinica IFM10152 to 73.4% in Corynebacterium glutamicum ATCC 13032, 71.9% in Corynebacterium diphtheriae NCTC 13129, and 71.3% in Corynebacterium efficiens YS-314. Although the organizations of the genes in these bacteria are different from that in mycobacteria, constituents of the ABC transport system (i.e., genes coding for nucleotide binding and membrane-spanning components) are always found in the vicinity of the glycosyltransferase.
In silico analysis did not show the presence of transmembrane domains in the Rv3782 protein; however, our data clearly show that the enzyme is associated with membranous structures. In general, it is anticipated that effective biosynthesis of polysaccharide structures in bacteria is a result of the coordinated action of membrane-associated GTs (24). Such a model has also been suggested for the biosynthesis of mycobacterial lipoarabinomannan (3). It is possible that Rv3782 is part of a complex, along with the proteins responsible for the translocation of the AG intermediates across the plasma membrane, in light of its link to the genes coding for components of ABC transporters.
Association of ABC transporters with GTs has been suggested for the buildup of lipopolysaccharide O antigen in several species of gram-negative bacteria (24). An ABC transporter-dependent pathway is one of the three suggested means of production of these structures (24) and involves chain extension by addition of glycosyl residues to the nonreducing terminus of an undecaprenyl-P-P-linked growing chain. Similar to the sequence of events involved in mycobacterial galactan biosynthesis, it is initiated by the ortholog of WecA, forming polyprenyl-P-P-GlcNAc. Next, in the case of O antigen, one or two glycosyl residues, called adapters, are attached, and their addition commits the lipid intermediate to the chain extension pathway. Polymerization can be catalyzed by a monofunctional enzyme or by transferases that can add one or more residues of a given linkage (24).
Data presented in this paper suggest that Rv3782 is the GalTr catalyzing addition of the first and/or second galactose on the lipid-associated linker region (decaprenyl-P-P-GlcNAc-Rha). Although bifunctionality of this enzyme has not been firmly proven, in this report there are indications that this could be the case. A bifunctional protein, WbbO, of a similar size (377 amino acids versus 304 amino acids in Rv3782) has been described for adapter formation in O-antigen synthesis of Klebsiella pneumoniae that takes part in the biosynthesis of d-galactan I (9). WbbO is a galactosyl transferase that initiates polymerization of galactan by attachment of α-linked Galp and β-linked Galf residues onto the glycolipid acceptor, undecaprenyl-P-P-GlcNAc, but also catalyzes the addition of Galf residues to the polymer (9). Although individual activities of the protein can be separated by limitation of the substrate, UDP-Galf, there are no indications of the existence of two separate active sites in the sequence of the protein, and the mechanism involved in the bifunctionality of WbbO is unknown (9).
Our attempts to perform the in vitro reaction with the pure enzyme and pure substrate to unequivocally resolve the question of bifunctionality of the enzyme have so far been unsuccessful. Although the precise activity of Rv3782 has yet to be demonstrated, it is clear that it may represent an attractive target for drug development due to its role in the assembly of the mycobacterial AG, leading eventually to cell wall biogenesis. Moreover, based on the study of Sassetti and Rubin (27), who identified the genes that can be interrupted by transposon mutagenesis in M. tuberculosis, Rv3782 is most probably an essential gene.
Acknowledgments
This work was supported by the grants NIH NIAID AI-18357 (P.J.B.) and NIH NIAID AIDS-FIRCA TW 006487 (P.J.B. and K.M.) and by a grant from the Slovak Grant Agency, VEGA 1/2324/05 (K.M.).
We thank Stefan Berg for helpful discussions.
REFERENCES
- 1.Angus, W. W., and R. L. Lester. 1972. Turnover of inositol and phosphorus containing lipids in Saccharomyces cerevisiae; extracellular accumulation of glycerophosphorylinositol derived from phosphatidylinositol. Arch. Biochem. Biophys. 151:483-495. [DOI] [PubMed] [Google Scholar]
- 2.Belanger, A. E., and J. M. Inamine. 2000. Genetics of cell wall biosynthesis. In J. G. F. Hatfull and W. R. Jacobs (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 3.Berg, S., J. Starbuck, J. B. Torrelles, V. D. Vissa, D. C. Crick, D. Chatterjee, and P. J. Brennan. 2005. Roles of conserved proline and glycosyltransferase motifs of EmbC in biosynthesis of lipoarabinomannan. J. Biol. Chem. 280:5651-5663. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daffe, M., P. J. Brennan, and M. McNeil. 1990. Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J. Biol. Chem. 265:6734-6743. [PubMed] [Google Scholar]
- 7.Dhiman, R. K., M. C. Schulbach, S. Mahapatra, A. R. Baulard, V. Vissa, P. J. Brennan, and D. C. Crick. 2004. Identification of a novel class of omega,E,E-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J. Lipid Res. 45:1140-1147. [DOI] [PubMed] [Google Scholar]
- 8.Folch, J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 9.Guan, S., A. J. Clarke, and C. Whitfield. 2001. Functional analysis of the galactosyltransferases required for biosynthesis of d-galactan I, a component of the lipopolysaccharide O1 antigen of Klebsiella pneumoniae. J. Bacteriol. 183:3318-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 11.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 12.Kremer, L., L. G. Dover, C. Morehouse, P. Hitchin, M. Everett, H. R. Morris, A. Dell, P. J. Brennan, M. R. McNeil, C. Flaherty, K. Duncan, and G. S. Besra. 2001. Galactan biosynthesis in Mycobacterium tuberculosis. Identification of a bifunctional UDP-galactofuranosyltransferase. J. Biol. Chem. 276:26430-26440. [DOI] [PubMed] [Google Scholar]
- 13.Lee, R., D. Monsey, A. Weston, K. Duncan, C. Rithner, and M. McNeil. 1996. Enzymatic synthesis of UDP-galactofuranose and an assay for UDP-galactopyranose mutase based on high-performance liquid chromatography. Anal. Biochem. 242:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Ma, Y., J. A. Mills, J. T. Belisle, V. Vissa, M. Howell, K. Bowlin, M. S. Scherman, and M. McNeil. 1997. Determination of the pathway for rhamnose biosynthesis in mycobacteria: cloning, sequencing and expression of the Mycobacterium tuberculosis gene encoding alpha-d-glucose-1-phosphate thymidylyltransferase. Microbiology 143:937-945. [DOI] [PubMed] [Google Scholar]
- 15.McNeil, M. 1999. Arabinogalactan in mycobacteria: structure, biosynthesis, and genetics, p. 207-223. In J. B. Goldberg (ed.), Genetics of bacterial polysaccharides. CRC Press, Washington, D.C.
- 16.McNeil, M., M. Daffe, and P. J. Brennan. 1990. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem. 265:18200-18206. [PubMed] [Google Scholar]
- 17.McNeil, M. R., K. G. Robuck, M. Harter, and P. J. Brennan. 1994. Enzymatic evidence for the presence of a critical terminal hexa-arabinoside in the cell walls of Mycobacterium tuberculosis. Glycobiology 4:165-173. [DOI] [PubMed] [Google Scholar]
- 18.Meier-Dieter, U., K. Barr, R. Starman, L. Hatch, and P. D. Rick. 1992. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J. Biol. Chem. 267:746-753. [PubMed] [Google Scholar]
- 19.Meier-Dieter, U., R. Starman, K. Barr, H. Mayer, and P. D. Rick. 1990. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J. Biol. Chem. 265:13490-13497. [PubMed] [Google Scholar]
- 20.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 21.Mikusova, K., T. Yagi, R. Stern, M. R. McNeil, G. S. Besra, D. C. Crick, and P. J. Brennan. 2000. Biosynthesis of the galactan component of the mycobacterial cell wall. J. Biol. Chem. 275:33890-33897. [DOI] [PubMed] [Google Scholar]
- 22.Mills, J. A., K. Motichka, M. Jucker, H. P. Wu, B. C. Uhlik, R. J. Stern, M. S. Scherman, V. D. Vissa, F. Pan, M. Kundu, Y. F. Ma, and M. McNeil. 2004. Inactivation of the mycobacterial rhamnosyltransferase, which is needed for the formation of the arabinogalactan-peptidoglycan linker, leads to irreversible loss of viability. J. Biol. Chem. 279:43540-43546. [DOI] [PubMed] [Google Scholar]
- 23.Nassau, P. M., S. L. Martin, R. E. Brown, A. Weston, D. Monsey, M. R. McNeil, and K. Duncan. 1996. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J. Bacteriol. 178:1047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rush, J. S., J. G. Shelling, N. S. Zingg, P. H. Ray, and C. J. Waechter. 1993. Mannosylphosphoryldolichol-mediated reactions in oligosaccharide-P-P-dolichol biosynthesis. Recognition of the saturated alpha-isoprene unit of the mannosyl donor by pig brain mannosyltransferases. J. Biol. Chem. 268:13110-13117. [PubMed] [Google Scholar]
- 26.Sambrook, J., F. E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]