Abstract
The sequences of the terminal inverted repeats (TIRs) ending the linear chromosomal DNA of two Streptomyces ambofaciens strains, ATCC23877 and DSM40697 (198 kb and 213 kb, respectively), were determined from two sets of recombinant cosmids. Among the 215 coding DNA sequences (CDSs) predicted in the TIRs of strain DSM40697, 65 are absent in the TIRs of strain ATCC23877. Reciprocally, 45 of the 194 predicted CDSs are specific to the ATCC23877 strain. The strain-specific CDSs are located mainly at the terminal end of the TIRs. Indeed, although TIRs appear almost identical over 150 kb (99% nucleotide identity), large regions of DNA of 60 kb (DSM40697) and 48 kb (ATCC23877), mostly spanning the ends of the chromosome, are strain specific. These regions are rich in plasmid-associated genes, including genes encoding putative conjugal transfer functions. The strain-specific regions also share a G+C content (68%) lower than that of the rest of the genome (from 71% to 73%), a percentage that is more typical of Streptomyces plasmids and mobile elements. These data suggest that exchanges of replicon extremities have occurred, thereby contributing to the terminal variability observed at the intraspecific level. In addition, the terminal regions include many mobile genetic element-related genes, pseudogenes, and genes related to adaptation. The results give insight into the mechanisms of evolution of the TIRs: integration of new information and/or loss of DNA fragments and subsequent homogenization of the two chromosomal extremities.
Streptomyces chromosomal DNA is linear and is among the largest described for bacteria, typically 8 to 10 Mb (6, 19). Streptomyces linear replicons (chromosomes and plasmids) share an invertronic structure including the presence of terminal inverted repeat sequences (TIRs) ended by bacterial telomeres covalently linked to terminal proteins (35). The lengths and sequences of the TIRs are extremely variable, and their sizes are not correlated to that of the replicon. The terminal duplications can be as large as several hundreds of kilobases, e.g., in the chromosome of Streptomyces coelicolor M600 (∼1 Mb) (43), or can be restricted to the telomeric palindromes, e.g., in the chromosome of Streptomyces avermitilis (167 bp) (19). Two Streptomyces annotated genomes have been released so far: that for S. coelicolor A3(2), with 7,825 predicted coding DNA sequences (CDSs) (6), and that for Streptomyces avermitilis MA-4680, with 7,577 putative CDSs (19). Comparison of the two genomes revealed a common general organization with a conserved central region of about 5 Mb and terminal regions (or “arms”) carrying mainly nonessential variable genes (6, 19). The terminal regions appear poorly conserved at the level of gene content and organization, which contrasts with the strong synteny observed for the central region. The fact that the terminal regions are dispensable for vegetative growth in laboratory conditions was revealed earlier by the characterization of instability phenomena (25). In S. ambofaciens, up to 2.5 Mb located at the ends of the chromosome can be lost and is not essential for vegetative growth in laboratory growth conditions. In addition, large DNA rearrangements, such as duplications, deletions, and amplifications, frequently occur in the subtelomeric regions. One of the most spectacular rearrangements affects the size of the TIRs, which can vary from 5 kb to 1.4 Mb in spontaneous mutant strains (46), while the wild-type strain DSM40697 harbors 210-kb TIRs (26). This variation implies nonreciprocal translocations of chromosomal extremities that in some cases result from homologous recombination between duplicated genes (14). Another phenomenon triggering TIR variation implies exchanges of replicon extremities between plasmids and chromosomes. This was demonstrated for a strain of S. coelicolor in which chimeric chromosomes can be generated by crossover of the wild-type chromosome and the linear plasmid SCP1 (47). This phenomenon was also shown for Streptomyces rimosus by interaction between plasmid pZG101 and the chromosome (30) and strongly suggested by the analysis of the terminal structure of the Streptomyces lividans chromosome, which could result from partial integration of the linear plasmid SLP2 (17).
In order to gain further insight into the mechanisms of chromosomal end diversification in Streptomyces, we analyzed the variability of the TIRs of two isolates belonging to the S. ambofaciens species. It was previously shown by cross-hybridization experiments that they could be distinguished by the terminal regions (13). These two independent soil isolates, ATCC23877 (32) and DSM40697 (18), were assigned to the same species according to classical morphological and physiological traits, e.g., antibiotic production. These two isolates indeed share the same antibiotic synthesis profile, both producing the three known antibiotic compounds spiramycin, congocidin, and alpomycin (31, 32). In contrast, the closely related species S. coelicolor (1.1% divergence of 16S rDNA sequence from that of S. ambofaciens ATCC23877) shows a completely different secondary metabolite profile.
This phenetic classification was supported by more-recent molecular analyses. First, at the whole-genome scale, pulsed-field gel electrophoresis analysis, which is a highly sensitive approach to distinguishing and identifying bacterial isolates, showed that the two strains differ only slightly, whereas the S. coelicolor chromosome diverges much more (26). Further, mapping experiments with large chromosomal DNA fragments and linking clones showed that most of them were common to both strains, leading to the conclusion that the two chromosomes show a colinear organization (26). An additional piece of evidence is the presence of a recent gene duplication affecting a sigma factor-encoding gene (has gene). This duplication is common to the two S. ambofaciens isolates, while a single copy of the has gene is present in the two complete genome sequences of S. coelicolor and S. avermitilis (34).
Furthermore, the most convincing evidence for their close relationships comes from the analysis of their 16S-23S internal transcribed spacer sequences, whose evolution is the most effective (among the rrn operon) to infer close phylogenetic relationships. The six internal transcribed spacer regions were isolated from the two strains, and their sequences were compared to those of S. coelicolor (45). While the two isolates share identical sequences, these sequences differ from that of S. coelicolor, thereby showing the divergence of the strains. Although gene conversion between the rrn loci could be identified, no crossover has occurred between them, maintaining the colinearity of the two chromosomes (45).
Altogether, these data show that the two isolates are closely related strains and that their assignment to the same species is supported by a multicriterion analysis. In this work, we undertook a detailed sequence analysis of the TIRs of these two S. ambofaciens strains to determine the strain-specific gene content and to gain a deeper insight into the mechanisms important for the evolution of chromosomal extremities.
MATERIALS AND METHODS
Sequencing.
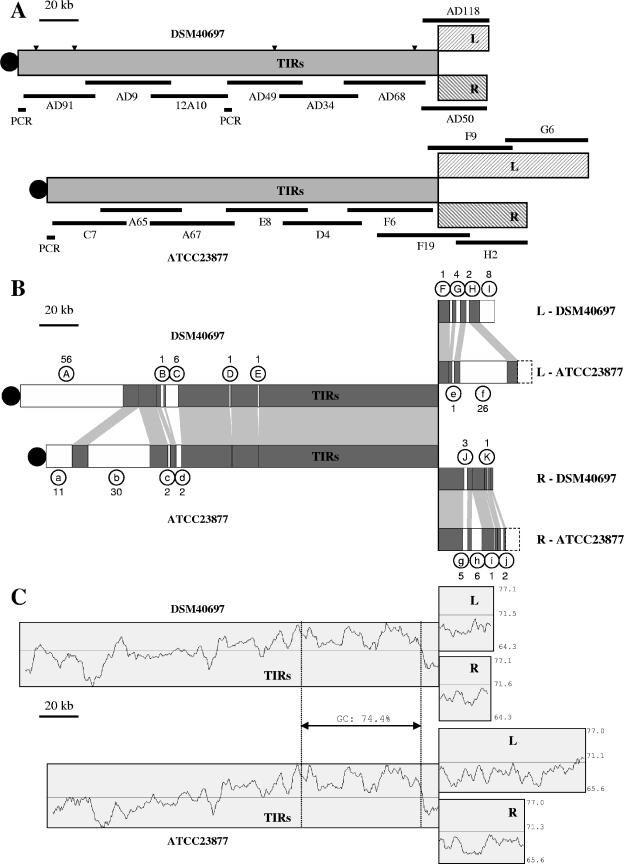
For each strain, ATCC23877 and DSM40697, a cosmid library was constructed from partially BamHI-digested S. ambofaciens genomic DNA cloned into the Supercos1 (Stratagene) vector. As the size of the S. ambofaciens TIRs greatly exceeds that of a fragment readily clonable into a cosmid vector, each copy of the TIRs cannot be isolated as a single recombinant molecule. Consequently, the sequences were obtained from a set of ordered recombinant cosmids (Fig. 1A). Note that for cosmids with insert sequences entirely corresponding to the TIRs, e.g., from cosmid C7 to F6 for strain ATCC23877, the chromosomal origin (i.e., from the right or left arm) of the recombinant cosmids cannot be deduced. Therefore, the TIR sequence is likely to consist of a chimera between the left and right repeats.
FIG. 1.
(A) Schematic representation of the S. ambofaciens ATCC23877 and DSM40697 TIRs and adjacent regions (L, left arm; R, right arm) and of the cosmids and PCR products used for the sequencing. Gap positions are represented by triangles (see Materials and Methods). (B) Comparison of the TIR sequences and flanking regions of S. ambofaciens ATCC23877 and DSM40697. Strain-specific regions are represented by white rectangles, whereas conserved regions are represented in gray. The strain-specific regions are named as follows: from A to K for strain DSM40697 and from a to j for strain ATCC23877, and the number of genes (including pseudogenes) carried by each specific region is indicated. Since the regions sequenced outside of the TIRs are larger for strain ATCC23877, sequences which cannot be compared because they were not sequenced in the second strain are represented by dashed rectangles. The terminal protein, covalently bound to the telomere, is represented by a black circle. (C) G+C content of TIRs and adjacent regions for the two strains (displayed with a 5,000-bp window size). Minimum, maximum, and mean (represented by a straight line) values of G+C percentages are indicated at the right side. These values are calculated for the whole contigs including the TIRs and the left or right adjacent region.
For sequencing, cosmids were mechanically fragmented and cloned with a BstXI adaptor into either pcDNA2.1 vector (Invitrogen) or pCNS (a derivate of pSU18 [4]). Ligation products were then introduced into Escherichia coli DH10B. It was not possible to obtain a cosmid clone containing the terminal fragment that includes the telomeres, and a PCR strategy was therefore used to amplify and sequence this region in both strains. Primers were designed from the cosmid C7 sequence (5′CACCCAGCGAGCCCAGCA3′) for strain ATCC23877 and from the cosmid AD91 sequence (5′AGCTGCAACGGTGCGTTCTATTGGG3′) for strain DSM40697, and a third primer was designed according to a consensus of the telomere sequences derived from several Streptomyces species (5′CGGAGCGGGTACCACATCGCTG3′). PCR was performed using 50 ng of DNA, with 800 μM deoxynucleoside triphosphates, 2 units of LA Taq polymerase (Takara), 2.5% dimethyl sulfoxide, and 20 pmol of each primer in a 50-μl final volume. After a denaturation step (95°C, 5 min), 30 cycles of denaturation (95°C, 30 s), annealing (58°C, 30 s), and polymerization (68°C, 3 min) were used to amplify the terminal fragment.
The 10 cosmids of strain ATCC23877 were completely sequenced (mean coverage, 10×). For strain DSM40697 (mean coverage, 10×), 17 gaps were remaining after the shotgun sequencing of the eight cosmids. PCR products were obtained for each gap and were cloned into pGEM-T Easy vector (Promega) for production of single-stranded DNA. For each gap, sequencing reactions were performed on all available templates, i.e., PCR product, double-stranded recombinant pGEM-T Easy vector, and the single-stranded DNA. In addition, sequencing reactions were carried out using two different reagents, CEQ Dye terminator (Beckman) and BigDye Terminator (Applied Biosystems). Four gaps remained, all localized in intergenic regions, suggesting they probably resulted from the formation of intrastrand secondary structures (e.g., terminators). Gap 1 (between DSMT0010 and DSMT0011) and gap 2 (between DSMT0032 and DSMT0033) are included in a strain-specific region (cosmid AD91) (Fig. 1A), and they are estimated to be less than 50 bp in length. Gap 3 (between DSMT0146 and DSMT00147) and gap 4 (between DSMT0204 and DSMT0205) belong to regions highly conserved between the two strains, and their sizes can be estimated as less than 10 bp and 200 bp, respectively, by comparison to the sequence of strain ATCC23877.
Annotation.
The gene finder Glimmer2.10 (11) was used for CDS prediction, with a minimum size of 40 codons arbitrarily chosen as the threshold. Results were then refined by RBSfinder (39). The Basic Local Alignment Search Tool (BLAST 2.2.6) was used to find similarities (1), and the Interpro package was used to describe protein domains (48). CDSs were assigned a functional category where their best cluster of orthologous groups (COG) homologue is classified (40). Then, BLASTX translations were realized for each intergenic region in order to detect initially unpredicted CDSs and pseudogenes. While comparing the predicted protein sequences with BLASTP, proteins sharing more than 30% identity over at least 80% of the length of the query sequence were considered to be homologous.
For strain ATCC23877, duplicated genes in the TIRs were named SAMTnnnn, whereas those specific only to either the left or right arm were annotated as SAMLnnnn or SAMRnnnn, respectively (underlining indicates the type of specificity). A similar principle was adopted for strain DSM40697, with the prefix “DSM” used instead of “SAM.” Sequences of left and right contigs for the two strains and their corresponding annotations are available through the SAMDB web server at http://www.weblgm.scbiol.ambofaciens.uhp-nancy.fr/.
Nucleotide sequence accession numbers.
The sequences were deposited in EMBL under the following accession numbers: AJ937740 (ATCC23877 left TIR), AJ937741 (ATCC23877 right TIR), AM279694 (DSM40697 left TIR), and AM279695 (DSM40697 right TIR).
RESULTS
Intraspecific variability at the chromosomal ends.
The large TIR sequences of two S. ambofaciens strains were determined using sets of recombinant cosmids spanning the terminal regions of the chromosome (Fig. 1A). Chimeric sequences of 197,936 bp and 212,655 bp (see Materials and Methods) were produced for strains ATCC23877 and DSM40697, respectively (see Table 1 for general features).
TABLE 1.
General features of the TIR sequences of the S. ambofaciens strains ATCC23877 and DSM40697
| Strain | Size of TIR (bp) | G+C content (%) | No. of predicted CDSs (no. of pseudogenes) | No. of proteins with assigned function (%) | No. conserved with proteins of unknown function (%) | No. of orphans (%) |
|---|---|---|---|---|---|---|
| ATCC23877 | 197,936 | 71.8 | 194 (3) | 125 (65) | 49 (25) | 20 (10) |
| DSM40697 | 212,655 | 71.9 | 215 (10) | 135 (63) | 54 (25) | 26 (12) |
Considering a single genome, the 100% nucleotide identity of the two TIR copies was supported by two lines of evidence. First, hybridization of DNA probes corresponding to the TIRs onto genomic DNA did not reveal any polymorphism (26). Second, no mismatch was found within the overlaps of sequenced cosmids, despite the fact that they can originate from either of the chromosomal arms.
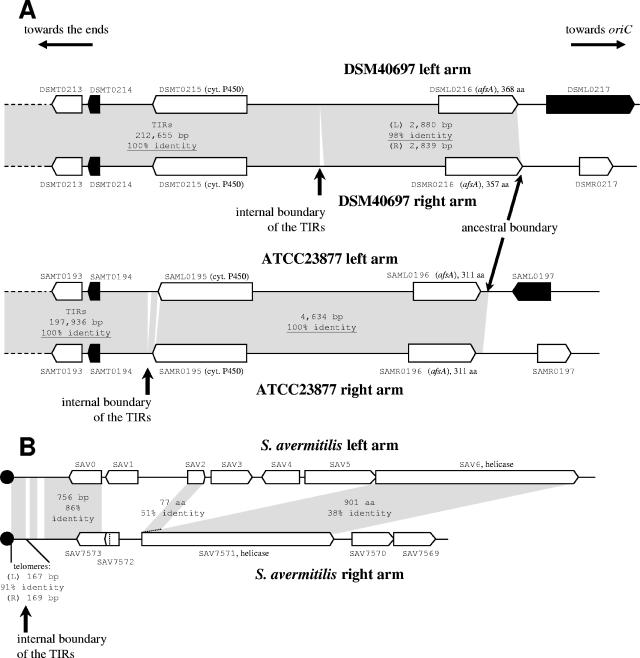
The ends of the TIRs will be defined as the first nucleotide of divergence (noted as “internal boundary of the TIRs” in Fig. 2A). Consequently, the terminal duplication does not end at exactly the same nucleotide position in the two strains. However, this situation results from point mutations and small insertions/deletions causing minor differences between the arms in both strains in the flanking small region (over about 5 kb). These regions show near identity and contain two CDSs, a probable cytochrome P450 (DSMT0215, SAML/R0195) and a homologue of afsA (DSML/R0216, SAML/R0196), separated by a large intergenic region that includes stretches of short repeated C/A-rich motifs (Fig. 2A). Another stretch of C/A-rich motifs is located in the 3′ part of afsA, and the variation in the number of motifs results in differences between the four afsA copies (two copies in each strain). Consequently, the afsA coding sequences have different sizes (three different alleles for four copies). In addition, these open reading frames (ORFs) might be pseudogenes, since only the 5′ end (about 200 codons) shows similarity (between 38% and 46%) with the afsA-like gene of Streptomyces rochei (28).
FIG. 2.
Intrachromosomal comparisons of the left and right arm regions surrounding the ends of the TIRs of S. ambofaciens strains ATCC23877 and DSM40697 (A) and S. avermitilis (B). Duplicated regions between both arms are shaded in gray, and their sizes and percentages of identity are indicated. Black arrows represent ORFs similar to genes encoding transposases (or truncated transposases), and the terminal proteins are represented by the black circles. The SAV0 putative CDS was not predicted in reference 19 and was added in this work. cyt., cytochrome. aa, amino acid.
The two arm sequences diverge totally at the same nucleotide proximal to afsA in the two strains. Thus, the two strains share the same ancestral boundaries of TIRs (Fig. 2A).
Thus, 194 CDSs (from SAMT0001 to SAMT0194) belong to the TIRs of strain ATCC23877. This last ORF is similar to that encoding a truncated transposase. In strain DSM40697, the end of the sequences assumed to be identical occurs within the long intergenic region separating DSMT0215 (putative cytochrome P450) and DSML/R0216 (afsA homologues) (Fig. 2A).
When the TIRs of the two strains are compared, two regions can be distinguished: a terminal strain-specific region and a conserved region with a more internal location. In total, the syntenic regions extend over about 150 kb and include 149 genes sharing an average 99% nucleotide identity (Fig. 1B).
Within the whole TIRs, seven strain-specific segments resulting from DNA rearrangements such as insertions, deletions, and/or gene replacements involving from 1 to 56 CDSs can be delimited (Fig. 1B). For strain DSM40697, five specific regions (A to E) represent 62 kb and contain 65 CDSs (including probable pseudogenes). Region A on its own includes 56 of the 65 strain-specific CDSs and represents a quarter of the TIR size. For strain ATCC23877, the specific loci are scattered into four regions (a to d) spanning 49 kb and including 45 strain-specific CDSs. Again, 41 of these 45 specific genes are clustered into two loci, one of which constitutes the chromosomal end (region a, 14 kb).
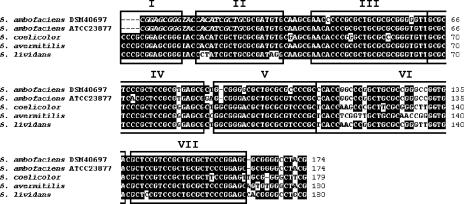
Although the chromosomal ends are strain specific, highly similar telomere sequences were found in the two strains (96.1% nucleotide identity over 180 bp) (Fig. 3). The seven typical palindromes predicted to form the secondary structures found in other known Streptomyces telomeres are present in both strains (7, 17) (Fig. 3).
FIG. 3.
Sequence alignment of the telomeres of the two S. ambofaciens strains with the chromosomal telomeres of S. coelicolor A3(2), S. avermitilis MA-4680, and S. lividans ZX7. The palindromes are labeled, and the primer sequence used for the amplification of the telomeres (see Materials and Methods) is represented in italic. This PCR strategy explains the missing nucleotides at the end of the S. ambofaciens telomeres. The boxes labeled I to VII indicate the positions of palindromes starting from the end of the chromosomal DNA. Numbers to the right of the sequences indicate the cumulative length for each of the aligned sequences.
Outside the TIRs, intraspecific variability can also be noticed. Within the areas sequenced, at least four and five DNA rearrangements have occurred in the left and right arms, respectively (Fig. 1B; also see below).
Altogether, these results suggest that, despite their variability in gene content, the current TIRs of the two S. ambofaciens strains appear to derive from a single ancestral event of terminal duplication, since they share the same ancestral internal boundary.
Low G+C content.
Despite the differences in gene content observed between the two strains, profiles of G+C percentages are quite similar (Fig. 1C). For each strain, a decrease in G+C content characterized the strain-specific extremities as well as the sequences outside of the TIRs. Thus, the terminal 50 kb of the ATCC23877 and DSM40697 chromosomes show G+C contents of 68.8% and 69.2%, respectively. Similar values are observed for the regions sequenced outside of the TIRs (69.1% in strain ATCC23877 and 68.9% in strain DSM40697). In contrast, G+C contents of Streptomyces chromosomes are 72.1% for S. coelicolor and 70.7% for S. avermitilis. In fact, the TIRs of S. ambofaciens seem to be present in regions of even lower G+C content, but the presence of a 61-kb cluster (which includes the alpomycin cluster [31]) showing a G+C content of 74.4% skews the data by causing a local increase in G+C content (Fig. 1C).
The low G+C content of the chromosomal extremities is reminiscent of that of Streptomyces plasmids (SLP2, 68.4% [17]; pSV2, 69.7% [38]; SCP1, 69.0% [5]; and SAP1, 69.2% [19] [see “Plasmid-associated genes” below]). The lower G+C content observed in the regions outside of the TIRs is related to a remarkable abundance of insertion sequences (ISs) and related genes in the strain-specific regions (found in regions F, G, H, and K in strain DSM40697 and in regions f, h, and j in strain ATCC23877) (Fig. 1B; also see “Mobile genetic element-related genes” below). The lower G+C content of mobile genetic elements in bacteria has been discussed previously (33), and their presence, together with an observed low G+C content, suggests acquisition by horizontal gene transfer.
Plasmid-associated genes.
A large proportion of the strain-specific CDSs included in the TIRs may have a plasmid origin (see Table 2) . This situation is particularly striking for strain DSM40697, for which 16 of the 56 CDSs located in the strain-specific region A show best similarity with plasmid-associated genes. For half of them, the level of identity is particularly high, i.e., more than 80% amino acid identity. Five gene products share best similarity with proteins encoded by linear plasmid SAP1 from S. avermitilis (19) and eight with linear plasmid SCP1 from S. coelicolor (5). Interestingly, one gene fragment (DSMT0048) is similar to tpg from S. lividans linear plasmid SLP2, which encodes the terminal protein involved in the replication of the telomeres (17). Some of the best hits were also with plasmids from other Actinomycetales spp., i.e., pNF1 (circular) from Nocardia farcinica (20) and pREL1 (linear) from Rhodococcus erythropolis (37). Furthermore, syntenic clusters between these strain-specific regions and linear plasmids (Table 2) were found, which strongly supports the hypothesis of integration of plasmid DNA into the terminal regions (9, 42). For example, the DSMT0042-45 cluster is conserved with the cluster SCP1.199-202 from linear plasmid SCP1.
TABLE 2.
Strain-specific genes predicted in the TIRs of S. ambofaciens strains DSM40697 and ATCC23877
| Strain | Specific region | Gene | Producta | Identity (%)b | Overlap (%)c | Gene name | Plasmid | Organism |
|---|---|---|---|---|---|---|---|---|
| DSM40697 | A | DSMT0001 | Unknown | |||||
| DSMT0002 | Putative transcriptional regulator | |||||||
| DSMT0003 | Putative phage integrase | 35 | 93 | NocaDRAFT_4522 | Nocardioides sp. strain JS614 | |||
| DSMT0004 | Unknown | |||||||
| DSMT0005 | Putative serine/threonine protein kinase | |||||||
| DSMT0006 | Unknown | |||||||
| DSMT0007 | Unknown | |||||||
| DSMT0008 | Putative helicase | 83 | 88 | SAV7571 | Streptomyces avermitilis | |||
| DSMT0009 | Putative ATP-dependent DNA ligase | 64 | 98 | SAP1_90 (lig) | SAP1 | Streptomyces avermitilis | ||
| DSMT0010 | Putative integral membrane transport protein | 72 | 100 | SCO6809 | Streptomyces coelicolor | |||
| DSMT0011 | Putative secreted protein | 75 | 100 | SCO6811 | Streptomyces coelicolor | |||
| DSMT0012 | Putative FAD-dependent oxidoreductase | 58 | 98 | pREL1_0108 | pREL1 | Rhodococcus erythropolis | ||
| DSMT0013 | Putative transcriptional regulator | 95 | 96 | pFQ25.11 | Streptomyces sp. strain F2 | |||
| DSMT0014 | Putative phosphinothricin N-acetyltransferase | 77 | 99 | pFQ25.10 | Streptomyces sp. strain F2 | |||
| DSMT0015 | Unknown | |||||||
| DSMT0016 | Conserved hypothetical protein | 85 | 99 | pnf1840 | pNF1 | Nocardia farcinica | ||
| DSMT0017 | Putative membrane protein | 76 | 100 | SCO3280 | Streptomyces coelicolor | |||
| DSMT0018 | Putative glycosyl hydrolase, BNR repeat | 36 | 98 | ArthDRAFT_2101 | Arthrobacter sp. strain FB24 | |||
| DSMT0019 | Putative lipoprotein | 72 | 100 | SCO4458 | Streptomyces coelicolor | |||
| DSMT0020 | Putative membrane protein | 60 | 96 | SCO4459 | Streptomyces coelicolor | |||
| DSMT0021 | Putative monooxygenase | 78 | 94 | SCO6838 | Streptomyces coelicolor | |||
| DSMT0022 | Putative arsenic resistance membrane transport protein | 84 | 99 | SCO6837 | Streptomyces coelicolor | |||
| DSMT0023 | Putative transcriptional regulator | 82 | 100 | SCO3699 | Streptomyces coelicolor | |||
| DSMT0024 | Putative arsenate reductase | 87 | 95 | SCO6835 | Streptomyces coelicolor | |||
| DSMT0025 | Putative thioredoxin reductase | 79 | 99 | SCO6834 | Streptomyces coelicolor | |||
| DSMT0026 | Conserved hypothetical protein | 25 | 75 | Tfu_2935 | Thermobifida fusca | |||
| DSMT0027 | Unknown | |||||||
| DSMT0028 | Conserved hypothetical protein | 87 | 100 | SCP1.257 | SCP1 | Streptomyces coelicolor | ||
| DSMT0029 | Conserved hypothetical protein | 36 | 83 | SAP1_88 | SAP1 | Streptomyces avermitilis | ||
| DSMT0030 | Putative RNA polymerase sigma factor | 37 | 97 | SAP1_87 (sig) | SAP1 | Streptomyces avermitilis | ||
| DSMT0031 | Conserved hypothetical protein | 35 | 93 | SAP1_86 | SAP1 | Streptomyces avermitilis | ||
| DSMT0032 | Putative secreted protein | 84 | 100 | SCP1.323c | SCP1 | Streptomyces coelicolor | ||
| DSMT0033 | Putative secreted protein | 89 | 100 | SCP1.261c | SCP1 | Streptomyces coelicolor | ||
| DSMT0034 | Conserved hypothetical protein | 84 | 94 | SCP1.262 | SCP1 | Streptomyces coelicolor | ||
| DSMT0035 | Ttra helicase fragment (pseudogene) | 80 | 95 | SAV7571 | Streptomyces avermitilis | |||
| DSMT0036 | Conserved hypothetical protein DUF1099 | 63 | 95 | ShewDRAFT_1466 | Shewanella sp. strain PV-4 | |||
| DSMT0037 | Unknown | |||||||
| DSMT0038 | Conserved hypothetical protein (pseudogene) | 70 | 99 | SCO0085 | Streptomyces coelicolor | |||
| DSMT0039 | Putative ATP-dependent DNA ligase | 59 | 100 | SAP1_90 (lig) | SAP1 | Streptomyces avermitilis | ||
| DSMT0040 | Conserved hypothetical protein | 71 | 100 | SCO0048 | Streptomyces coelicolor | |||
| DSMT0041 | Unknown | |||||||
| DSMT0042 | Conserved hypothetical protein | 29 | 95 | SCP1.202 | SCP1 | Streptomyces coelicolor | ||
| DSMT0043 | Conserved hypothetical protein | 84 | 100 | SCP1.201 | SCP1 | Streptomyces coelicolor | ||
| DSMT0044 | Putative secreted protein | 94 | 100 | SCP1.200c | SCP1 | Streptomyces coelicolor | ||
| DSMT0045 | Putative secreted esterase | 93 | 100 | SCP1.199c | SCP1 | Streptomyces coelicolor | ||
| DSMT0046 | Putative transposase | 55 | 71 | Francci3_3385 | Frankia sp. strain CcI3 | |||
| DSMT0047 | Putative transporter | 55 | 93 | nfa29650 | Nocardia farcinica | |||
| DSMT0048 | Tpg protein fragment (pseudogene) | 75 | 97 | tpgSLP2 | SLP2 | Streptomyces lividans | ||
| DSMT0049 | Putative AraC-family transcriptional regulator (pseudogene) | 82 | 97 | SCO3804 | Streptomyces coelicolor | |||
| DSMT0050 | Conserved hypothetical protein | 73 | 48 | SCO3803 | Streptomyces coelicolor | |||
| DSMT0051 | Putative nourseothricin acetyltransferase | 69 | 100 | nat1 | Streptomyces noursei | |||
| DSMT0052 | Conserved hypothetical protein | 30 | 93 | Francci3_1866 | Frankia sp. strain CcI3 | |||
| DSMT0053 | Conserved hypothetical protein | 47 | 76 | Francci3_1863 | Frankia sp. strain CcI3 | |||
| DSMT0054 | Unknown | |||||||
| DSMT0055 | Putative peptidase | 38 | 98 | SCO3610 | Streptomyces coelicolor | |||
| DSMT0056 | Putative major facilitator superfamily | 29 | 93 | NocaDRAFT_1725 | Nocardioides sp. strain JS614 | |||
| B | DSMT0083 | Putative 3-oxoacyl-ACP synthase III | 40 | 100 | ArthDRAFT_2448 | Arthrobacter sp. strain FB24 | ||
| C | DSMT0085 | Putative ABC transport system ATP-binding protein | 64 | 93 | SCO0121 | Streptomyces coelicolor | ||
| DSMT0086 | Putative integral membrane protein | 44 | 93 | SCO0120 | Streptomyces coelicolor | |||
| DSMT0087 | Conserved hypothetical protein | 46 | 76 | Tfu_1509 | Thermobifida fusca | |||
| DSMT0088 | Conserved hypothetical protein | 31 | 98 | SCO0123 | Streptomyces coelicolor | |||
| DSMT0089 | Putative polyprenyl synthetase | 61 | 98 | SCO0568 | Streptomyces coelicolor | |||
| DSMT0090 | Putative geranylgeranyl diphosphate synthase | 48 | 100 | ggdps | Streptomyces sp. strain KO-3988 | |||
| D | DSMT0118 | Conserved hypothetical protein | 84 | 79 | SCP1.218c | SCP1 | Streptomyces coelicolor | |
| E | DSMT0134 | Unknown | ||||||
| ATCC23877 | a | SAMT0001 | Conserved hypothetical protein | 62 | 92 | SAV7573 | Streptomyces avermitilis | |
| SAMT0002 | Putative helicase | 84 | 100 | ttrA | SLP2 | Streptomyces lividans | ||
| SAMT0003 | Unknown | |||||||
| SAMT0004 | Conserved hypothetical protein | 42 | 100 | pFRL1.57 | pFRL1 | Streptomyces sp. strain FR1 | ||
| SAMT0005 | Unknown | 64 | 59 | pFRL1.57 | pFRL1 | Streptomyces sp. strain FR1 | ||
| SAMT0006 | Putative NTP pyrophosphohydrolase | 39 | 67 | PFL_4894 | Pseudomonas fluorescens | |||
| SAMT0007 | Conserved hypothetical protein | 52 | 95 | nfa38470 | Nocardia farcinica | |||
| SAMT0008 | Putative glyoxalase | 64 | 96 | nfa38460 | Nocardia farcinica | |||
| SAMT0009 | Putative ferredoxin NADPH reductase | 55 | 97 | nfa38450 | Nocardia farcinica | |||
| SAMT0010 | KilB-like protein, role in intramycelial spread | 48 | 95 | pRL2.23 | pRL2 | Streptomyces sp. strain 44414 | ||
| SAMT0011 | Putative acetyltransferase | 50 | 100 | SAV2967 | Streptomyces avermitilis | |||
| b | SAMT0023 | Unknown | ||||||
| SAMT0024 | Putative hydrolase | 53 | 90 | Adeh_0548 | Anaeromyxobacter dehalogenans | |||
| SAMT0025 | Conserved hypothetical protein | 38 | 100 | SCO7248 | Streptomyces coelicolor | |||
| SAMT0026 | Putative phosphotransferase | 54 | 91 | ard | Streptomyces avermitilis | |||
| SAMT0027 | Unknown | |||||||
| SAMT0028 | Putative secreted protein | 47 | 80 | SCO0072 | Streptomyces coelicolor | |||
| SAMT0029 | Putative secreted protein | 61 | 89 | SCO0072 | Streptomyces coelicolor | |||
| SAMT0030 | Unknown | 30 | 67 | Francci3_0808 | Frankia sp. strain CcI3 | |||
| SAMT0031 | Unknown | |||||||
| SAMT0032 | Putative truncated transposase | 71 | 80 | orf118 | pSLA2-L | Streptomyces rochei | ||
| SAMT0033 | Putative secreted protein | 58 | 97 | SCO0072 | Streptomyces coelicolor | |||
| SAMT0034 | Putative secreted protein | 86 | 100 | SCO0072 | Streptomyces coelicolor | |||
| SAMT0035 | Conserved hypothetical protein | 60 | 100 | SCO0073 | Streptomyces coelicolor | |||
| SAMT0036 | Putative SAM-dependent methyltransferase | 67 | 98 | SCO2653 | Streptomyces coelicolor | |||
| SAMT0037 | Conserved hypothetical protein | 82 | 100 | SCO0031 | Streptomyces coelicolor | |||
| SAMT0038 | Putative transmembrane restriction endonuclease | 87 | 100 | SCO7763 | Streptomyces coelicolor | |||
| SAMT0039 | Unknown | |||||||
| SAMT0040 | Putative stress response protein | 77 | 100 | SCO3763 | Streptomyces coelicolor | |||
| SAMT0041 | Unknown | 54 | 48 | Francci3_1126 | Frankia sp. strain CcI3 | |||
| SAMT0042 | Putative membrane protein | 37 | 100 | SAV3190 | Streptomyces avermitilis | |||
| SAMT0043 | Unknown | |||||||
| SAMT0044 | Putative regulator | 39 | 81 | SAV1103 | Streptomyces avermitilis | |||
| SAMT0045 | Putative anti-sigma factor antagonist (pseudogene) | 42 | 81 | SCO3692 | Streptomyces coelicolor | |||
| SAMT0046 | Putative regulator | 71 | 96 | prpC3 | Streptomyces avermitilis | |||
| SAMT0047 | Putative hydrolase | 79 | 100 | SAV923 | Streptomyces avermitilis | |||
| SAMT0048 | Putative urease beta/gamma subunit | 50 | 94 | DR_A0319 | Deinococcus radiodurans | |||
| SAMT0049 | Putative urease alpha subunit | 65 | 98 | SCO1234 | Streptomyces coelicolor | |||
| SAMT0050 | Putative ureF-like urease accessory protein | 76 | 100 | ureF | Streptomyces avermitilis | |||
| SAMT0051 | Putative ureG-like urease accessory protein | 75 | 98 | ureG | Streptomyces avermitilis | |||
| SAMT0052 | Putative ureD-like urease accessory protein | 54 | 81 | SCO1231 | Streptomyces coelicolor | |||
| c | SAMT0065 | Putative truncated transposase | 41 | 100 | SAV18 | Streptomyces avermitilis | ||
| SAMT0066 | Putative haloacid dehalogenase | 89 | 99 | SAV737 | Streptomyces avermitilis | |||
| d | SAMT0071 | Putative transcriptional regulator | 41 | 96 | Saccharopolyspora erythraea | |||
| SAMT0072 | Putative esterase | 38 | 100 | SCO4392 | Streptomyces coelicolor |
FAD, flavin adenine dinucleotide; BNR, bacterial neuraminidase repeat; ACP, acyl carrier protein; NTP, nucleoside triphosphate.
Results for the best BLASTP hit are summarized.
“Overlap” corresponds to the ratio between the length of the BLASTP alignment and the length of the query protein (for cases in which this ratio was >100%, 100% overlap was indicated).
The same conclusion can be inferred from the analysis of region a from strain ATCC23877. First, the probable DNA helicase TtrA (SAMT0002) shows the best BLASTP hit (84% amino acid identity) with that of plasmid SLP2 of S. lividans. This helicase is implicated in the conjugal transfer of the SLP2 plasmid (8). The ttrA gene is present at the extremities of the chromosome of each S. ambofaciens strain, although it is located in the strain-specific regions. It should be pointed out that a ttrA homologue is present close to the telomeres in almost all Streptomyces replicons. In fact, TtrA proteins of the two S. ambofaciens strains, which share 45% identity, may have a different origin: TtrA from strain ATCC23877 shares 84% identity with TtrA from plasmid SLP2, whereas that of strain DSM40697, which is truncated, shares 83% identity with TtrA from the chromosome of S. avermitilis. In addition, the SAMT0010 protein is similar to the KilB protein from different Streptomyces plasmids, pRL2 (linear) and pIJ101 (circular), in which it is implicated in conjugal transfer and intramycelial spread (36).
Mobile genetic element-related genes.
Genomic islands often carry genes implicated in mobility, such as those encoding integrases, recombinases, and transposases (15). Thus, the presence of such genes in variable regions supports the idea of acquisition by horizontal gene transfer. In the conserved part of the TIRs, 149 pairs of orthologues between the two S. ambofaciens strains were predicted. Among them, three gene products showing similarity with transposases (DSMT0057/SAMT0012, DSMT0058/SAMT0013, and DSMT0214/SAMT0194) and one with an integrase/recombinase (DSMT0060/SAMT0015) were annotated. For three of them, the best similarity is found with transposase (or integrase) from Frankia species (Actinomycetales). Two transposase ORFs are located in a conserved region of the TIRs, just at the borders of the strain-specific regions A and a. The third one is located at the internal boundary of the TIRs as described above. However, none of the transposase-encoding genes seem to constitute a functional IS, the transposase being either inactivated by frameshift mutations or truncated, with no detectable flanking inverted repeats. The terminal strain-specific region A (DSM40697) contains a truncated IS (DSMT0046) similar to an IS from Frankia sp. strain Cci3 and a phage integrase (DSMT0003), close to the telomeres, sharing no homology to Streptomyces but with homology to Nocardioides species (see Table 2 for more details). Reciprocally, in the strain ATCC23877, one truncated IS (SAMT0032) is present in strain-specific region b.
Interestingly, many transposase-encoding genes are found close to the ancestral boundary of the TIRs (four in strain DSM40697 and three in strain ATCC23877), and this is a common feature of Streptomyces replicons. In S. coelicolor A3(2), an IS constitutes the ends of the chromosomal TIRs (6), while in plasmid SCP1, Tn5714 is located 3 kb outside the left TIR and IS466 is located at the end of the right one (5).
Given the close relationship between S. ambofaciens strains, it is even possible to spot recent IS- or transposon-mediated rearrangements. Indeed, outside of the TIRs, a putative composite transposon that is absent from the DSM40697 strain was detected in strain ATCC23877 (SAMR0213 to SAMR0218 [complete region h in Fig. 1B]). This transposon (5.1 kb) consists of two almost identical IS elements (99% nucleotide identity) flanking four CDSs, the products of two of which are also related to IS transposases, plus two orphans. Interestingly, the flanking ISs do not share any homology with Streptomyces sequences but do share homology with a transposase from Frankia sp. strain EAN1pec (67% amino acid identity).
Coding density and pseudogenes.
Acquisition of DNA by horizontal gene transfer and loss of useless genes are the main causes of intraspecific variability, and an equilibrium between these two phenomena leads to genomic flux that shapes bacterial genomes (24). The first step of gene loss is the creation of pseudogenes either by point mutations or by truncations.
In S. ambofaciens, the TIRs are characterized by a low coding density. When considering pseudogenes as noncoding DNA, the coding densities of the sequenced regions are 77.1% and 75.7% for strains DSM40697 and ATCC23877, respectively. These values are not biased significantly by the presence of pseudogenes, since the coding densities are 80.1% (DSM40697) and 77.1% (ATCC23877) when pseudogenes are included as CDSs. These values contrast with values of 88.9% and 86.2% predicted for the complete chromosomes of S. coelicolor (6) and S. avermitilis (19), respectively. This is again a common trait of Streptomyces plasmids, such as SAP1 from S. avermitilis, for which the density falls to 79% (19).
Another striking feature of the terminal regions is the strong presence of pseudogenes, notably in the TIRs of strain DSM40697 (10, including two truncated transposases, representing 5% of the gene content). In contrast, pseudogenes represent less than 1% of the CDSs predicted in the whole genome of S. coelicolor (6). Eight out of the 10 pseudogenes are carried by the strain-specific region A. In addition to the truncation of the ttrA gene (DSMT0008; 219 amino acid residues out of 835 in S. avermitilis), two additional TtrA fragments are encoded in strain-specific regions A (DSMT0035; 44 residues [Table 2]) and G (DSML0224; 31 residues), outside of the TIRs. The first two fragments are homologous to different parts of the same gene (ttrA from S. avermitilis). However, the latter pseudogene is not related to them, suggesting acquisitions of extra copies by different integrations of parts of linear replicons. This hypothesis is further supported by the finding, in the same region A, of a gene fragment (DSMT0048; 99 bp) showing best identity with tpgC encoding the terminal protein of the linear plasmid SLP2 and of three gene fragments (DSMT0009, DSMT0038, and DSMT0039) similar to the lig gene encoding a ligase from the S. avermitilis linear plasmid SAP1 (SAP1_90).
In the regions sequenced outside of the TIRs, 10 additional probable pseudogenes were predicted for strain DSM40697, of which 6 belong to the strain-specific regions.
The number of pseudogenes is probably underestimated, and the low coding density could be a consequence of a high mutation rate. Altogether, these data support the hypothesis that the TIRs and the terminal regions of the genome constitute a hot spot for horizontal gene transfer events mediated by linear plasmids.
Horizontal transfer of accessory genes.
Horizontal transfer mostly involves accessory genes that are able to confer a selective advantage to the recipient cell. Housekeeping genes are more recalcitrant to transfer (23). In S. ambofaciens, functions of many genes predicted for the strain-specific regions, such as resistance to toxic compounds, are related to adaptation to the environment. They are more particularly abundant in the terminal specific region A of strain DSM40697 (Fig. 1B). A good example is the five-gene cluster DSMT0021-25, which is highly similar (from 78% to 87% amino acid identity) to the S. coelicolor cluster SCO6838-34, which is implicated in the transport of and resistance to arsenate.
Antibiotic resistance genes are also present. For example, the DSMT0051 gene product shows best similarity (69%) with the Streptomyces noursei nourseothricin acetyltransferase, Nat1 (22). In the same way, a homologue of the Ard2-encoding gene (SAMT0026; 54% identity) from Saccharothrix mutabilis subsp. capreolus (Actinomycetales), which confers resistance to A201A antibiotic (3), is present in the ATCC23877 strain-specific chromosomal end.
The DSM40697 strain-specific regions B and C also carry functions related to adaptation. Indeed, the seven specific genes identified here (DSMT0083, DSMT0085-90) all have associations with secondary metabolism: DSMT0083 is a probable 3-oxoacyl-ACP synthase III-encoding gene having homologues in different Actinomycetales spp.; DSMT0085/86/88 are conserved, with three membrane and transport protein-encoding genes of S. coelicolor adjacent to the eicosapentaenoic acid cluster (SCO0124-0129); DSMT0087 shares similarity with a CDS of unknown function located in the fredericamycin biosynthesis gene cluster from Streptomyces griseus (44); and DSMT0089/90 are similar to genes encoding a putative polyprenyl synthase and a geranylgeranyl diphosphate synthase, respectively, from Streptomyces sp. strain KO-3988 (21). Interestingly, although these genes all seem to be implicated in secondary metabolism, they do not correspond to a whole conserved cluster but rather show similarity to genes from many different clusters among Actinomycetales. This could therefore be an example of a locus created by the association of genes derived from different pathways. Examples of transfer of secondary metabolism clusters have already been discussed in reference 27. The alp polyketide synthase cluster present in the TIRs of the two S. ambofaciens strains could also constitute an example of a chimeric cluster (31). While its left part is mostly similar to the kinamycin-biosynthetic cluster of S. murayamaensis (accession number AY228175), most of the right part shows high similarity to and the same genetic organization as a locus identified in the S. rochei linear plasmid pSLA2-L (28).
In addition, the variable region C is replaced by region d in strain ATCC23877 (Fig. 1B), in which one of the two specific CDSs identified encodes a probable transcriptional regulator sharing best identity with the OrfD regulator belonging to a cluster involved in the biosynthesis of a pigment in Saccharopolyspora erythraea (10).
However, a significant part of the strain-specific genes have no known function and are, in some cases, orphans.
Species specificity of the S. ambofaciens TIR conserved genes.
The part of the TIRs conserved at the intraspecific level is highly variable at the interspecific level. No synteny can be observed the other Streptomyces with genomes. The regions conserved with S. coelicolor are limited to eight small syntenic clusters comprising two to seven CDSs (e.g., a urea degradation cluster). In contrast to what is found for S. avermitilis, only one cluster of four CDSs is syntenic.
Seventeen percent of the 149 pairs of orthologues between the two strains do not have any homologue in the NR database. In addition, among the 37 proteins showing best similarity with those from organisms other than Streptomyces, the majority (19/37) show highest similarity with those from other Actinomycetales genera (especially Frankia, Arthrobacter, Nocardia, Nocardioides, and Kineococcus). Many of these organisms are fellow soil-dwelling bacteria, which plausibly suggests lateral gene transfer events.
DISCUSSION
Acquisition of new functions by exchange of extremities between linear replicons.
The S. ambofaciens TIRs contain a large proportion of strain- and species-specific genes as well as sequences potentially involved in genome plasticity. Genetic organization of the TIRs is consistent with the idea that DNA rearrangements of endogenous sets of genes (duplication and translocation), integration of exogenous information, and/or deletions have been fixed during evolution from their common ancestor.
Several lines of evidence corroborate the acquisition of the terminal strain-specific regions by exchange of replicon extremities with linear plasmids: the presence of gene clusters homologous to plasmid-associated ones; the presence of genes implicated in conjugal transfer (kilB and ttrA); and their low G+C content (68.8% and 69.2%), which is characteristic of many Streptomyces plasmids. In addition, many similarities are found with genes carried by the extremities of the linear plasmids (ttrA, tpgC, lig, and kilB). Since the last telomere-proximal ORF conserved in the two strains is a truncated IS, it is plausible that exchange of extremities could have happened by homologous recombination involving two IS copies.
This comparison made by use of S. ambofaciens describes for the first time such exchanges at the origin of intraspecific variability in natural isolates. Insertion of DNA extremities of linear replicons may be a favored mechanism for gene acquisition in Streptomyces. Indeed, exchange of the terminal parts requires a single crossover event, the success of which would be guaranteed by the presence of the telomeres in the two replicons. In addition, the presence of a helicase-like gene (ttrA) closely associated with the telomere is a trait common to almost all Streptomyces linear replicons. However, it is truncated in strain DSM40697, and no deleterious effect could be assigned in laboratory growth conditions to the mutation of both copies of ttrA in S. lividans (17). The strong conservation of a gene in a variable region appears paradoxical, but it suggests a role for conferring some long-term advantage, e.g., conjugal transfer, as suggested by C.W. Chen in his “end-first” model (8). This model predicts that this probable helicase would be involved in conjugal transfer by acting on the DNA terminus which would correspond to an origin of transfer (7, 8).
Many of the functions predicted for the strain-specific regions are potentially related to adaptation, which consolidates the idea that horizontal transfer is at the origin of the S. ambofaciens terminal variability. In general, genes successfully transferred are responsible for adaptation to the environment. The presence in the DSM40697 strain chromosomal extremities of multiple resistance genes may have conferred advantages responsible for their maintenance in the bacterial population.
In addition to exchange of extremities, multiple events of insertions/deletions have occurred recently both inside and outside of the TIRs. For example, specific region b of the strain ATCC23877 chromosome could be the result of an integration event, but the absence of a detectable target site suggests either integration by illegitimate recombination or deletion of this locus in strain DSM40697. The specific regions C (DSM40697) and d (ATCC23877), which are two different variable regions located at the same locus, could be the consequence of a DNA exchange by double crossover or by different deletions at the same locus in the two strains.
All these multiple rearrangements are posterior to the formations of the TIRs that are themselves totally different from those of the very close S. coelicolor genome. Thus, genome plasticity is extremely strong in the terminal regions, which could be privileged targets for environmental adaptation by loss, acquisition, and creation of new functions. They might be considered as a vector for genetic transfer and also as the “Swiss army knife” of the “boy scout” Streptomyces (6, 16).
Origin and evolution of the size of the TIRs.
Although the presence of two telomeres is known to be essential for the maintenance of chromosome linearity (2), no role has so far been attributed to the TIRs. Indeed, some replicons, such as the S. avermitilis chromosome, have TIRs restricted to a part of the telomeres, while others carry very large TIRs (up to 1.4 Mb [46]). In addition, the duplication of the genes located in the TIRs does not appear to be a mechanism for gene regulation. Thus, there is no obvious difference between the transcriptional levels of the genes duplicated in the S. coelicolor M600 TIRs (1 Mb) and those for the same set of genes present in single copy in S. coelicolor M145 (22-kb TIRs [43]).
The formation of TIRs could be the consequence of terminal recombination induced either by dysfunction of a telomere or by formation of double-strand breaks (DSBs). Chromosomal rescue could then be achieved by recombination with a DNA fragment including a telomere similar to the endogenous one. This fragment may be a broken daughter chromatid (46) or a plasmidic or chromosomal extremity either endogenously present or resulting from horizontal gene transfer (e.g., conjugal transfer). DSB repair might also result from the break-induced replication as described previously for Saccharomyces cerevisiae (29). Finally, circularization can also trigger chromosomal rescue (reviewed in reference 25).
Analysis of the S. ambofaciens TIR boundary reveals a preliminary step in TIR shortening (Fig. 2A). Thus, while the strict border of the TIRs is located at different positions in the two strains, the imperfect duplication includes the same genes. Point mutations and variability in tandem repeated motifs contribute to the decrease in length of the TIRs. In S. avermitilis, the terminal repetitions are restricted to the telomeres (19), which suggests the loss of ancestral TIRs in this species. Indeed, detailed analysis of the subtelomeric sequences reveals putative traces of ancestral TIRs (Fig. 2B). The last predicted ORF, SAV7573, is in fact imperfectly duplicated at the other end of the chromosome (Fig. 2B), and a putative CDS, which we tentatively called SAV0, can be detected between the telomere and SAV1. SAV0 (372 bp) and SAV7573 (411 bp) share 87% nucleotide identity over most of the CDS length (271/311 nucleotides). Furthermore, although only weakly similar (38% amino acid identity), a duplicated helicase gene is also present at both ends of the chromosome (SAV6/SAV7571). SAV2 also shows similarity with the helicase-like gene (Fig. 2B) and could correspond to the duplication of the 5′ end of SAV6 (69% nucleotide identity). These data suggest that once formed, the TIRs may accumulate point or insertional mutations that result in divergence which might progressively reduce and even prevent the frequent homogenization between TIRs and consequently accelerate the rate of divergence. The TIRs would then tend to degenerate by progressive shortening. The divergence of the duplicated genes present in the TIRs may result in pseudogene formation or leave functional coding sequences evolving toward new functions (34). Alternatively, the loss of TIRs in S. avermitilis could result from a single recombination event between the ancestral chromosome and an exogenous DNA molecule.
The reduction in TIR size may be balanced by recombination events leading to expansion. Enlarged TIRs have been reported for mutants of S. griseus and S. ambofaciens (12, 41). In both cases, recombination events occurring between duplicated genes (each copy located on a different arm and in a divergent orientation) led to a dramatic increase of the TIR size (from 210 kb to 480 kb and 850 kb in S. ambofaciens and from 24 kb to 450 kb in S. griseus). For S. coelicolor, TIR length variation has been shown to be associated with strain lineage (43). The formation of large TIRs could be ascribed to homologous recombination between transposed IS110 copies. TIR shortening from 1.06 Mb to 22 kb was observed (43). It therefore seems possible that TIR formation corresponds to a by-product of chromosomal rescue mechanisms. However, once formed, terminal duplications would provide an appropriate substrate for homologous recombination and thus for DNA repair of DSBs occurring in the terminal parts of the Streptomyces linear chromosome. The fact that the S. ambofaciens strain-specific regions are duplicated on both arms in a given chromosome strongly suggests a homogenization mechanism. In other words, these data show that DNA rearrangements occurring within a copy of TIRs can be followed by homogenization of both arms, probably by intrachromosomal recombination between duplicated sequences.
.
Acknowledgments
F.C. and A.G. were recipients of a grant from the “Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche” (M.E.N.E.S.R.). This research was supported by the PAI (ALLIANCE), the “ACI Microbiologie 2003” programs funded by the M.E.N.E.S.R., and the VIth PCRDT (“ActinoGen”).
Many thanks are due to K. Chater, G. Chandra, and T. Kieser (John Innes Centre, Norwich, United Kingdom) for their warm welcome and their help in the development of the computational methods. Many thanks to B. Segurens (Génoscope, CNS) for her help. We are grateful to A. Hesketh (John Innes Centre, Norwich, United Kingdom) and Eriko Takano (University of Groningen, Groningen, The Netherlands) for critical reading of the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrasa, M. I., J. A. Tercero, and A. Jimenez. 1997. The aminonucleoside antibiotic A201A is inactivated by a phosphotransferase activity from Streptomyces capreolus NRRL 3817, the producing organism. Isolation and molecular characterization of the relevant encoding gene and its DNA flanking regions. Eur. J. Biochem. 245:54-63. [DOI] [PubMed] [Google Scholar]
- 4.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., S. Brown, L. D. Murphy, D. E. Harris, M. A. Quail, J. Parkhill, B. G. Barrell, J. R. McCormick, R. I. Santamaria, R. Losick, M. Yamasaki, H. Kinashi, C. W. Chen, G. Chandra, D. Jakimowicz, H. M. Kieser, T. Kieser, and K. F. Chater. 2004. SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Mol. Microbiol. 51:1615-1628. [DOI] [PubMed] [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Bey, S. J., M. F. Tsou, C. H. Huang, C. C. Yang, and C. W. Chen. 2000. The homologous terminal sequence of the Streptomyces lividans chromosome and SLP2 plasmid. Microbiology 146:911-922. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. W. 1996. Complications and implications of linear bacterial chromosomes. Trends Genet. 12:192-196. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. W., C. H. Huang, H. H. Lee, H. H. Tsai, and R. Kirby. 2002. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18:522-529. [DOI] [PubMed] [Google Scholar]
- 10.Cortes, J., J. Velasco, G. Foster, A. P. Blackaby, B. A. Rudd, and B. Wilkinson. 2002. Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol. Microbiol. 44:1213-1224. [DOI] [PubMed] [Google Scholar]
- 11.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, G., B. Decaris, and P. Leblond. 1997. Occurrence of deletions, associated with genetic instability in Streptomyces ambofaciens, is independent of the linearity of the chromosomal DNA. J. Bacteriol. 179:4553-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, G., A. Kyriacou, B. Decaris, and P. Leblond. 1997. Genetic instability and its possible evolutionary implications on the chromosomal structure of Streptomyces. Biochimie 79:555-558. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, G., T. Wenner, B. Decaris, and P. Leblond. 1998. Chromosomal arm replacement generates a high level of intraspecific polymorphism in the terminal inverted repeats of the linear chromosomal DNA of Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 95:14296-14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopwood, D. A. 2003. The Streptomyces genome—be prepared! Nat. Biotechnol. 21:505-506. [DOI] [PubMed] [Google Scholar]
- 17.Huang, C. H., C. Y. Chen, H. H. Tsai, C. Chen, Y. S. Lin, and C. W. Chen. 2003. Linear plasmid SLP2 of Streptomyces lividans is a composite replicon. Mol. Microbiol. 47:1563-1576. [DOI] [PubMed] [Google Scholar]
- 18.Hütter, R. 1967. Systematik der Streptomycete. Karger Verlag, Basel, Switzerland.
- 19.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki, T., T. Kuzuyama, Y. Kuwamori, N. Matsuura, N. Itoh, K. Furihata, H. Seto, and T. Dairi. 2004. Presence of copalyl diphosphate synthase gene in an actinomycete possessing the mevalonate pathway. J. Antibiot. (Tokyo) 57:739-747. [DOI] [PubMed] [Google Scholar]
- 22.Krugel, H., G. Fiedler, C. Smith, and S. Baumberg. 1993. Sequence and transcriptional analysis of the nourseothricin acetyltransferase-encoding gene nat1 from Streptomyces noursei. Gene 127:127-131. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. G., and H. Hendrickson. 2005. Genome evolution in bacteria: order beneath chaos. Curr. Opin. Microbiol. 8:572-578. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence, J. G., and J. R. Roth. 1999. Genomic flux: genome evolution by gene loss and acquisition in bacterial genomes, p. 263-289. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington D.C.
- 25.Leblond, P., and B. Decaris. 1999. “Unstable” linear chromosomes: the case of Streptomyces, p. 235-261. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 26.Leblond, P., G. Fischer, F. X. Francou, F. Berger, M. Guérineau, and B. Decaris. 1996. The unstable region of Streptomyces ambofaciens includes 210 kb terminal inverted repeats flanking the extremities of the linear chromosomal DNA. Mol. Microbiol. 19:261-271. [DOI] [PubMed] [Google Scholar]
- 27.Metsa-Ketela, M., L. Halo, E. Munukka, J. Hakala, P. Mantsala, and K. Ylihonko. 2002. Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various Streptomyces species. Appl. Environ. Microbiol. 68:4472-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mochizuki, S., K. Hiratsu, M. Suwa, T. Ishii, F. Sugino, K. Yamada, and H. Kinashi. 2003. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48:1501-1510. [DOI] [PubMed] [Google Scholar]
- 29.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandza, S., G. Biukovic, A. Paravic, A. Dadbin, J. Cullum, and D. Hranueli. 1998. Recombination between the linear plasmid pPZG101 and the linear chromosome of Streptomyces rimosus can lead to exchange of ends. Mol. Microbiol. 28:1165-1176. [DOI] [PubMed] [Google Scholar]
- 31.Pang, X., B. Aigle, J. M. Girardet, S. Mangenot, J. L. Pernodet, B. Decaris, and P. Leblond. 2004. Functional angucycline-like antibiotic gene cluster in the terminal inverted repeats of the Streptomyces ambofaciens linear chromosome. Antimicrob. Agents Chemother. 48:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinnert-Sindico, S. 1954. Une nouvelle espèce de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n. sp. caractères culturaux. Ann. Inst. Pasteur (Paris) 87:702-707. [PubMed] [Google Scholar]
- 33.Rocha, E. P., and A. Danchin. 2002. Base composition bias might result from competition for metabolic resources. Trends Genet. 18:291-294. [DOI] [PubMed] [Google Scholar]
- 34.Roth, V., B. Aigle, R. Bunet, T. Wenner, C. Fourrier, B. Decaris, and P. Leblond. 2004. Differential and cross-transcriptional control of duplicated genes encoding alternative sigma factors in Streptomyces ambofaciens. J. Bacteriol. 186:5355-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaguchi, K. 1990. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol. Rev. 54:66-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schully, K. L., and G. S. Pettis. 2003. Separate and coordinate transcriptional control mechanisms link expression of the potentially lethal KilB spread locus to the upstream transmission operon on Streptomyces plasmid pIJ101. J. Mol. Biol. 334:875-884. [DOI] [PubMed] [Google Scholar]
- 37.Sekine, M., S. Tanikawa, S. Omata, M. Saito, T. Fujisawa, N. Tsukatani, T. Tajima, T. Sekigawa, H. Kosugi, Y. Matsuo, R. Nishiko, K. Imamura, M. Ito, H. Narita, S. Tago, N. Fujita, and S. Harayama. 2006. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 8:334-346. [DOI] [PubMed] [Google Scholar]
- 38.Spatz, K., H. Kohn, and M. Redenbach. 2002. Characterization of the Streptomyces violaceoruber SANK95570 plasmids pSV1 and pSV2. FEMS Microbiol. Lett. 213:87-92. [DOI] [PubMed] [Google Scholar]
- 39.Suzek, B. E., M. D. Ermolaeva, M. Schreiber, and S. L. Salzberg. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123-1130. [DOI] [PubMed] [Google Scholar]
- 40.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchida, T., M. Miyawaki, and H. Kinashi. 2003. Chromosomal arm replacement in Streptomyces griseus. J. Bacteriol. 185:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volff, J. N., and J. Altenbuchner. 2000. A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol. Lett. 186:143-150. [DOI] [PubMed] [Google Scholar]
- 43.Weaver, D., N. Karoonuthaisiri, H. H. Tsai, C. H. Huang, M. L. Ho, S. Gai, K. G. Patel, J. Huang, S. N. Cohen, D. A. Hopwood, C. W. Chen, and C. M. Kao. 2004. Genome plasticity in Streptomyces: identification of 1 Mb TIRs in the S. coelicolor A3(2) chromosome. Mol. Microbiol. 51:1535-1550. [DOI] [PubMed] [Google Scholar]
- 44.Wendt-Pienkowski, E., Y. Huang, J. Zhang, B. Li, H. Jiang, H. Kwon, C. R. Hutchinson, and B. Shen. 2005. Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J. Am. Chem. Soc. 127:16442-16452. [DOI] [PubMed] [Google Scholar]
- 45.Wenner, T., V. Roth, B. Decaris, and P. Leblond. 2002. Intragenomic and intraspecific polymorphism of the 16S-23S rDNA internally transcribed sequences of Streptomyces ambofaciens. Microbiology 148:633-642. [DOI] [PubMed] [Google Scholar]
- 46.Wenner, T., V. Roth, G. Fischer, C. Fourrier, B. Aigle, B. Decaris, and P. Leblond. 2003. End-to-end fusion of linear deleted chromosomes initiates a cycle of genome instability in Streptomyces ambofaciens. Mol. Microbiol. 50:411-425. [DOI] [PubMed] [Google Scholar]
- 47.Yamasaki, M., and H. Kinashi. 2004. Two chimeric chromosomes of Streptomyces coelicolor A3(2) generated by single crossover of the wild-type chromosome and linear plasmid SCP1. J. Bacteriol. 186:6553-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]