Abstract
Conditions perturbing protein homeostasis are known to induce cellular stress responses in prokaryotes and eukaryotes. Here we show for the first time that expression and assembly of a functional type IV secretion (T4S) machinery elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. After induction of T4S genes by a nutritional upshift and assembly of functional DNA transporters encoded by plasmid R1-16, host cells activated the CpxAR envelope stress signaling system, as revealed by induction or repression of downstream targets of the CpxR response regulator. Furthermore, we observed elevated transcript levels of cytoplasmic stress genes, such as groESL, with a concomitant increase of σ32 protein levels in cells expressing T4S genes. A traA null mutant of plasmid R1-16, which lacks the functional gene encoding the major pilus protein pilin, showed distinctly reduced stress responses. These results corroborated our conclusion that the activation of bacterial stress networks was dependent on the presence of functional T4S machinery. Additionally, we detected increased transcription from the rpoHp1 promoter in the presence of an active T4S system. Stimulation of rpoHp1 was dependent on the presence of CpxR, suggesting a hitherto undocumented link between CpxAR and σ32-regulated stress networks.
For translocation of macromolecular substrates, gram-negative bacteria are endowed with protein assemblies termed type I to type V secretion systems, which are localized in the cell envelope (28, 55). Among these machineries, type IV secretion (T4S) systems are considered most versatile, since they mediate conjugative DNA transfer, uptake and secretion of DNA, and export of virulence factors (reviewed in references 12, 13, and 51). T4S proteins forming the transporter complex localize to the inner and outer membranes and to the periplasm and possess typical signal sequences suggesting Sec-dependent export.
The T4S genes present on F-like plasmids (which are predominant in the Enterobacteriaceae), such as F, the resistance plasmid R1, or pSLT, the virulence plasmid of Salmonella enterica serovar Typhimurium, promote the exchange of genetic material between bacteria in a cell-to-cell contact-dependent manner (31, 35). The R1 plasmid has been extensively studied regarding T4S and DNA transfer functions (in this respect, R1 is highly similar to F). Moreover, genetic and biochemical characterization of its replication (for a recent review, see reference 45) and plasmid stability functions (19) has an impressive history.
Some of the T4S proteins encoded by the F-like plasmids have been shown to interact with each other (6, 17, 27, 37), indicating that these proteins form an envelope-spanning protein complex similarly to the VirD4/VirB machinery of Agrobacterium tumefaciens (4, 56). We assume that the DNA to be transported travels through this complex, as was demonstrated recently for the T-DNA which is transported through the VirD4/VirB complex (11).
Expression of functional T4S systems encoded by conjugative plasmids F, R100, or R1 compromises the integrity of Escherichia coli cells, as shown by increased sensitivity towards sodium dodecyl sulfate (SDS) or bile salts (1, 7) and by increased leakage of cytoplasmic proteins into hyperosmotic environments (D. Zahrl and G. Koraimann, unpublished data). These observations imply that these bacteria are sensitized, presumably by perturbations in the cell envelope. Furthermore, it has been reported that expression and assembly of other envelope structures, such as P pili or bundle-forming pili, elicit extracytoplasmic stress responses which were found to be mediated by the CpxAR two-component system and the alternative sigma factor σE (29, 44). Interestingly, the first phenotype associated with a gain-of-function mutation in the cpxA (conjugative plasmid expression) gene was a greatly reduced transfer of the F plasmid (38). This phenotype could later be explained by a reduced level of the positive regulator of the tra operon TraJ caused by a hyperactivated CpxA sensor kinase (25, 48, 52). These observations led us to scrutinize the response of E. coli cells upon expression and assembly of T4S machineries.
For our investigations on T4S-induced stress genes, we used the T4S machinery encoded by plasmid R1-16, in which transfer gene expression is not subject to repression by FinOP (32, 33). It can therefore be deduced that a large proportion of cells harboring this plasmid expresses the transfer genes and is conjugation proficient. This is reflected by high DNA transfer rates, typically between 20 and 80 DNA transfer events per 100 donor cells during a 30-min mating time. Derepression therefore means that in a given population of growing bacteria, 20 to 80 percent of bacterial cells express the transfer functions. In the cae of repressed plasmids only 0.1% of the cells express the transfer functions. However, expression of tra genes in individual cells carrying R1-16 is not deregulated and still subject to negative regulation by plasmid- and host-encoded proteins that serve to limit the expression level of Tra proteins.
We show here that R1-16 transfer gene expression and DNA transfer are profoundly reduced in stationary phase and that upon a nutritional upshift these functions are quickly resumed. The transition from non-T4S gene-expressing cells to cells that do express T4S genes provided the basis for the present work and allowed us to study how E. coli cells respond to the assembly of T4S complexes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains used in this study were the K-12 derivatives MG1655 (8), MC4100 (10), and J5 (F− Lac+ λ+ pro met; Institut für Molekulare Biowissenschaften collection). For β-galactosidase assays, the E. coli strains SP556 (same as PND2000; MC4100, λRS88[degP-lacZ]) (14) and SP559 (SP556, cpxR::spec) (29) were used. For construction of R1-16/A0, the E. coli strain DY330 was used (60). Plasmids used in this study were the IncFII plasmid R1 (Institut für Molekulare Biowissenschaften collection), the derepressed variant R1-16 (24), and R1-16/A0 (this study).
Media and growth conditions.
Media used in this study were 2× tryptone-yeast extract (TY) medium (16 g tryptone liter−1, 10 g yeast extract liter−1, and 5 g NaCl liter−1) and for β-galactosidase assays MinA minimal medium [10.5 g K2HPO4 liter−1, 4.5 g KH2PO4 liter−1, 1 g (NH4)SO4 liter−1, 3 g Casamino Acids liter−1, 1 mM MgSO4, 4 g glucose liter−1, and 20 mg thiamine liter−1]. The following antibiotics, when appropriate, were added at the indicated final concentrations: 8 μg tetracycline ml−1 and 40 μg kanamycin ml−1. Spent 2× TY medium was prepared by centrifugation of overnight cultures of E. coli MC4100[R1-16] grown at 37°C at 2,600 × g for 4 min at 25°C. The supernatant was collected, filtered through a sterile 0.2-μm nitrocellulose filter, and immediately used for mating assays. Cultures (30 ml total volume for time course experiments and for β-galactosidase assays; 18 ml total volume for mating assays) were grown in 300-ml Erlenmeyer flasks in a shaking water bath to provide aeration. For time course experiments, E. coli cultures grown to stationary phase at 37°C were diluted 1:10 into prewarmed medium and incubated at 37°C. From cultures in stationary phase as well as after dilution into fresh medium, 3 × 108 CFU were harvested by centrifugation at 2,600 × g for 4 min at 4°C, quickly frozen in liquid nitrogen, and stored at −70°C until RNA and protein isolation procedures were performed.
Preparation of RNA and Northern blot analyses.
Total RNA was isolated from E. coli cells using the RNeasy Mini kit (QIAGEN) following the manufacturer's protocol (available from http://www1.qiagen.com/literature/). RNA concentrations were determined by measuring the absorption at 260 nm. Northern blot analysis was performed as described previously (50). Briefly, 5 μg of total RNA from each sample was denatured and separated in a 1.2% agarose gel containing 1.2 M formaldehyde. RNA was transferred onto a nylon membrane (Hybond-N; Amersham Biosciences) by capillary transfer and cross-linked by UV radiation. For hybridization and detection, reagents of the DIG High Prime DNA labeling and detection starter kit II (Roche Applied Science) were used according to the manufacturer's protocol (http://www.roche-applied-science.com/pack-insert/1585614a.pdf). DNA probes were amplified from chromosomal DNA of E. coli MG1655 or from R1-16 in the case of the traA probe with the oligonucleotides listed in Table 1 and labeled with digoxigenin-11-2′-deoxyuridin-5′-triphosphate (Roche Applied Science). Digoxigenin-labeled probes were detected with anti-digoxigenin-11-dUTP alkaline phosphatase conjugate and CSPD substrate. Chemiluminescence was visualized by X-ray films. Developed films were scanned densitometrically using a Personal Densitometer (Molecular Dynamics). The obtained signal intensities of specific bands were normalized to the amount of ethidium bromide-stained 23S RNA from each sample using ImageQuant 5.1 software (Molecular Dynamics). Analyses were performed at least twice with independently isolated RNA samples; one representative result is shown.
TABLE 1.
Oligonucleotides used for preparation of DNA probes, primer extensions, and site-specific mutagenesis
| Oligonucleotide | Sequence |
|---|---|
| traA_fw | CGTCTGAATATGCTTCGCC |
| traA_rev | AGAACCGCTGCACCAATAC |
| degP_fw | CTATGTCGTCACCAACAACC |
| degP_rev | CGTCAACTTTCATCGCTTTC |
| cpxA_fw | TTCAACAATCGCCAGCCC |
| cpxA_rev | AAACTTACGCCAGCACCC |
| motA_fw | TGAACGACCCCCATTACAG |
| motA_rev | GCAAAGCGATTAAAGGCAC |
| groEL_fw | CCATCTCCGCTAACTCCGAC |
| groEL_rev | GCAACCACGCCTTCTTCTAC |
| dnaK_fw | GACGAACCCGCAAAACAC |
| dnaK_rev | TGACCACCAACGAGGATAAC |
| hslU_fw | AACAGTGTGTAAACGACGAG |
| hslU_rev | GGAAGAGATGACCAGCCAG |
| groESL_fw_ext | GACCCAGAATCGCAAACAC |
| groESL_rev_ext | TCTTTACGCTTGACGATCAC |
| Cy5groESL_rev_ext | Cy5-TCTTTACGCTTGACGATCAC |
| rpoH_fw | CCCAGTTATCATCTTCAATGCC |
| rpoH_rev | GCAGCTAAAACGCTGATCC |
| rpoH_fw_ext | CGCCTGAATAATAAAAGCGTG |
| rpoH_rev_ext | CAACTGGGGCTAAAGCTAAAC |
| Cy5rpoH_rev_ext | Cy5-CAACTGGGGCTAAAGCTAAAC |
| Ucas | CAAGAATTGCCGGCGGAT |
| Dcas | GGTATTTCACACCGCATAGC |
| fw_traAhom | TGAAGAAATATTCAGGGAGGTGATCGAAGAGAACGAGTCAACAAAGAATTGCCGGCGGAT |
| rev_traAhom | ACCGCTTGCCATCAGGTCCTGGCCCTGTGCTGCCATTGCCAGTTGGTATTTC ACACCGCATAGC |
| traAmut_au_fw | GGCGGGATAAGCAGAACAG |
| traAmut_au_rev | AATAAATACAGAGATGATGGCAAAAC |
| traAmut_in_fw | TAAACAGAGGAATCATTAGTAGACTGa |
| traAmut_in_rev | CAGTCTACTAATGATTCCTCTGTTTAa |
Underlined letters indicate the two stop codons which were introduced into the traA gene for construction of R1-16/A0.
Primer extension analyses.
For primer extension analyses, 5 μg (analyses of the groESL transcripts) or 10 μg (analyses of the rpoH transcripts) of total RNA isolated as described above was incubated with 4.2 pmol of the Cy5-labeled oligonucleotide Cy5groESL_rev_ext or Cy5rpoH_rev_ext (Table 1) in a total volume of 16.5 μl for 15 min at 70°C. After cooling of the mixture to 4°C, 2.5 μl of 2 mM deoxynucleoside triphosphates (final concentration of each, 0.2 mM), 5 μl of reverse transcriptase buffer, and 1 μl (200 U) of Moloney murine leukemia virus reverse transcriptase (RNase H Minus Point Mutant; Promega) were added. Reverse transcription was performed for 90 min at 43°C followed by 30 min at 45°C. After addition of 7.5 μl of 0.5 M EDTA (final concentration, 108 mM), the RNA was hydrolyzed with 2.3 μl of 3 M NaOH (final concentration, 200 mM) for 20 min at 65°C. After neutralization with 6.9 μl of 1 M HCl, the DNA fragments were precipitated with 4.2 μl of 3 M sodium acetate and 104 μl of 96% ethanol overnight at −20°C. The precipitates were pelleted by centrifugation and washed with 100 μl of 70% ethanol. After removal of ethanol, the DNA was resuspended in 5 μl H2O and 4.5 μl formamide loading dye (Thermo Sequenase fluorescent-labeled primer cycle sequencing kit; Amersham Biosciences) and denatured for 5 min at 95°C. A total of 4.5 μl of each sample was loaded onto a 7% acrylamide DNA sequencing gel (ReproGel TM Long Read, ALFexpress system; Amersham Biosciences). To determine the transcription start sites, sequence ladders comprising the regulatory region of the groESL operon as well as the rpoH gene were run alongside the primer extension products on the same polyacrylamide gel. For sequencing, the regulatory regions of the groESL operon and the rpoH gene were PCR amplified using the oligonucleotides groESL_fw_ext and groESL_rev_ext as well as rpoH_fw_ext and rpoH_rev_ext (Table 1). The resulting PCR fragments were sequenced using the Thermo Sequenase fluorescent-labeled primer cycle sequencing kit (Amersham Biosciences) and the same Cy5-labeled oligonucleotide that was used for the primer extension analyses. Quantification of the signals corresponding to the transcription start sites of the respective promoter were performed using Fragment Manager software (version 1.2; Pharmacia Biotech). Experiments were performed in duplicate; one representative result is shown.
β-Galactosidase assays.
β-Galactosidase activities were determined as described previously (40) with the following modifications to facilitate kinetic measurements in 96-well microplates. E. coli cultures were grown as described above in MinA minimal medium. Fifteen and 30 min after dilution of the overnight cultures, 0.5 optical density at 600 nm (OD600) unit was harvested by centrifugation at 20,000 × g for 3 min at 4°C. Subsequently, the cell pellets were resuspended in 300 μl of 0.9% NaCl and split into two 150-μl aliquots. Z-buffer (850 μl; 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0), (50 μl), chloroform and 0.1% SDS (25 μl) were added to each aliquot and vigorously shaken for 30 s to disrupt the cells. Two 125-μl aliquots of each sample were transferred into 96-well microtiter plates and preincubated for 5 min at room temperature. The enzyme reaction was started by addition of 25 μl ONPG (2-nitrophenyl-β-d-galactopyranoside, 4 mg per ml in 0.1 M phosphate buffer, pH 7.2) as a substrate. The change in absorbance at 420 nm per minute (ΔmOD420/min) at 420 nm was determined with a Bio-Rad 550 microplate reader using a kinetic protocol which measured the OD420 every 120 s for 20 min at 25°C. The ΔmOD420/min was calculated using OD420 values obtained while the absorption increased linearly.
Western blotting.
Western blotting was essentially performed as described previously (47). For determination of steady-state levels of TraM, cultures of E. coli grown and harvested as described above were lysed by sonication with four 30-s pulses with a Branson sonifier 250 using a microtip. After removal of cell debris by centrifugation for 10 min at 2,600 × g and 4°C, the protein concentration for each sample was determined using a Bio-Rad protein assay. Twenty micrograms of total protein per lane was separated on 15% SDS-polyacrylamide gels. After electrophoresis, proteins were electrotransferred onto nitrocellulose membranes (Immobilon-P; Millipore). The immunological detection of TraM was performed using an affinity-purified polyclonal antiserum raised against purified TraM (47), which was diluted 1:2,000 in TST buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 80) containing 1% dry milk (Bio-Rad) and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (diluted 1:15,000 in TST buffer containing 1% dry milk; Sigma-Aldrich). For determination of σ32 steady-state levels, proteins of whole-cell lysates corresponding to 0.3 OD600 units were separated on 12.5% SDS-polyacrylamide gels. The immunological detection of σ32 was performed using a monoclonal anti-σ32 antibody (Neoclone), which was diluted 1:2,000 in TST buffer containing 1% dry milk and horseradish peroxidase-conjugated anti-mouse immunoglobulin G (diluted 1:15,000 in TST buffer containing 1% dry milk; Amersham Biosciences). Steady-state levels of maltose binding protein were determined as previously described (61). Chemiluminescent detection was performed using the ECL system (Amersham Biosciences). The chemiluminescent bands were visualized on an X-ray film and scanned densitometrically using a Personal Densitometer (Molecular Dynamics). For quantification, ImageQuant 5.1 software (Molecular Dynamics) was used. Experiments were performed in duplicate, and one representative result is shown.
Mating assays.
To determine the conjugation frequency of donor cells in stationary phase, overnight cultures of E. coli MC4100[R1-16] were diluted 1:22 into 450 μl of spent 2× TY medium in 1.5-ml reaction tubes, and 50 μl of recipient E. coli J5 from an overnight culture grown at 37°C was added. DNA transfer was interrupted after 15 min of incubation at 37°C by vigorously vortexing for 1 min and placing the tubes on ice. Serial dilutions were prepared in 0.9% NaCl, and aliquots were plated out on MacConkey agar plates containing kanamycin and incubated at 37°C to allow the formation of colonies. To determine the conjugation frequency after dilution into fresh medium, overnight cultures of the donor strain E. coli MC4100[R1-16] were diluted 1:22 into 18 ml of fresh, prewarmed 2× TY medium and further incubated at 37°C. Directly after dilution as well as 15 and 30 min after dilution (preincubation time), 450 μl of the cultures was transferred into prewarmed 1.5-ml reaction tubes, and 50 μl of the recipient strain E. coli J5 was added. To allow mating, the suspensions were further incubated at 37°C for 15 min and processed further as described above. After DNA transfer was interrupted, serial dilutions were plated out as described above. The conjugation frequency was calculated in each case and is expressed as the number of transconjugants per donor cell.
Construction of R1-16/A0.
The wild-type traA gene of R1-16 was replaced by a mutant allele which contains two subsequent stop codons instead of the initiation codon and the codon following the start codon. Allelic exchange was performed by homologous recombination mediated by the λ Red recombination proteins as described previously (42, 49). The oligonucleotides used to construct R1-16/A0 are listed in Table 1. Briefly, a tetRA sacB cassette was amplified in two steps. First, the selection cassette from pAR48 (kindly provided by A. Reisner) was amplified using the oligonucleotides Ucas and Dcas. Methylated template plasmid DNA was digested with DpnI. Flanking arms homologous to traA were introduced in a second PCR using the oligonucleotides fw_traAhom and rev_traAhom. The PCR product was directly electroporated into E. coli DY330 harboring R1-16. Recombinants harboring R1-16/traA::tetRAsacB were selected on 2× TY agar plates containing tetracycline and kanamycin. Integration of the selection cassette was verified by PCR. For site-specific PCR mutagenesis of the traA gene, the oligonucleotides traAmut_in_fw and traAmut_in_rev in combination with the outer primers traAmut_au_rev and traAmut_au_fw were used. The resulting PCR product was used for electroporation of E. coli DY330[R1-16/traA::tetRAsacB] to replace the selection cassette with the mutant traA allele by homologous recombination. The cells were plated on NaCl-free LB plates containing 5% sucrose and kanamycin. Recombinants were tested for tetracycline sensitivity, and replacement of the selection cassette by the mutant traA allele was confirmed by PCR using the oligonucleotides traA_fw and traA_rev. To confirm the presence of the two stop codons introduced into traA, the PCR product was digested with NdeI to ensure the loss of the restriction site within the start codon region.
RESULTS
T4S genes of plasmid R1-16 are induced at the onset of cell growth.
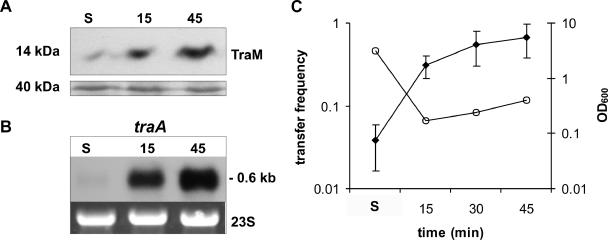
In our experimental setup, we exploited the observation that in the derepressed F plasmid which, in terms of DNA transfer, is closely related to R1, transfer gene expression and conjugation become repressed as cells enter stationary phase (22, 59). We tested whether this was also true for the derepressed variant of plasmid R1, R1-16. For that purpose, we determined the steady-state levels of the essential transfer protein TraM as well as steady-state levels of traA mRNA encoding the major sex pilus protein pilin. In early stationary phase (OD600 ≈ 3), TraM levels were fourfold lower than 15 and 45 min after dilution of the cells into fresh medium (OD600 ≈ 0.3), indicating an induction of TraM expression after reinitiation of cell growth (Fig. 1A). In stationary phase, cells harboring R1-16 showed low traA mRNA levels which we interpreted as growth phase-dependent repression of tra operon transcription (Fig. 1B). Fifteen and 45 min after dilution into fresh medium, the traA signal increased more than 10-fold, with a concomitant approximately 10-fold increase in the conjugation frequency (Fig. 1C). When late-stationary-phase cells (OD600 ≈ 7) were examined, the conjugation frequency dropped to 10−4 (not shown). These results demonstrated that conjugative DNA transfer of plasmid R1-16 and the expression of the transfer genes are reduced in stationary phase and that repression is relieved when cells are provided with fresh nutrients. The strong increases of the TraM protein levels and the traA transcript levels imply an increase in the percentage of cells that possess a functional T4S system when cells exit stationary phase. As a consequence, cells become fully transfer competent within 15 to 30 min after dilution into fresh medium. We concluded that the induction of tra gene expression upon dilution into fresh medium represents the time window of the highest metabolic burden for the host bacterium, since Tra proteins are expressed and functional transporters are assembled. The following experiments were designed to investigate how host cells reacted to the growth phase-dependent derepression of conjugative DNA transfer and whether a specific stress response was elicited following this natural induction of tra genes.
FIG. 1.
A nutritional upshift leads to rapid induction of T4S genes. TraM protein and traA mRNA levels as well as DNA transfer frequencies were determined in different phases of bacterial growth. Analyses were performed in stationary phase as well as at the indicated time points after providing E. coli MG1655 cells harboring plasmid R1-16 with fresh media. (A) Analysis of the 14-kDa TraM protein by Western blotting. A part of the Coomassie-stained SDS gel with a 40-kDa protein is shown to demonstrate equal loading. (B) Northern blot analysis of traA mRNA levels. 23S rRNA is shown as a loading control. (C) Mating assays. The transfer frequencies (filled diamonds) and culture densities (OD600; open circles) are shown. Values represent means of results from three independent experiments. No error bar indicates that the standard deviation was below the graphical resolution of the drawing program. Numbers on the x axis indicate minutes after dilution of the cultures into fresh medium. S, stationary phase.
Expression of the R1-16 T4S machinery activates a CpxAR-dependent extracytoplasmic stress response.
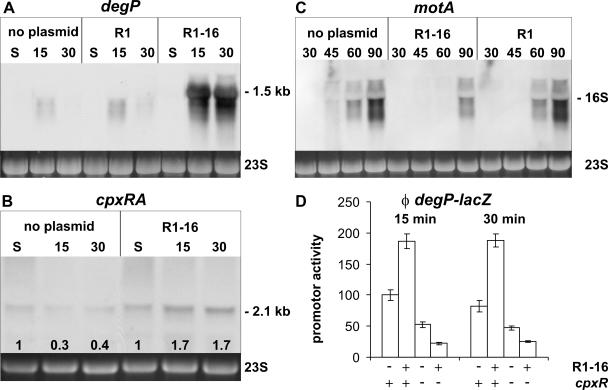
To determine whether T4S gene expression and transporter assembly during the exit from stationary phase elicit stress in the bacterial envelope, we first investigated a possible activation of envelope stress sensing systems, such as CpxAR and/or σE. As suitable indicators of this activation, we chose the degP (htrA) gene and the cpxRA operon, since transcription of these genes is known to be activated by envelope distress. The degP gene encodes the periplasmic serine protease DegP and is transcriptionally induced by both the CpxAR and σE systems but not by σ32 (14, 20, 36). The cpxRA operon is positively autoregulated when the CpxAR system is activated (15). Northern blot analyses revealed similar low degP transcript levels in plasmid-free cells and in cells harboring R1-16 in stationary phase. In contrast, 15 and 30 min after dilution into fresh medium, cells harboring plasmid R1-16 showed markedly increased degP mRNA levels compared to plasmid-free cells (Fig. 2A). Similarly, after addition of nutrients, the cpxRA mRNA levels increased approximately four- to sixfold in cells expressing the T4S components compared to plasmid-free cells, whereas in stationary phase the transcript levels were similar (Fig. 2B). To exclude the possibility that plasmid replication or stability function was the cause of the observed extracytoplasmic stress response, we tested cells harboring the repressed plasmid R1. As expected but in contrast to cells containing the derepressed plasmid R1-16, when cells harbored the repressed plasmid R1, no traA transcript was detectable by Northern blotting at any time point after feeding the cells with nutrients (data not shown). The degP transcript levels in cells harboring R1 were similar to plasmid-free cells (Fig. 2A), indicating that the sole presence of a plasmid in the cells and its replication do not trigger an extracytoplasmic stress response. We interpreted these data to mean that in cells harboring plasmid R1-16, an extracytoplasmic stress response is elicited as a result of growth-dependent induction of the T4S components. In order to corroborate that the CpxAR system was activated in cells expressing T4S functions, we also tested for transcript levels of flagellar synthesis genes which are known to be repressed by the phosphorylated response regulator CpxR-P (15). For that purpose, we used a probe detecting motA mRNA in the motABcheAW operon. As can be seen in Fig. 2C, the appearance of motA mRNA in cells harboring plasmid R1-16 is delayed (90 min) when compared to cells without plasmid (45 min) or cells containing plasmid R1 (60 min). In this case, later time points were used for the Northern blot analyses because no signals could be detected even in plasmid-free strains in stationary phase and 15 min after the nutritional upshift.
FIG. 2.
Induction of a CpxAR-mediated extracytoplasmic stress response in E. coli cells expressing an active T4S system. E. coli cells without plasmid or harboring the repressed plasmid R1 or the derepressed plasmid R1-16 were analyzed by Northern blotting (A to C) or β-galactosidase assays (D). Analyses were performed in different phases of bacterial growth as indicated. Numbers indicate minutes after dilution of the cultures into fresh medium. S, stationary phase. (A to C) The visualized transcripts are indicated. The apparent size of the major mRNA species is given, when appropriate. 23S rRNA is shown as a loading control. Numbers at the bottom of the Northern blot shown in panel B indicate the relative signal intensities (n-fold) compared to the signal intensity in stationary phase. (D) The extracytoplasmic stress response mediated by the T4S system is abolished in a cpxR mutant genetic background. The activity of a transcriptional degP-lacZ fusion was measured 15 and 30 min after feeding stationary-phase cells with fresh medium. Shown are the results of two independent measurements.
A different experimental approach was used to confirm the role of the CpxAR two-component system in the T4S-dependent activation of degP. We performed β-galactosidase assays with extracts of cells carrying a chromosomally integrated transcriptional degP-lacZ fusion allowing investigations of the degP promoter activity in cpxR+ and cpxR mutant genetic backgrounds (14, 29). Supporting the data obtained by Northern blotting, we found that the β-galactosidase activity in the case of the cpxR+ E. coli strain SP556 harboring R1-16 increased twofold compared to the plasmid-free strain. A threefold-lower basal promoter activity was observed in β-galactosidase assays with cell lysates of the plasmid-free cpxR mutant strain, and no induction of the degP promoter could be observed when these cells harbored plasmid R1-16 (Fig. 2D). Taken together, our data demonstrate that the CpxAR two-component system is activated as a response to the expression and the assembly of T4S machineries in the cell envelope.
Expression of the plasmid R1-16-encoded T4S machinery elicits a stress response in the cytoplasm of E. coli.
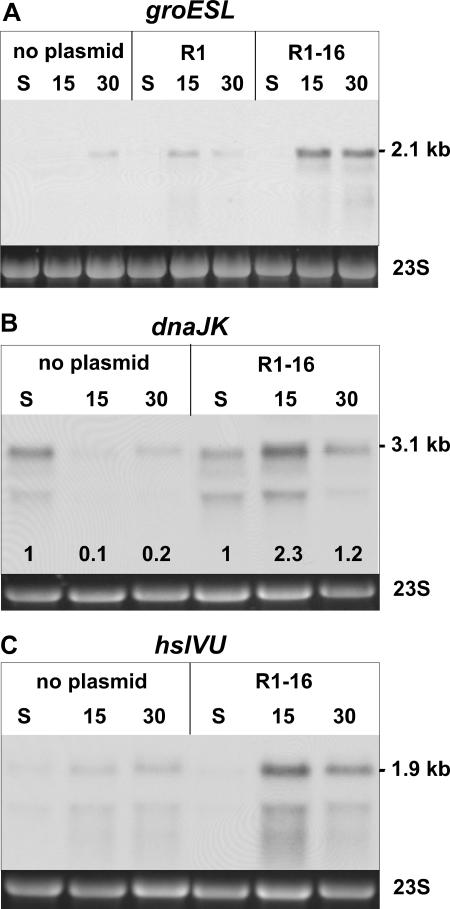
The data presented above demonstrate that expression of the plasmid R1-encoded T4S machinery activates the CpxAR extracytoplasmic stress monitoring system. This observation and the recently reported interaction of GroEL (Hsp60) with the specialized lytic transglycosylase encoded by plasmid R1 (61) prompted us to investigate whether the expression of the Tra proteins also stimulated the expression of genes encoding cytoplasmic heat shock proteins, such as GroEL. To address this question, we measured the steady-state levels of the groESL operon mRNA in plasmid-free cells and in cells harboring plasmid R1-16 after dilution of stationary-phase cells into fresh medium. Comparable to the results obtained for degP, groESL transcripts in cells with or without R1-16 were barely detectable in stationary phase. In contrast, in cells harboring R1-16, the groESL mRNA abundance strongly increased 15 and 30 min after dilution into fresh medium compared to plasmid-free cells (Fig. 3A). The presence of the repressed plasmid R1 did not result in such elevated transcript levels. In these cells, however, some activation of groESL transcription could be observed (compare signals at 15 min). This residual activation does not contradict the results above but rather reflects the fact that even in the population with cells harboring plasmid R1, a small proportion actively expresses the T4S system (approximately 1 out of 1,000 cells is transfer proficient).
FIG. 3.
Expression of the plasmid R1-encoded T4S system elicits a cytoplasmic stress response. E. coli MG1655 cells without plasmid or harboring the repressed plasmid R1 or the derepressed plasmid R1-16 were analyzed by Northern blotting. groESL (A), dnaJK (B), and hslVU (C) transcript levels are increased in cells expressing the R1-16-encoded T4S genes. 23S rRNA is shown as a loading control. Numbers above the blots indicate minutes after dilution. Numbers at the bottom of the Northern blot shown in panel B indicate the relative signal intensity compared to the signal intensity in stationary phase. S, stationary phase.
We next considered the possibility that the cytoplasmic stress response elicited by the expression of the T4S genes involved the entire heat shock regulon. In order to test this hypothesis, Northern blot analyses with specific probes for different heat shock genes were performed. The dnaKJ operon and the hslVU operon were chosen as representative members of the heat shock regulon. In cells harboring R1-16, the steady-state levels of the dnaKJ and hslVU mRNAs were increased 15 min and 30 min after fresh nutrients were provided (Fig. 3B and C, respectively). These results indicated that in cells harboring R1-16, the complete set of classical heat shock genes is induced as a result of the expression of T4S genes.
The stress responses are suppressed in an R1-16 traA null mutant.
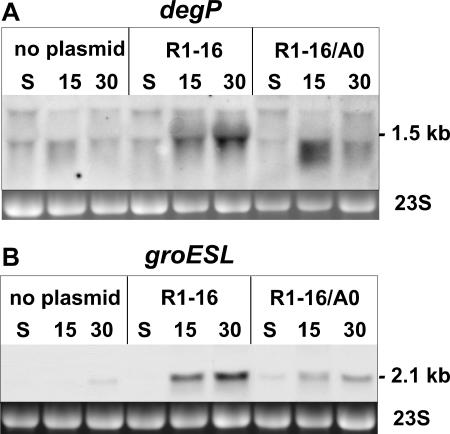
In order to confirm that the assembly of a functional T4S machinery into the cell envelope of E. coli is directly involved in the activation of the above-reported bacterial stress responses, we constructed a derivative of R1-16 that lacks a functional traA gene. The traA gene encodes pilin, the major structural component of sex pili, which is a conserved core element found in many T4S systems (13). As expected, the mutant R1-16/A0 plasmid was completely transfer negative because of the lack of the pilin protein. The mutation could be complemented by a plasmid expressing traA in trans and had no polar effects on the transcription of downstream genes, as revealed by investigation of traK transcription by quantitative reverse transcription-PCR. Also, stationary-phase repression and induction of transfer gene expression upon a nutritional upshift were similar to the R1-16 plasmid (data not shown). However, when mRNA levels of genes induced by extracytoplasmic or cytoplasmic stress were investigated, a response pattern completely different from R1-16 was observed. In the case of the steady-state levels of the degP mRNA, a much weaker signal corresponding to the full-length transcript was detected 15 and 30 min after supplying cells with fresh medium when cells harboring R1-16/A0 were compared to cells harboring R1-16 (Fig. 4A). Similar results were obtained when groESL mRNA levels were investigated (Fig. 4B). Compared to plasmid-free cells, only a weak residual activation of the investigated extracytoplasmic and cytoplasmic stress responses could be observed in cells carrying plasmid R1-16/A0 (Fig. 4). The residual stress responses in the case of the traA null mutant could result from partially or incompletely assembled T4S complexes in the cell envelope which lack a sex pilus. However, these results unambiguously demonstrated that activation of both extracytoplasmic and cytoplasmic stress responses is dependent on the presence of a core element of the plasmid-encoded T4S system and also indicate that pilin encoded by the traA gene plays a key role in the induction of the stress responses reported in this study.
FIG. 4.
Both extracytoplasmic and cytoplasmic stress responses are dependent on the presence of a functional pilin gene. E. coli MG1655 cells without plasmid or harboring plasmid R1-16 or the traA null variant R1-16/A0 were analyzed by Northern blotting. (A) Analysis of degP mRNA levels. (B) Analysis of groESL mRNA levels. 23S rRNA is shown as a loading control. Numbers indicate minutes after dilution. S, stationary phase.
Expression of T4S genes induces heat shock genes in a σ32-dependent manner.
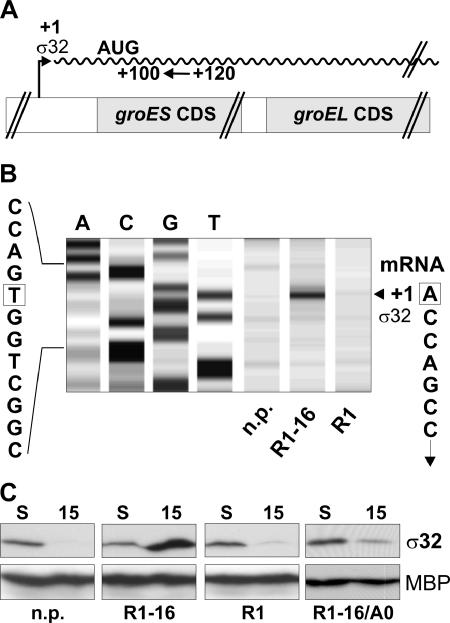
To determine the molecular mechanism that led to the activation of the observed T4S-induced cellular stress response, we performed further experiments aimed to clarify the role of the alternative sigma factor σ32 in the observed activation of cytoplasmic stress genes. Upon applying stress conditions to bacterial cells, such as a sudden increase of the growth temperature (heat shock), the transcription of stress genes is induced due to an increase of the cellular level of the alternative sigma factor σ32 (9, 26, 54). First, we determined whether the induction of heat shock genes in cells expressing the components of the T4S system is mediated by the alternative sigma factor σ32. For that purpose, primer extension analyses of the groESL operon transcription in plasmid-free cells, cells harboring R1-16, and cells harboring the repressed plasmid R1 were performed. Figure 5A shows a schematic representation of the experimental setup used for these studies. Primer extension analyses with RNA isolated from cells harboring R1-16 which were incubated for 15 min in fresh medium revealed a signal corresponding to the transcription start site of the σ32 promoter of the groESL operon (Fig. 5B). No detectable signals at the σ32-dependent transcription start site were observed with plasmid-free cells, and a very weak band was observed with cells harboring R1. From this experiment, we concluded that expression of the T4S genes induced the transcription of heat shock genes in a σ32-dependent manner. Next, we examined whether the protein steady-state levels of the heat shock transcription factor σ32 are increased in cells expressing the T4S machinery encoded by plasmid R1-16. As can be seen in Fig. 5C, 15 minutes after dilution the steady-state level of σ32 was increased more than 10-fold in cells harboring plasmid R1-16 compared to plasmid-free cells and cells harboring plasmid R1. Furthermore, when cells harbored the traA null mutant plasmid R1-16/A0 and therefore did not assemble functional T4S machineries, only a weak, residual increase of σ32 steady-state levels was observed. Thus, transcriptional activation of cytoplasmic stress genes as shown above can be unequivocally attributed to increased σ32 levels in cells expressing functional T4S machineries including the sex pilus.
FIG. 5.
The T4S system-dependent cytoplasmic stress response is mediated by σ32. (A) Schematic representation of the experimental setup used for primer extension analyses of the groESL transcript which is represented by the waved line. For reverse transcription, a Cy5-labeled oligonucleotide (indicated by an arrow) complementary to the sequence downstream from the σ32 transcription start site was used. AUG indicates the translation start site for the GroES protein within the groESL transcript. (B) Primer extension analyses with RNA isolated 15 min after dilution into fresh medium. The signal indicated by an arrowhead corresponds to transcripts originating from the transcription start site (+1) of the groESL σ32 promoter. The four lanes on the left show the DNA sequence surrounding the σ32-dependent transcription start site. Corresponding nucleotides in the DNA and RNA sequences are boxed. (C) Steady-state levels of σ32 protein are elevated when cells express a functional T4S system. Western blot analyses detecting σ32 (32 kDa) and maltose binding protein (45 kDa) are shown. S, stationary phase; 15, number of minutes after feeding cells with fresh medium.
rpoH mRNA levels are increased in cells expressing the T4S proteins via CpxR-dependent activation of rpoHp1.
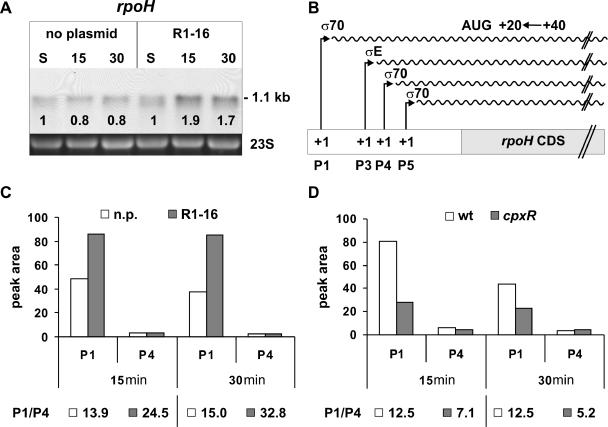
Besides regulation of σ32 at the protein level by a chaperone network (26), one mechanism to increase the cellular level of σ32 during heat shock in E. coli cells is the transcriptional induction of the rpoH gene encoding this alternative sigma factor (21, 23, 30, 43). Hence, we tested whether transcription of rpoH is increased in cells expressing the T4S machinery. Northern blot analysis showed that in cells harboring R1-16, the steady-state levels of the rpoH mRNA were 2.1- to 2.3-fold increased compared to plasmid-free cells, whereas in stationary phase the transcript levels were not distinguishable (Fig. 6A). From these data, we inferred that the expression of transfer proteins and/or assembly of the T4S complex in the cell envelope generated a signal which resulted in the transcriptional activation of the rpoH gene from one of its four promoters (for a schematic representation of rpoH promoters, see Fig. 6B). A transcriptional rpoHp3-lacZ fusion (39) was tested in β-galactosidase assays in the presence or absence of plasmid R1-16. Since we found no activation of σE-dependent rpoHp3 in the presence of R1-16 (data not shown), we excluded a transcriptional activation of rpoHp3, leaving p1, p4, and p5 as possible candidates for a transcriptional activation of the rpoH gene. To distinguish between the remaining possibilities, we performed primer extension analyses of rpoH mRNA. No signals corresponding to the rpoHp3 and p5 transcription start sites were detectable. In contrast, clear signals were detectable at the transcription start sites of the p1 and p4 promoters. Quantification revealed no difference in transcription of rpoH from the σ70-dependent p4 promoter in cells with or without plasmid (Fig. 6C). Since the p4 promoter activity remained constant, it was used to normalize p1 promoter activity in subsequent analyses. The abundance of transcripts starting at the p1 promoter was approximately twofold higher in cells expressing the T4S components than in cells without plasmid (Fig. 6C). This increase corresponds well to the data obtained by Northern blot analysis and indicates that the increase in rpoH transcript levels as described above arises mainly from increased transcription from rpoHp1.
FIG. 6.
Analyses of rpoH mRNA levels and promoter activities. (A) rpoH mRNA levels are increased in cells expressing the T4S system. E. coli MG1655 cells without plasmid or harboring plasmid R1-16 were analyzed by Northern blotting. 23S rRNA is shown as a loading control. Numbers at the top indicate minutes after dilution. Numbers at the bottom of the Northern blot indicate the relative signal intensity compared to the signal intensity in stationary phase. S, stationary phase. (B) Schematic representation of the experimental setup used for primer extension analyses of the rpoH transcripts (waved lines). For reverse transcription, a Cy5-labeled oligonucleotide (indicated by an arrow) complementary to the sequence downstream from the initiation codon (indicated by AUG) of the rpoH coding sequence was used. The promoters p1, p3, p4, and p5 as well as the sigma factors which are known to stimulate transcription from these promoters are indicated. The transcription start site is at +1. (C and D) Quantitative representation of primer extension analyses of rpoH transcripts isolated from cells at the indicated time points after feeding cells with fresh media. The relative abundance (reflected by the peak area) of transcripts starting at the indicated rpoH promoters p1 and p4 is shown. P1/P4, p1 promoter activity normalized to the unregulated promoter p4. (C) Comparison of specific rpoH transcripts from E. coli MG1655 cells without plasmid (n.p.) or harboring plasmid R1-16. (D) Comparison of specific rpoH transcripts from wild-type (wt) and cpxR mutant cells (cpxR).
Transcription from rpoHp1 is stimulated by CpxR-P.
The T4S-dependent activation of rpoHp1 as shown in the previous experiments suggested a possible involvement of CpxR-P in positive regulation of this promoter. In addition a CpxR-P binding site overlapping the rpoHp1 promoter has been previously identified, and in vitro binding of CpxR-P to a fragment containing rpoHp1 has been reported (16). However, how CpxR-P contributed to the regulation of this promoter was unknown, and it has been proposed that CpxR-P rather negatively regulates transcription from rpoHp1 (16). To resolve this issue and unravel the in vivo role of CpxR-P in regulation of the rpoHp1 promoter, we used the primer extension method described above and investigated the abundance of transcripts originating from the p1 promoter in cpxR+ and cpxR mutant cells. The results showed an approximately twofold reduced p1 promoter activity in the cpxR mutant background (Fig. 6D). Based on this observation and the results presented above, we propose that CpxR-P positively regulates transcription from rpoHp1. The positive regulatory role of CpxR-P in transcription from rpoHp1 has not been shown before and points to a link between extracytoplasmic and cytoplasmic stress regulons (Fig. 7).
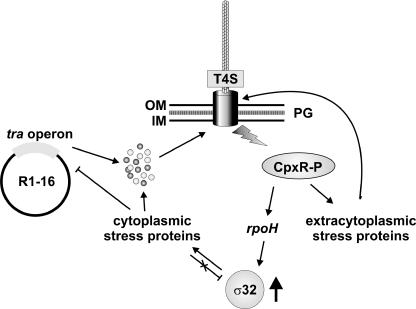
FIG. 7.
Sex is stress. In the depicted model, expression and assembly of the conjugative T4S complex activate extracytoplasmic and cytoplasmic stress regulons. The assembly of a functional T4S complex leads to extracytoplasmic stress that is sensed by the CpxAR system. The phosphorylated CpxR response regulator (CpxR-P) stimulates the transcription of periplasmic folding factors and proteases. These proteins may play a protective role or may be needed for efficient assembly of the T4S machinery or for degradation of excessive Tra proteins. A cytoplasmic stress response which induces the transcription of classical heat shock genes is mediated via increased steady-state levels of the alternative sigma factor σ32 (encoded by the rpoH gene). We assume that chaperons such as DnaJK-GrpE and/or GroEL/S bind to Tra proteins in the cytoplasm, thereby increasing the half-life of σ32. The induced proteins might be involved in degradation of regulatory Tra proteins and in conferring export competence to precursors of T4S proteins. Our data suggest that the two stress networks are connected via CpxR-P-dependent transcriptional activation of rpoH. OM, outer membrane; IM, inner membrane; PG, peptidoglycan.
DISCUSSION
Activation of the CpxAR system by the R1-encoded T4S machinery.
In the work presented here we demonstrate that in bacteria expressing T4S genes, extracytoplasmic and cytoplasmic stress networks are activated. The extracytoplasmic stress response was found to depend on CpxR, the response regulator of the CpxAR two-component system. This and the fact that known target genes of CpxR are either up- or down-regulated as predicted imply that in this case, CpxAR is central to the extracytoplasmic stress response induced by expression and assembly of functional DNA transporters encoded by the IncF plasmid R1. This conclusion is supported by the finding that the envelope stress response is greatly diminished in a traA null mutant. Pilin, the product of the traA gene and the major component of sex pili, is the most abundant of all Tra proteins and represents an essential core component of F-like T4S systems. However, in cells harboring the R1-16/A0 mutant plasmid, other T4S genes were still expressed, and their products were probably inserted into the cell envelope, resulting in the observed residual stress responses. Based on our data, we propose that the main signal which activates the CpxAR system emerges during the assembly of the sex pilus. Since mature pilin accumulates in the inner membrane before the assembly of the sex pilus (41), it is very likely that these pilin subunits—or other components of the T4S complex in the case of the R1-16/A0 mutant—might sequester the inhibitory CpxP protein, and thus autophosphorylation of CpxA in the inner membrane and subsequent phosphorylation of the response regulator CpxR are stimulated. Similarly, NlpE overexpression (53), P pilus assembly (29), and the assembly of bundle-forming pili (44) also have been reported to activate CpxAR.
Biological role of the observed periplasmic stress response.
The activation of the CpxAR two-component system leads to a transcriptional induction of genes encoding periplasmic folding factors, including disulfide bond catalysts and isomerases, peptidyl-prolyl cis/trans isomerases, and periplasmic chaperones which can also act as proteases, such as DegP (for a recent review, see reference 18). Periplasmic folding factors may support correct folding of T4S components in the periplasm and assembly of the T4S complex, including elaboration of a sex pilus. The finding that periplasmic disulfide oxidoreductase DsbA, which is a downstream target of the CpxAR system, is required for F pilus assembly indicates that this could indeed be the case (3). Since a functional cpxR gene is not essential for efficient DNA transfer of F (25) or R1-16 (our unpublished data), it is possible, however, that the basic expression of periplasmic folding factors is sufficient for F-type T4S assembly. Why then is CpxAR activated? It seems possible that CpxAR activation serves primarily a protective role for bacterial cells assembling the envelope-spanning DNA transporter and the sex pilus. A higher sensitivity of cells expressing F plasmid T4S genes toward bile salts (7) and enhanced leakiness of cells harboring R1-16 (our unpublished results) indicate that such protective effects may be beneficial for T4S gene-expressing bacteria. We are currently performing experiments to resolve this issue. Additionally, suppression of mot genes by CpxR-P is a likely cause for the observed decreased motility of E. coli in the presence of plasmid R1-19 (5). We dare to speculate that rapid flagella-driven movement in liquids is somehow incompatible with the formation of stable mating pairs between donor and recipient bacteria.
Activation of the σ32 stress regulon by the R1-encoded T4S machinery.
In addition to activation of the CpxAR system indicative of extracytoplasmic stress, we observed elevated transcript levels of cytoplasmic stress genes, such as groESL, in response to the expression and assembly of the T4S system of R1. These elevated transcript levels arise from transcriptional induction of these genes via the alternative “heat shock” sigma factor σ32, as demonstrated by transcripts initiating at the σ32-dependent groESL promoter and by high σ32 levels. How can this typical heat shock response be generated by the expression and assembly of the R1 plasmid-encoded T4S system? One possibility is the transcriptional activation of the rpoH gene encoding σ32 by the CpxAR signal-transducing cascade which originates in the cell envelope. It has been proposed that the CpxAR system regulates the transcription of the rpoH gene negatively (15, 16), but a direct proof of an in vivo effect on this promoter has not been provided so far. In this report, we conclusively showed that rpoHp1 activation is dependent on CpxR. In fact, rpoHp1 promoter activation is the cause of the elevated rpoH transcript levels, as demonstrated by primer extension analyses. Our data strongly indicate that CpxR-P acts as a positive regulator of rpoH transcription and reveals a hitherto undiscovered link between the periplasmic CpxAR system and the σ32 regulon. Thus, both the Cpx and σE pathways converge on rpoH transcription to transmit the information of envelope distress to the σ32-dependent stress regulon. In the model we propose (Fig. 7), the stress signal generated by the assembly of a functional T4S system leads to phosphorylation of CpxR and in turn to a transcriptional induction of extracytoplasmic stress genes and of the σ32-encoding gene rpoH. However, the observed twofold-enhanced steady-state levels of rpoH mRNA cannot explain the more than 10-fold elevated levels of σ32. We therefore suggest that σ32, which normally has a very short half-life of about 60 seconds, is stabilized by titration of DnaJK-GrpE and/or GroEL/S to newly synthesized T4S proteins (presumably mainly prepilin, which is the most abundant of all synthesized T4S proteins) awaiting export into the periplasm. Indeed, the observed very low level of σ32 in the case of the R1-16 traA null mutant in comparison to R1-16 strongly supports this hypothesis. Mechanistically, this process would be comparable to the reported stabilization of σ32 by the accumulation of secretory protein precursors in a secB-deficient E. coli strain (58). Enhanced transcription of rpoH as discussed above and increased σ32 stability together can explain the observed increase of σ32 levels which ultimately lead to transcriptional induction of the “classical” heat shock genes.
Biological role of the observed cytoplasmic stress response.
Up-regulation of cytoplasmic stress genes encoding chaperones and proteases could fulfill a dual role. First, chaperones could be involved in binding to T4S proteins destined for export and in keeping these proteins in an export-competent conformation by preventing premature folding. Both GroEL/S and DnaJK-GrpE have been found to function in protein export of a subset of exported proteins in E. coli (34, 46, 57), and it has been reported that heat shock proteins other than these can substitute for SecB (2). Although we have no evidence for such a requirement of chaperones in export of R1-encoded T4S proteins, we recently showed that P19, the specialized lytic transglycosylase encoded by plasmid R1, is bound by GroEL (61). Second, data from our laboratory indicate that a negative feedback loop exists in which GroEL, which is one of the induced proteins, is involved in the regulation of transfer gene expression (D. Zahrl and G. Koraimann, unpublished data). Excessive T4S proteins could be the target of proteases that are induced by the σ32-mediated stress response system. In this context, it is interesting that a gain-of-function mutation in the sensor kinase gene cpxA (cpxA*) results in severe destabilization of the cytoplasmic TraJ protein, a key regulator of tra operon transcription of F and F-like plasmids (25). This finding would perfectly fit to our data considering the herein proposed link between the CpxAR system and the σ32 stress regulons. According to this model, TraJ would be destabilized by one of the proteases that are induced indirectly via transcriptional induction of rpoH. We are currently investigating this possibility in our laboratory.
Acknowledgments
We thank T. Silhavy for E. coli strains SP556 and SP559. We are grateful to A. Reisner for providing plasmid pAR48 and for helpful suggestions.
Research in our laboratory is supported by the EU project QLK22-CT-2001-01200 within the fifth framework (FP5) and by the Fonds zur Förderung der Wissenschaftlichen Forschung (FWF), grant no. P17857-B12.
REFERENCES
- 1.Adachi, H., M. Nakano, M. Inuzuka, and M. Tomoeda. 1972. Specific role of sex pili in the effective eliminatory action of sodium dodecyl sulfate on sex and drug resistance factors in Escherichia coli. J. Bacteriol. 109:1114-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman, E., C. A. Kumamoto, and S. D. Emr. 1991. Heat-shock proteins can substitute for SecB function during protein export in Escherichia coli. EMBO J. 10:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 4.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 5.Barrios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188-200. [DOI] [PubMed] [Google Scholar]
- 6.Beranek, A., M. Zettl, K. Lorenzoni, A. Schauer, M. Manhart, and G. Koraimann. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 186:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidlack, J. E., and P. M. Silverman. 2004. An active type IV secretion system encoded by the F plasmid sensitizes Escherichia coli to bile salts. J. Bacteriol. 186:5202-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 9.Bukau, B. 1993. Regulation of the Escherichia coli heat-shock response. Mol. Microbiol. 9:671-680. [DOI] [PubMed] [Google Scholar]
- 10.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 11.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 15.De Wulf, P., O. Kwon, and E. C. Lin. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181:6772-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 17.Disqué-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duguay, A. R., and T. J. Silhavy. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 1694:121-134. [DOI] [PubMed] [Google Scholar]
- 19.Ebersbach, G., and K. Gerdes. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39:453-479. [DOI] [PubMed] [Google Scholar]
- 20.Erickson, J. W., and C. A. Gross. 1989. Identification of the σE subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 21.Erickson, J. W., V. Vaughn, W. A. Walter, F. C. Neidhardt, and C. A. Gross. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1:419-432. [DOI] [PubMed] [Google Scholar]
- 22.Frost, L. S., and J. Manchak. 1998. F phenocopies: characterization of expression of the F transfer region in stationary phase. Microbiology 144:2579-2587. [DOI] [PubMed] [Google Scholar]
- 23.Fujita, N., and A. Ishihama. 1987. Heat-shock induction of RNA polymerase sigma-32 synthesis in Escherichia coli: transcriptional control and a multiple promoter system. Mol. Gen. Genet. 210:10-15. [DOI] [PubMed] [Google Scholar]
- 24.Goebel, W., W. Lindenmaier, H. Schrempf, R. Kollek, and D. Blohm. 1977. Dissociation and recombination of fragments with defined functions of the antibiotic resistance factor R1. In J. Drews and G. Högenauer (ed.), Topics in infectious diseases, vol. 2. Springer Verlag, Vienna, Austria. [Google Scholar]
- 25.Gubbins, M. J., I. Lau, W. R. Will, J. M. Manchak, T. L. Raivio, and L. S. Frost. 2002. The positive regulator, TraJ, of the Escherichia coli F plasmid is unstable in a cpxA* background. J. Bacteriol. 184:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guisbert, E., C. Herman, C. Z. Lu, and C. A. Gross. 2004. A chaperone network controls the heat shock response in E. coli. Genes Dev. 18:2812-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris, R. L., V. Hombs, and P. M. Silverman. 2001. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42:757-766. [DOI] [PubMed] [Google Scholar]
- 28.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallipolitis, B. H., and P. Valentin-Hansen. 1998. Transcription of rpoH, encoding the Escherichia coli heat-shock regulator σ32, is negatively controlled by the cAMP-CRP/CytR nucleoprotein complex. Mol. Microbiol. 29:1091-1099. [DOI] [PubMed] [Google Scholar]
- 31.Koraimann, G. 2004. Bacterial conjugation: cell-cell contact-dependent horizontal gene spread. In R. V. Miller and M. J. Day (ed.), Microbial evolution: gene establishment, survival, and exchange. ASM Press, Washington, D.C.
- 32.Koraimann, G., C. Koraimann, V. Koronakis, S. Schlager, and G. Högenauer. 1991. Repression and derepression of conjugation of plasmid R1 by wild-type and mutated finP antisense RNA. Mol. Microbiol. 5:77-87. [DOI] [PubMed] [Google Scholar]
- 33.Koraimann, G., K. Teferle, G. Markolin, W. Woger, and G. Högenauer. 1996. The FinOP repressor system of plasmid R1: analysis of the antisense RNA control of traJ expression and conjugative DNA transfer. Mol. Microbiol. 21:811-821. [DOI] [PubMed] [Google Scholar]
- 34.Kusukawa, N., T. Yura, C. Ueguchi, Y. Akiyama, and K. Ito. 1989. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 8:3517-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 36.Lipinska, B., S. Sharma, and C. Georgopoulos. 1988. Sequence analysis and regulation of the htrA gene of Escherichia coli: a σ32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 16:10053-10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, J., W. Zhao, and L. S. Frost. 2004. Mutational analysis of TraM correlates oligomerization and DNA binding with autoregulation and conjugative DNA transfer. J. Biol. Chem. 279:55324-55333. [DOI] [PubMed] [Google Scholar]
- 38.McEwen, J., and P. Silverman. 1980. Chromosomal mutations of Escherichia coli that alter expression of conjugative plasmid functions. Proc. Natl. Acad. Sci. USA 77:513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of σE, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Moore, D., B. A. Sowa, and K. Ippen-Ihler. 1981. Location of an F-pilin pool in the inner membrane. J. Bacteriol. 146:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 43.Nagai, H., R. Yano, J. W. Erickson, and T. Yura. 1990. Transcriptional regulation of the heat shock regulatory gene rpoH in Escherichia coli: involvement of a novel catabolite-sensitive promoter. J. Bacteriol. 172:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevesinjac, A. Z., and T. L. Raivio. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187:672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordström, K. 2006. Plasmid R1—replication and its control. Plasmid 55:1-26. [DOI] [PubMed] [Google Scholar]
- 46.Phillips, G. J., and T. J. Silhavy. 1990. Heat-shock proteins DnaK and GroEL facilitate export of LacZ hybrid proteins in E. coli. Nature 344:882-884. [DOI] [PubMed] [Google Scholar]
- 47.Pölzleitner, E., E. L. Zechner, W. Renner, R. Fratte, B. Jauk, G. Högenauer, and G. Koraimann. 1997. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol. Microbiol. 25:495-507. [DOI] [PubMed] [Google Scholar]
- 48.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reisner, A., S. Molin, and E. L. Zechner. 2002. Recombinogenic engineering of conjugative plasmids with fluorescent marker cassettes. FEMS Microbiol. Ecol. 42:251-259. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Schröder, G., and E. Lanka. 2005. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1-25. [DOI] [PubMed] [Google Scholar]
- 52.Silverman, P. M., L. Tran, R. Harris, and H. M. Gaudin. 1993. Accumulation of the F plasmid TraJ protein in cpx mutants of Escherichia coli. J. Bacteriol. 175:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Straus, D. B., W. A. Walter, and C. A. Gross. 1987. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature 329:348-351. [DOI] [PubMed] [Google Scholar]
- 55.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 56.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wild, J., P. Rossmeissl, W. A. Walter, and C. A. Gross. 1996. Involvement of the DnaK-DnaJ-GrpE chaperone team in protein secretion in Escherichia coli. J. Bacteriol. 178:3608-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wild, J., W. A. Walter, C. A. Gross, and E. Altman. 1993. Accumulation of secretory protein precursors in Escherichia coli induces the heat shock response. J. Bacteriol. 175:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Will, W. R., J. Lu, and L. S. Frost. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol. Microbiol. 54:769-782. [DOI] [PubMed] [Google Scholar]
- 60.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zahrl, D., M. Wagner, K. Bischof, M. Bayer, B. Zavecz, A. Beranek, C. Ruckenstuhl, G. Zarfel, and G. Koraimann. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455-3467. [DOI] [PubMed] [Google Scholar]