Abstract
IS1999 and a point mutant derivative, IS1999.2, have been described inserted upstream of emerging antibiotic resistance genes blaVEB-1 and blaOXA-48. 5′ Rapid amplification of cDNA ends experiments revealed that expression of these β-lactamase genes was driven by the outward-directed promoter, Pout, located in the IS1999 elements. These findings led us to study IS1999-mediated gene mobilization. Thus, the transposition properties of IS1999 and of IS1999-based composite transposons, made of two copies of IS1999 in different orientations, were investigated. IS1999 or IS1999-based composite transposons were capable of transposing onto the conjugative plasmid pOX38-Gen. Sequence analysis of the insertion sites revealed that IS1999 inserted preferentially into DNA targets containing the consensus sequence NGCNNNGCN. Transposition was more efficient when at least one left inverted repeat end was located at an outside end of the transposon. The transposition frequency of IS1999.2 was 10-fold lower than that of IS1999, and transposition frequencies of the putative natural transposon, Tn1999, were below detection limits of our transposition assay. This reduced transposition frequency of IS1999.2-based elements may result from a lower transcription of the transposase gene, as revealed by reverse transcription-PCR analyses.
Insertion sequences (ISs) are present in most bacterial genomes analyzed and may represent up to 2 to 5% of genomic DNA. They play an important role in assembling sets of “accessory” functions in bacteria and in dissemination of resistance genes (6, 15, 19, 22). Many IS elements can transmit these gene arrays in the form of composite transposons, where two flanking IS elements cooperate in mobilizing an intervening DNA segment.
ISs of the IS4 family are delimited by short imperfect inverted repeat sequences (IR). These ISs encode a transposase that is required for transposition and are capable of inserting into a target molecule, leading to the duplication of several base pairs (directly repeated sequences) at the site of insertion (6).
IS1999 was initially identified in clinical Pseudomonas aeruginosa isolates from Thailand (8, 17, 32). In these strains, IS1999 was inserted into the integron-specific recombination site, attI1, upstream of the integron-borne blaVEB-1 gene that encodes an extended-spectrum β-lactamase (17, 23, 25). IS1999 encodes a putative transposase of 402 amino acids (17) that shares 71% amino acid identity with IS10, the best-characterized IS4 family member (6). IS1999 is 1,328 bp long, has 21-bp imperfect terminal repeats, and generates a 9-bp target site duplication after transposition (17). An outward-directed promoter, Pout, located close to the left inverted repeat (IRL) of IS1999 was characterized (1). In previous work, Pout together with the promoter Pant of class 1 integrons was shown to increase the expression of the blaVEB-1 gene in P. aeruginosa (1).
Recently, IS1999.2, an isoform of IS1999, has been detected in Klebsiella pneumoniae (21). This IS was located upstream of another unrelated antibiotic resistance gene, blaOXA-48, that encodes a broad-spectrum β-lactamase of a different type (21). The present work describes the analysis of the genetic environment of blaOXA-48 that revealed the presence of two identical copies of the IS1999 isoform inserted on either side of the resistance gene. The frequent association of IS1999 with resistance genes of clinical interest led us to study IS1999-mediated antibiotic resistance gene mobilization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Clinical strains P. aeruginosa 1 (blaVEB-1), K. pneumoniae 11978 (blaOXA-48), and Escherichia coli DH10B (pA-1) harboring the natural plasmid of K. pneumoniae 11978 have been previously described (8, 21). E. coli DH10B (Life Technologies, Eragny, France) was used as the bacterial host in electroporation experiments. The recombination-deficient strain E. coli DH5α (pOX38-Gen) and the rifampin-resistant E. coli DH10B Rifr were used for transposition and conjugation experiments (12). The low-copy-number cloning vector pBBR1MCS.3 was used for cloning experiments (13). Bacterial cells were grown in trypticase soy (TS) broth or on TS agar plates (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France) with antibiotic solutions when required.
Antimicrobial agents and susceptibility testing.
Routine antibiograms were determined by the disk diffusion method on Mueller-Hinton agar (Sanofi Diagnostics Pasteur). The antimicrobial agents (and their sources) were the following: ticarcillin (GlaxoSmithKline, Marly-Le-Roi, France), gentamicin (Schering-Plough, Levallois-Perret, France), rifampin (Aventis, Paris, France), tetracycline (Sigma, Saint Quentin Fallavier, France), and kanamycin (Euromedex, Mundolsheim, France). The antibiotics, with abbreviations and concentrations used for selection, were the following: kanamycin (Kan; 30 μg/ml), ticarcillin (Tic; 50 μg/ml), tetracycline (Tet; 15 μg/ml), gentamicin (Gen; 7 μg/ml), and rifampin (Rif; 200 μg/ml).
Nucleic acid extraction.
Recombinant plasmids and pOX38-Gen-derivative plasmids were extracted using Plasmid Mini-Midi kits and the Very Low Copy Plasmids purification protocol, respectively (QIAGEN, Courtaboeuf, France). Extractions of whole-cell DNAs were as described elsewhere (18). Total RNAs from E. coli DH10B (pA-1) were extracted with an RNeasy Maxi kit (QIAGEN) according to the recommendations of the manufacturer.
The presence of episomal plasmids in the host strains was checked using a Plasmid Mini kit (QIAGEN). DNA extracted from agarose gel slices and PCR products were purified using QIAquick columns (QIAGEN).
PCR experiments and hybridization.
Taq DNA polymerase (Roche Diagnostics, Meylan, France) and Pfu DNA polymerase (Promega Corporation, Madison, Wis.) were used as recommended. Standard PCR amplification experiments (27) were performed with primers listed in Table 1.
TABLE 1.
Sequences of primers used in this studya
| Primer | Sequence (5′→3′) | Accession no. | Primer location (bp) | Reference or source |
|---|---|---|---|---|
| IS1999A | CAGCAATTCTTTCTCCGTG | AF133699 | 1205-1223 | 21 |
| IS1999B1 | CAAGCACAACATCAAGCGC | AF133699 | 2189-2171 | 21 |
| IS1999B2INV | TCGTTTTAGGTGAAGTTCTGG | AF133699 | 3475-3495 | This work |
| IS1999IRLext | TGCGCTTCCACCCTAATTTG | AF133699 | 3292-3611 | This work |
| IS1999IRRext | ggatccccggaATTCGCCA | AF133699 | 1190-1183 | This work |
| IS1999IRRext2 | TGGGATTGGAGGTACTCAGGC | AF133699 | 1260-1240 | This work |
| IS-1Kpn | TTAAggTACCGCTAACTTTGTTTTAGGG | AF133699 | 3695-3668 | This work |
| IS-2Eco | ctcTgaatTCATAAATCAGCCATAGCATAGC | AF133699 | 1142-1164 | This work |
| IS-3Eco | GTAgaATTCGCCAATCAGTTGCTC | AF133699 | 1195-1172 | This work |
| IS-4Pst | TGTcTGcaGTTATGGAGCAGCAACGATG | AF133699 | 1094-1121 | This work |
| IS-1Eco | TTgAATTcCCGCTAACTTTGTTTTAGGG | AF133699 | 3695-3668 | This work |
| IS-1Sac | TTAAATTACCGCggACTTTGTTTTAGGG | AF133699 | 3695-3668 | This work |
| IS-4Eco | TGaaTtcTGTTATGGAGCAGCAACGATG | AF133699 | 1094-1121 | This work |
| IS-stopXmn | TCACGAAAGGTTTCCTCatcaTTGCATACGTTTGG | AF133699 | 1461-1494 | This work |
| OXA-48A | TTGGTGGCATCGATTATCGG | AY236073 | 1280-1299 | 21 |
| OXA-48B | GAGCACTTCTTTTGTGATGGC | AY236073 | 2023-2003 | 21 |
| IS1999.2F | ATCCGCTTTTTTTACAGGCCGA | AY648695 | 4374-4395 | This work |
| blaOXA-48GSP1 | AAAGCATGTAGCATCTTGCTCATAC | AY236073 | 1674-1650 | This work |
| blaOXA-48GSP2 | ACAGGCACAACTGAATATTTCATC | AY236073 | 1614-1591 | This work |
| blaOXA-48GSP3 | TCTGTCCATCCCACTTAAAGACTT | AY236073 | 1544-1521 | This work |
| GapA1 | ATCAACGGTTTTGGCCGTAT | X02662 | 502-521 | Corvec |
| GapA2 | GTTGATAACTTTAGCCAGCGG | X02662 | 972-952 | Corvec |
| TnpA1999-GSP1 | GCTTAGCCAAAACCAACCAT | AF133697 | 649-630 | This work |
| TnpA1999-GSP2 | TAGGGGATAGGACTTCTCAT | AF133697 | 496-477 | This work |
| TnpA1999-GSP3 | ATGCGCTTGATGTTGTGCTT | AF133697 | 289-269 | This work |
Nucleotides that were not complementary to the sequence submitted to the GenBank database under the accession number AF133699 are shown by lowercase letters; the restriction sites introduced into the primer sequences are boldfaced; the two successive TGA stop codons that are located in the IS-stopXmn primer (inverse sequence) are underlined.
Southern blot hybridizations (27) were performed using whole-cell DNAs of P. aeruginosa 1, K. pneumoniae 11978, E. coli DH10B (pA-1), and E. coli DH10B. Four micrograms of each DNA was digested with 10 U of the restriction enzyme EcoRV. Hybridizations were performed under high-stringency conditions using the ECL nonradioactive labeling and detection kit (Amersham Pharmacia Biotech, Orsay, France). PCR-generated probes consisted of a 985-bp fragment internal to IS1999 using IS1999A and IS1999B1 as primers and whole-cell DNA of P. aeruginosa 1 as template as well as of a 744-bp fragment internal to blaOXA-48 using OXA-48A and OXA-48B as primers and whole-cell DNA of K. pneumoniae 11978 as template (Table 1).
Cloning experiments and sequencing.
T4 DNA ligase and restriction endonucleases were used according to the recommendations of the manufacturer (Amersham Biosciences). In order to study the transposition of IS1999, an omega fragment (ΩKm) from plasmid pHP45Ω-Km (24), made of a kanamycin resistance gene [aph(3′)-IIa] flanked by transcriptional and translational termination sequences, was introduced into IS1999 (Fig. 1). A 1.3-kb fragment containing the left IR up to the end of the transposase gene was amplified with primers tailed with restriction sites for KpnI (IS-1Kpn) and EcoRI (IS-2Eco) and genomic DNA from P. aeruginosa 1 as template (Table 1). The primers IS-3Eco and IS-4Pst were used to amplify a 102-bp fragment containing the right IR (IRR) of IS1999 (Table 1). The digested PCR products were mixed in a four-way ligation together with an EcoRI-restricted ΩKm fragment (2.2 kb) and the KpnI-PstI-restricted pBBR1MCS.3 vector in order to create the tagged insertion sequence IS1999.Kan, yielding plasmid pIS1999.Kan (Fig. 1).
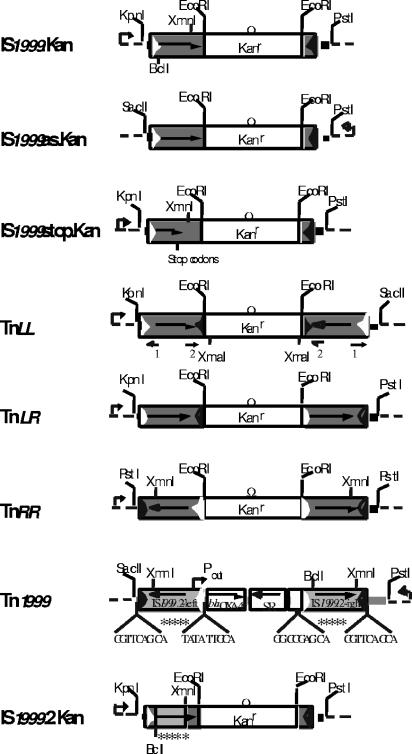
FIG. 1.
Schematic map of the constructs used in this study. All constructs were cloned into the multiple cloning site of the pBBR1MCS.3 shuttle vector. Restriction sites that were used for cloning are indicated. The coding regions are shown as boxes, with an arrow indicating the orientation of their transcription. The broken arrows indicate the Plac promoter. The IS1999 left and right inverted repeats are shown by empty and filled triangles, respectively. IS1999 and iso-IS1999 elements are represented as dark gray boxes and light gray boxes, respectively. Ω Kanr is an omega fragment conferring resistance to kanamycin. The environments of IS1999 in P. aeruginosa 1 and of Tn1999 on the plasmid pA-1 are shown by dark and gray lines, respectively. Dashed lines represent the cloning vector. The 9-bp sequences located on each side of IS1999.2-left and of IS1999.2-right are indicated on the Tn1999 representation. The five nucleotide substitutions in the IS1999.2 sequence are symbolized with five stars. The position of the primers IS1999B2INV (1) and IS1999IRRext2 (2), used in inverse PCR experiments, are symbolized by small arrows on the TnLL representation. The location of the Pout promoter responsible for the blaOXA-48 transcription is represented by a small broken arrow on the Tn1999 representation.
In order to obtain a construct in which the transposase gene was in opposite orientation relative to Plac, DNA fragments of 1.3 kb and of 102 bp were amplified by PCR with the pairs of primers IS-2Eco/IS-1Sac and IS-3Eco/IS-4Pst, respectively. These DNA fragments were subsequently restricted with the appropriate enzymes and mixed in a four-way ligation together with EcoRI-restricted ΩKm fragment and PstI-SacII-restricted pBBR1MCS.3 vector to create pIS1999as.Kan (Fig. 1).
A similar PCR-based cloning strategy was used for the following constructs (Fig. 1). Fragments of 1.3 kb were amplified with the pairs of primers IS-1Kpn/IS-4Eco and IS-4Eco/IS-1Sac, digested with the appropriate enzymes, and mixed in a four-way ligation with EcoRI-restricted ΩKm fragment and KpnI-SacII-restricted pBBR1MCS.3 vector to create pTnLL, carrying two copies of IS1999 in inward and opposite orientations. Fragments of 1.3 kb were amplified with the pairs of primers IS-1Kpn/IS-4Eco and IS-1Eco/IS-4Pst, digested with the appropriate enzymes, and mixed in a four-way ligation with EcoRI-restricted ΩKm fragment and KpnI-PstI-restricted pBBR1MCS.3 vector to create pTnLR, carrying two copies of IS1999 in the same orientation. Fragments of 1.3 kb were amplified with the pair of primers IS-1Eco/IS-4Pst, digested with the appropriate enzymes, and ligated with EcoRI-restricted ΩKm fragment and PstI-restricted pBBR1MCS.3 vector to create pTnRR, carrying two copies of IS1999 in outward and opposite orientations.
The introduction of two stop codons in the IS1999 transposase sequence was performed as follows. A 1.1-kb fragment was amplified with the primers IS-1Kpn and IS-stopXmn using the genomic DNA of P. aeruginosa 1 as template (Table 1). The purified PCR product was restricted using KpnI and XmnI prior to its introduction into the KpnI-XmnI-restricted plasmid pIS1999.Kan, yielding plasmid pIS1999stop.Kan (Fig. 1).
SacII-PstI-restricted whole-cell DNA of E. coli DH10B (pA-1) was cloned into a SacII-PstI-restricted pBBR1MCS.3 vector (Fig. 1), giving rise to plasmid pTn1999.
The IS1999 isoform (IS1999.2) was cloned in the same environment as pIS1999.Kan as follows. A 1.3-kb fragment was amplified with the primers IS1999.2F and IS1999A, with genomic DNA of E. coli DH10B (pA-1) as template (Table 1), and then it was digested with BclI-XmnI (this fragment contained five nucleotide substitutions relative to the sequence of IS1999). The BclI-XmnI fragment was purified and introduced into the BclI-XmnI-restricted pIS1999.Kan plasmid, generating pIS1999.2.Kan (Fig. 1).
Ligation products were electroporated into E. coli DH10B, as previously described (18), and selection was performed on TS agar plates containing the appropriate antibiotics.
Sequencing of the insert of each construct was performed using laboratory-designed primers on an ABI PRISM 3100 automated sequencer (Applied Biosystems, Les Ullis, France).
Mating-out assay and transposition experiments.
The transposition of single tagged ISs and composite transposons onto the conjugative plasmid pOX38-Gen was investigated by mating-out assay in liquid medium (18). The recombinant plasmids pIS1999.Kan, pIS1999as.Kan, pIS1999stop.Kan, pIS1999.2.Kan, pTnLR, pTnLL, pTnRR, and pTn1999 (Fig. 1) were electroporated into E. coli DH5α (pOX38-Gen) for transposition experiments. The experiments were performed in triplicate in three independent experiments. For each strain tested, one colony that grew on agar plates for 24 h was cultured under weak agitation in 1 ml TS broth at 37°C for 3 h. The inoculum was then used as a donor for mating assays with E. coli DH10B Rifr as recipient. Mating was done by incubating 800 μl of recipient and 200 μl of donor strains under low agitation at 37°C for an additional 3 h. Mating was stopped by vigorous vortexing and cooling on ice. One-hundred-microliter aliquots of serial 10-fold dilutions were then plated onto TS agar plates with gentamicin, with rifampin, and with or without ticarcillin for pTn1999 and with gentamicin, with rifampin, and with or without kanamycin for the other plasmids. The transposition frequency was calculated by dividing the number of Genr Kanr Rifr or Genr Ticr Rifr transconjugants by the number of Genr Rifr transconjugants. All the Genr Kanr Rifr or Genr Ticr Rifr colonies were screened for tetracycline susceptibility to exclude those that may have arisen from nontranspositional events.
Insertion site determination.
Plasmid pOX38-Gen carrying various insertions of either IS1999.Kan alone or IS1999 composite transposons was extracted from independent and randomly chosen E. coli DH10B Rifr transconjugants. Direct sequencing of the ends of IS1999 using the primers IS1999IRLext and IS1999IRRext (Table 1) was performed in order to determine the precise sites of insertion of IS1999.Kan elements, whereas the insertion site for composite transposons was determined by an inverse PCR technique as follows. Plasmid DNAs were digested with XbaI (Fig. 1). After inactivation of the endonuclease, an intramolecular ligation was performed. Two microliters of the ligation mixture was used as template with IS1999B2INV and IS1999IRRext2 primers for inverse PCR amplification, and then the PCR-generated products were sequenced (Table 1).
Transcription initiation.
RNA was extracted using the RNA Quick kit (QIAGEN) according to the recommendation of the manufacturer. Reverse transcription (RT) and rapid amplification of cDNA ends (RACE) were performed with the 5′RACE system, version 2.0 (Invitrogen, Life Technologies, Cergy Pontoise, France). Five micrograms of total RNAs extracted from cultures of E. coli DH10B (pA-1) and the blaOXA-48GSP1, blaOXA-48GSP2, and blaOXA-48GSP3 antisense blaOXA-48 gene-specific primers (Table 1) were used to determine the blaOXA-48 transcription initiation site. Similarly, five micrograms of total RNAs extracted from culture of E. coli DH10B (pIS1999.Kan) and E. coli DH10B (pIS1999.2.Kan) with TnpA1999-GSP1, TnpA1999-GSP2, and TnpA1999-GSP3 antisense gene-specific primers (Table 1) were used to determine the transcription initiation site of the transposase gene of IS1999 and IS1999.2.
RT-PCR conditions.
Five micrograms of total RNAs extracted from culture of E. coli DH10B (pIS1999.Kan) and E. coli DH10B (pIS1999.2.Kan) was DNase treated for 15 min at 25°C in a final volume of 16 μl containing 2 U of RNase-free DNase (Roche). EDTA was added to a final concentration of 2.5 mM, and the DNase was inactivated by a 10-min incubation at 65°C. Five microliters of DNase-treated RNA was reverse transcribed in a final volume of 25 μl using 200 U of the Moloney murine leukemia virus based SuperScript III reverse transcriptase as recommended by the manufacturer (Invitrogen) and 2 μM of the reverse primer (TnpA1999-GSP1 and GapA2; see below). The reaction mixture was incubated for 1 h at 42°C, followed by a 5-min incubation at 95°C. Ten microliters of the cDNA was used for amplification of specific IS1999-tnpA mRNA using primers IS1999B1 and TnpA1999-GSP2 (Table 1), yielding a 228-bp fragment, and 10 μl was used for amplification of gap (4, 7) (encoding d-glyceraldehyde-3-phosphate dehydrogenase, used as an external control) using primers GapA1 and GapA2 and yielding a 470-bp fragment in the following conditions: 12 min of initial denaturation at 94°C, followed by 35 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and 1 min of extension at 72°C, with a final extension step of 10 min at 72°C. Both PCR products were detected on a 2% agarose gel. Each RT-PCR was performed in triplicate. The band intensities were estimated using the Taxotron software (Institut Pasteur).
Nucleotide sequence accession number.
The nucleotide sequence of Tn1999 has been submitted to the EMBL/GenBank nucleotide sequence database under the accession number AY236073.
RESULTS
Transposition of IS1999 by mating-out assay.
The transposition ability of IS1999.Kan was investigated by mating-out assays with the E. coli DH5α strain, harboring pIS1999.Kan (Kanr) and the gentamicin-resistant plasmid pOX38-Gen (Genr) (a transfer-proficient F plasmid derivative) as the donor and E. coli DH10B Rifr as the recipient.
Transposition events that resulted from conjugation of pOX38-Gen:IS1999.Kan were selected as Genr Kanr Rifr colonies and screened for susceptibility to tetracycline (Tets) at a frequency of 3.7 × 10−6 (Table 2). Susceptibility of E. coli (pOX38-Gen::IS1999.Kan) transconjugants to tetracycline indicated that resistance to kanamycin was neither due to plasmid cointegration (pIS1999.Kan::pOX38-Gen) nor to selection of rifampin-resistant E. coli DH5α (pIS1999.Kan). This suggests that a transpositional event had occurred. As a control, no Genr Kanr Rifr Tets transconjugant was obtained when IS1999stop.Kan was used (Table 2).
TABLE 2.
Frequency of the transposition of IS1999 and its derivatives
| Transposon | Transposition frequency (±SDa) |
|---|---|
| IS1999.Kan | 3.7 × 10−6 (1.7 × 10−6) |
| IS1999as.Kan | 1.7 × 10−6 (2.1 × 10−7) |
| IS1999stop.Kan | <1.0 × 10−7 |
| IS1999.2.Kan | 2.6 × 10−7 (8.7 × 10−8) |
| TnLR | 4.0 × 10−6 (9.2 × 10−7) |
| TnLL | 3.6 × 10−6 (1.0 × 10−6) |
| TnRR | 3.7 × 10−7 (2.6 × 10−7) |
| Tn1999 | <1.0 × 10−7 |
Standard deviations calculated from three independent cultures are indicated in parentheses.
Plasmids pIS1999.Kan and pIS1999as.Kan were isogenic, except for the Plac promoter orientation. The plasmid pIS1999.Kan carries the Plac promoter in a sense orientation with the transposase gene and might lead to its overexpression, whereas pIS1999as.Kan carries the Plac promoter in an antisense orientation. Compared to pOX38-Gen::IS1999.Kan, transposition events that resulted from conjugation of pOX38-Gen::IS1999as.Kan were screened as Genr Kanr Rifr Tets colonies at a lower frequency (1.7 × 10−6). The similarity between the transposition frequencies obtained with pIS1999.Kan and pIS1999as.Kan suggested that the transposition frequency of a single IS1999 element is independent of its orientation within the cloning vector. Moreover, it suggested that the Plac promoter in pIS1999.Kan (located in the cloning vector) had no significant effect on the transposase expression (Table 2).
Target site preference of insertion element IS1999.
Several randomly chosen Genr Kanr Rifr Tets transconjugants isolated from independent experiments were analyzed. The insertion sites of IS1999.Kan were determined by DNA sequencing of the external neighboring regions of the inverted repeats. A 9-bp target site duplication, consistent with a transposition event, was observed in all the studied insertion events (Fig. 2).
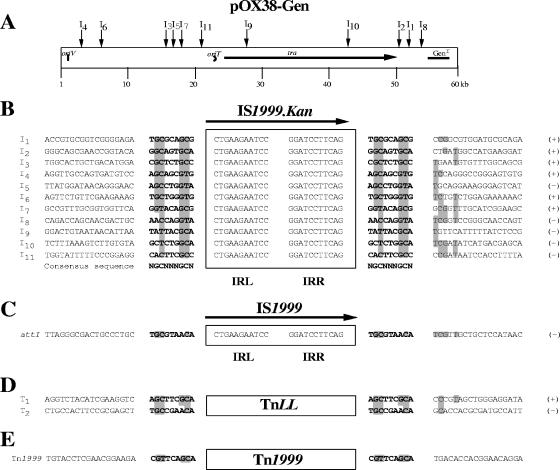
FIG. 2.
Target sites of IS1999 insertions. (A) Map positions of IS1999.Kan and TnLL insertions in plasmid pOX38-Gen. Insertions of the tagged insertion sequence (I1 to I11) and of the composite transposons (T1 and T2) are indicated by a vertical arrow. The origin of replication (oriV), the origin of transfer (oriT), the tra genes required for plasmid transfer, and the gentamicin resistance gene (Genr) are indicated on the pOX38-Gen representation. Nucleotide sequence alignment of the 11 pOX38-Gen::IS1999.Kan transconjugants (B), of the P. aeruginosa 1 attI1::IS1999 junctions (C), of two representative pOX38-Gen::TnLL insertions (D), and of the Tn1999 environment (E) are shown. Nucleotide sequences of the end regions of IS1999.Kan, IS1999, TnLL, and Tn1999 are boxed. Target site sequences duplicated after transposition are indicated by boldface letters. Gray boxes indicate conserved nucleotides in the environment of IS1999.Kan and IS1999 insertions. Orientation of the insertion sequences of IS1999.Kan and TnLL in plasmid pOX38-Gen are indicated by pluses and minuses.
To determine whether IS1999 had a target site preference, locations of 11 insertion events were mapped onto plasmid pOX38-Gen. IS1999.Kan insertions occurred in 11 different sites, and alignment of the insertion site sequences (I1 to I11) revealed a consensus motif (NGCNNNGCN) (Fig. 2). Indeed, most target sites had a purine at positions 2 and 7 with a preference for a guanosine (G) residue. The 11 target sites had a pyrimidine at positions 3 and 8, with C favored over T (Fig. 2).
Characterization of a putative natural transposon, Tn1999.
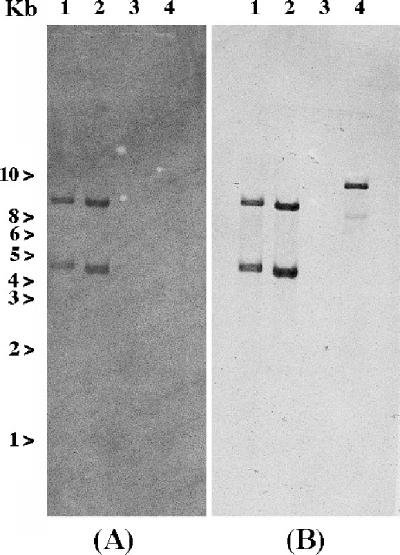
Recently, IS1999 was found upstream of the blaOXA-48 gene that was located on the natural plasmid (pA-1) (21). Southern hybridization experiments were performed with whole-cell DNA of E. coli DH10B (pA-1) that was digested with EcoRV (EcoRV cuts in blaOXA-48 and not in IS1999). Fragments internal to IS1999 or to blaOXA-48 were used as probes. Two fragments hybridizing with both probes were detected, suggesting the existence of two copies of IS1999 located on each side of blaOXA-48 that may be part of a composite transposon (Fig. 3).
FIG. 3.
Hybridizations of EcoRV-digested whole-cell DNAs of E. coli DH10B (pA-1) (lane 1), K. pneumoniae 11978 (lane 2), E. coli DH10B (lane 3), and P. aeruginosa 1 (lane 4). (A) blaOXA-48-specific hybridization. (B) IS1999-specific hybridization.
Whole-cell DNA of E. coli DH10B (pA-1) was restricted with several enzymes that cut at neither IS1999 nor the blaOXA-48 sequence. Hybridization with an IS1999-specific probe revealed that a SacII-PstI restriction generated a single fragment of 6.1 kb (data not shown). This SacII-PstI-restricted fragment was cloned and sequenced, identifying the structure of a 4.9-kb putative transposon named Tn1999. Tn1999 was made of two copies of IS elements in outward and opposite orientations that flanked a region originating from a Shewanella sp. genome (20). Tn1999 included two open reading frames encoding (i) a class D β-lactamase (OXA-48) that confers resistance to most β-lactams, including carbapenems, and (ii) a transcriptional regulator of the LysR family that shares 98% amino acid identity with that identified in the genome of Shewanella oneidensis MR-1 as well as a portion of an open reading frame encoding an acetyl-coenzyme A carboxylase multifunctional enzyme, AccADC (Fig. 1).
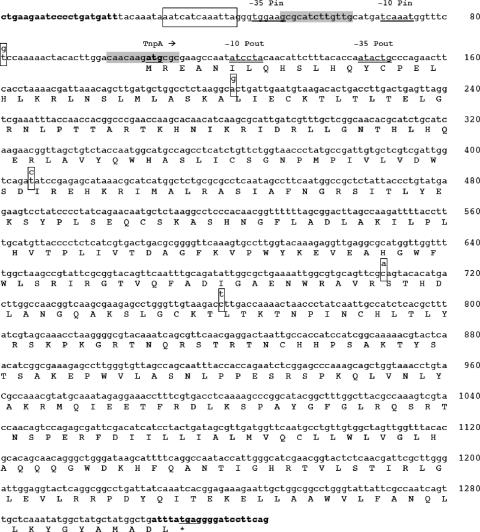
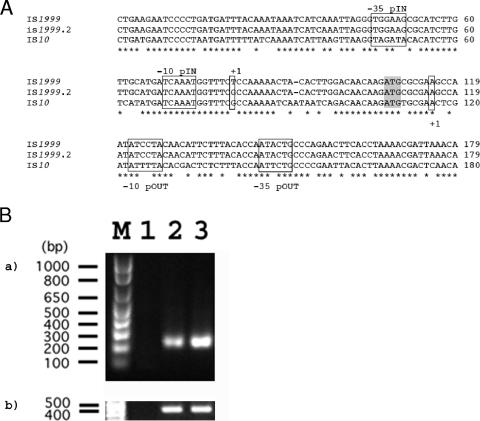
The IS elements of Tn1999 had identical sequences but were isoforms of IS1999 and were named IS1999.2. Indeed, IS1999.2-left and IS1999.2-right differed from the sequence of IS1999 (GenBank accession number AF133697) by five nucleotide substitutions. However, none of these substitutions led to changes of the amino acid sequence of the IS1999 transposase, and one of them was located next to the Pin +1 transcription start site (Fig. 4).
FIG. 4.
Nucleotide sequence of IS1999. The deduced amino acid sequence is designated in a single-letter code below the nucleotide sequence. The start and stop codons of the transposase gene are underlined. The transposase gene name, followed by an arrow indicating its translational orientation, is indicated above the initiation codon. IS1999 left and right inverted repeats (IRL and IRR) are boldfaced. The boxes represent the putative IHF binding site (bp 30 to 42). The complementary inverted repeat sequences are shown by gray boxes. The promoter sequences −10 and −35 of Pout and Pin promoters are double underlined and indicated above the sequence (GenBank accession no. AF133697). Nucleotide changes relative to IS1999.2 are indicated above each nucleotide and are boxed.
Using the 5′RACE technique, the promoter responsible for blaOXA-48 expression was mapped. Transcription initiation occurred at a thymidine residue (nucleotide position 115 from the inner terminus of the IS1999.2-left element). As expected, the deduced promoter region named Pout was identical to that previously identified for IS1999 (1). Pout is made of a −35 promoter region (CAGTAT) separated by 17 bp from a −10 region (TAGGAT), and it is similar to the outward-directed promoter identified in IS10 (−35 [CAGAAT] and −10 [TAAAAT]) (30).
The genetic environment of Tn1999 was analyzed. Tn1999 was inserted in a Tir gene encoding a transfer inhibition protein that shares 93% amino acid identity with that of plasmid pCTXM-3 in a Citrobacter freundii strain (GenBank accession number AF550415). The 9-bp sequences that were located on each side of IS1999.2-left (CGTTCAGCA/TATATTGCA) or IS1999.2-right (GGCCGAGCA/CGTTCAGCA) matched the consensus target sequence (Fig. 1 and 2). The 9-bp duplication (CGTTCAGCA) located on each side of the structure suggested that insertion into the Tir gene may have occurred by a transposition event.
Transposition of IS1999-based composite transposons.
The ends of IS1999 are not identical. (i) IS1999 contains two IR (IRL and IRR) that share 81% nucleotide identity. (ii) Moreover, analysis of the IS1999 nucleotide sequence revealed that IS1999 carries a putative IHF (integration host factor) binding site (bp 30 to 42) adjacent to the IRL (Fig. 4).
In order to evaluate whether two IS1999 elements were able to transpose in the form of various composite transposons, different constructs were made using two copies of IS1999 (i) in inward and opposite orientations (TnLL), (ii) in the same orientation (TnLR), or (iii) in outward and opposite orientations (TnRR) (Fig. 1). TnLL transposition involves the action of transposase at two IRL, TnRR transposition involves two IRR, and TnLR transposition involves one IRL and one IRR, as for a single IS1999 transposition. In these constructs, each insertion sequence could transpose either independently of each other or together under a composite transposon structure.
The composite transposons TnLL and TnLR transposed at similar frequencies (ca. 4 × 10−6) (Table 2). However, the TnRR transposition frequency was at least 10-fold lower than that of TnLL or of TnLR. This result suggested that transposition of a composite transposon flanked by two outside IRR ends (e.g., which do not contain any IHF binding site) is less efficient than other transposition events which involve at least one IRL end (containing an IHF binding site).
Analysis of the genetic environment of several insertions of composite transposons using IS1999 elements revealed in all cases a duplication of 9 bp, confirming that a transposition event had occurred. Sequence of the insertion sites matched to the consensus target site and two representative insertions (T1 and T2) are shown in Fig. 2.
Transposition assay using the putative transposon, Tn1999.
The transposition ability of Tn1999 was investigated using plasmid pTn1999. However, despite several attempts, no Genr Ticr Rifr Tets transconjugants were obtained, suggesting either that the Tn1999 transposition frequency might be lower than 10−7 (limit of detection of our transposition assay) or that this structure, flanked by two IS1999.2 elements, may not be able to transpose.
We have shown previously that transposition of TnRR but not of Tn1999 was detectable. Tn1999 and TnRR had similar structures. Both structures were made of two IS elements in outward and opposite directions located on each side of a 2.2-kb central region. However, the structures of Tn1999 and TnRR have two major differences that may affect their transposition frequency: the nature of the central region and the nature of the IS elements. Indeed, one structure was made of two copies of IS1999.2 (isoform of IS1999), whereas the other was made of two copies of IS1999.
The transposition ability of IS1999.2.Kan was investigated (Fig. 1). Transconjugants that resulted from conjugation of pOX38-Gen::IS1999.2.Kan were screened as Genr Kanr Rifr Tets colonies at the frequency of 2.6 × 10−7 (Table 2). Thus, the transposition efficiency of IS1999.2 was lower than that of IS1999.
Using the 5′RACE technique, the promoter responsible for the IS1999-tnpA expression was precisely mapped. Transcription initiation occurred at a thymidine residue for IS1999-tnpA and at a guanosine residue for IS1999.2-tnpA (both located at nucleotide position 81) (Fig. 5A). Pin is made of a −35 promoter region (TGGAAG) separated by 17 bp from a −10 region (TCAAAT) and is similar to the Pin promoter identified in IS10 (−35 [CAGAAT] and −10 [TAAAAT]) (30).
FIG. 5.
A. Nucleotide sequence alignments of the promoter regions of IS1999, IS1999.2, and IS10. The start codon of the transposase gene is boldfaced. The boxes represent the conserved boxes of Pout and Pin. The complementary inverted repeated sequences are shown by gray boxes. The promoter sequences −10 and −35 of Pout and Pin promoters are boxed and indicated above the sequence. B. RT-PCR results. M, molecular size marker (100 bp; Invitrogen). Lane 1, no RNA (negative control); lane 2, RNA from IS1999.2-containing cells; lane 3, RNA from IS1999-containing cells. Panel a represents IS1999-specific primers, and panel b represents the gap-specific primers.
RT-PCR experiments were performed, and the band intensities were normalized according to the gap gene amplification results (Fig. 5B). A factor of three to four differences in band intensities was observed for Pin of IS1999 versus that of IS1999.2, suggesting that IS1999-tnpA is more efficiently transcribed.
DISCUSSION
The association of IS1999 with emerging antibiotic resistance genes triggered our interest in the study of IS1999-mediated gene mobilization. We showed that IS1999 was capable of transposition into DNA targets containing a consensus sequence. Another important feature for IS1999 is the inclusion of strong outward-directed promoter sequences (Pout) involved in the expression of downstream-located genes. This promoter was similar to, and at the same location as, that previously identified for IS10 (30).
IS1999 and IS10 sequences share significant nucleotide identity (67%) and conserved genetic features. (i) IS1999 and IS10 possess a similar Pin promoter driving the transposase expression and a similar outward-directed promoter, Pout (Fig. 4). When cloned on a multicopy plasmid, IS10 is subjected to a phenomenon called multicopy inhibition (31). Indeed, an increase of the copy number of IS10 led to an increase in the intracellular amount of the antisense RNA generated from the Pout promoter. This antisense RNA, by interacting with the sense RNA generated from Pin, inhibits the transposase expression (31). The IS1999 Pout transcript is antisense to the transposase promoter Pin, and IS1999 elements were cloned on a low-copy plasmid. Thus, the multicopy inhibition is likely to occur in our study and may lead to an underestimation of the transposition frequency of IS1999. Another point known to be responsible for a transposition frequency decrease is the length of dependence. Indeed, as has been shown for other transposons, the transposition frequency decreases with the length of the DNA fragment inserted within the IR (5-6, 16, 26, 29). In our study, the kanamycin resistance gene was inserted in order to tag IS1999 and led to an additional 2.2-kb increase of the space between the IR. However, this increase likely has little effect on IS1999 transposition, since the length dependence is only valid for longer DNA fragments.
(ii) Both IS1999 and IS10 carry two complementary inverted repeats in the IRL region, one of which overlaps the translation initiation signal of the transposase gene (Fig. 4). As described for IS10 (14), once transcribed from an external promoter these sequences were able to form a hairpin RNA structure which sequesters translation initiation signals and results in termination of transcription. This mechanism of “protection from outside transcription” avoids overexpression of the transposase and consequently an excess of transposition events that may be detrimental to the bacterial host. Analysis of the IS1999 sequence revealed that such a potential structure could be formed when transcribed from an external promoter. The mechanism of protection from outside activation is likely to occur in IS1999, as suggested by our data. In pIS1999.Kan, the transcription of the IS1999 transposase seems to be protected from the Plac promoter. Indeed, the orientation of the tagged IS1999 (IS1999.Kan, sense; IS1999as.Kan, antisense) compared to the strong Plac promoter from the cloning vector did not lead to significant change of IS1999 transposition frequency.
(iii) The consensus DNA target of IS1999 (NGCNNNGCN) resembled that of IS10 (NGCTNAGCN) (9). The consensus sequence for IS1999 insertions was determined by alignment of target sequences of randomly chosen insertion sites (hotspots and non-hotspots). IS10 insertion sites that are not hotspots deviate more from the consensus sequence, especially at positions 4 and 6 (9). By extension, this fact may explain why no obvious consensus sequence was detected at positions 4 and 6 in the consensus target sequence of IS1999. However, regarding all the insertion sites analyzed (Fig. 2), the T is favored at position 4 (6T, 3A, 4C, 2G) and the A is favored at position 6 (5A, 4G, 4C, 2T).
Moreover, several base pairs of the DNA sequences flanking the target sites of IS1999.Kan were conserved (Fig. 2). Indeed, homology seemed to extend several base pairs beyond and to one side of the target sequence, suggesting that flanking sequences may influence the target site recognition, as is known to occur for IS10 (2).
The target site located in the attI1 site of class 1 integrons (Fig. 2) is located downstream of promoter sequences (Pant promoter located in the 5′ conserved sequence region of class 1 integrons [25]), and insertion of transposons such as Tn10 has been shown to be inhibited by heavily transcribed target sequences (3). Thus, we hypothesize that the attI1 site may be less efficient for IS1999 insertion.
(iv) IS1999 and IS10 possess a similar IHF binding site adjacent to the IRL end (Fig. 4). Tn10 is flanked by two identical IRL ends, both of which have IHF binding sites. The DNA bending activity of IHF stimulates assembly of an intermediate with folded transposon ends in which the transposase has additional subterminal contacts. These contacts are required to activate the chemical steps during the cleavage reaction (28).
We have shown that two copies of IS1999 were also capable of mobilizing intervening DNA sequence in the form of many composite transposons. However, a transposon made of two copies of IS1999 in outward and opposite orientations (TnRR) (involving the action of the transposase at two IRR ends, both ends lacking an IHF binding site) was less efficient for transposition. Transposition seems to be more efficient when at least one IRL end (containing an IHF binding site [TnLR, TnLL]) is present at the outside boundaries of the transposon. Thus, we hypothesize that IHF may also play an important role in IS1999 transposition and may explain the difference of transposition frequencies measured between TnRR, TnLR, and TnLL.
As Tn1999 and TnRR had similar structures, both were expected to have similar transposition frequencies. However, transposition of Tn1999 was not detected in our assay. Our study revealed that IS1999.2 elements were less efficient in transposition than IS1999. Compared to IS1999, the IS1999.2 sequence contained only one nucleotide substitution occurring outside of the transposase gene (G instead of T; bp 81), which corresponds to the +1 transcriptional initiation site of the Pin promoter that is responsible for transcription of the transposase gene (30). The Pin promoter is a weak promoter, and single changes occurring at the −35 or −10 promoter sequence of Pin have been shown to influence the IS10 transposition frequency (10, 30). However, sequences located in the immediate region of the transcription initiation site have been shown to influence the transcriptional initiation (11). This could be the case for IS1999.2, since the promoter is less efficient. An alternative explanation might be that the silent mutations replace frequently used codons by rarely used codons, thus slowing down the translation of tnpA (6). However, codon usage is likely to contribute very moderately, if at all.
The duplication of the 9-bp (CGTTCAGCA) sequence that is located on each side of the Tn1999 strongly suggested that it was a transposon that may be able to transpose. If this structure is indeed a transposon, the combination of the presence of two IS1999.2 elements, the lower level of IS1999-tnpA expression, and their outward and opposite orientations might be a part of the factors responsible for the low-transposition frequency of Tn1999. However, further experiments will be necessary to clearly demonstrate this issue.
Acknowledgments
This work was funded by grants from the Ministère de la Recherche (grant UPRES-EA 3539), Université Paris XI, Paris, France, and from the European Community (6th PCRD, LSHM-CT-2003-503-335).
REFERENCES
- 1.Aubert, D., T. Naas, and P. Nordmann. 2003. IS1999 increases expression of the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. J. Bacteriol. 185:5314-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, J., and N. Kleckner. 1992. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc. Natl. Acad. Sci. USA 89:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branlant, G., and C. Branlant. 1985. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of D-glyceraldehyde-3-phosphate dehydrogenase. Eur. J. Biochem. 150:61-66. [DOI] [PubMed] [Google Scholar]
- 4.Casadesus, J., and J. R. Roth. 1989. Transcriptional occlusion of transposon targets. Mol. Gen. Genet. 216:204-209. [DOI] [PubMed] [Google Scholar]
- 5.Chandler, M., M. Clerget, and D. J. Galas. 1982. The transposition frequency of IS1-flanked transposons is a function of their size. J. Mol. Biol. 154:229-243. [DOI] [PubMed] [Google Scholar]
- 6.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 7.Corvec, S., N. Caroff, E. Espaze, J. Marraillac, H. Drugeon, and A. Reynaud. 2003. Comparison of two RT-PCR methods for quantifying ampC specific transcripts in Escherichia coli strains. FEMS Microbiol. Lett. 228:187-191. [DOI] [PubMed] [Google Scholar]
- 8.Girlich, D., T. Naas, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum β-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 9.Halling, S. M., and N. Kleckner. 1982. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell 28:155-163. [DOI] [PubMed] [Google Scholar]
- 10.Huisman, O., P. R. Errada, L. Signon, and N. Kleckner. 1989. Mutational analysis of IS10's outside end. EMBO J. 8:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong, W., and C. Kang. 1994. Start site selection at lacUV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res. 22:4667-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, R. C., and W. S. Reznikoff. 1984. Copy number control of Tn5 transposition. Genetics 107:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. PBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 14.Ma, C. K., T. Kolesnikow, J. C. Rayner, E. L. Simons, H. Yim, and R. W. Simons. 1994. Control of translation by mRNA secondary structure: the importance of the kinetics of structure formation. Mol. Microbiol. 14:1033-1047. [DOI] [PubMed] [Google Scholar]
- 15.Mahillon, J., C. Leonard, and M. Chandler. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. Mol. Biol. Rev. 150:675-687. [DOI] [PubMed] [Google Scholar]
- 16.Morisato, D., J. C. Way, H. J. Kim, and N. Kleckner. 1983. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell 32:799-807. [DOI] [PubMed] [Google Scholar]
- 17.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 18.Philippon, L. N., T. Naas, A.-T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel, L., C. Héritier, and P. Nordmann. 2004. Chromosome-encoded ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., C. Héritier, V. Tolün, and P. Nordmann. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel, L., M. F. Lartigue, J. W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 25.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile elements. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 26.Roberts, D., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43:117-130. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sewitz, S., P. Crellin, and R. Chalmers. 2003. The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. Nucleic Acids Res. 31:5868-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen, M. M., E. A. Raleigh, and N. Kleckner. 1987. Physical analysis of Tn10- and IS10-promoted transpositions and rearrangements. Genetics 16:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons, R. W., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1983. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell 34:673-682. [DOI] [PubMed] [Google Scholar]
- 31.Simons, R. W., and N. Kleckner. 1983. Translational control of IS10 transposition. Cell 34:683-691. [DOI] [PubMed] [Google Scholar]
- 32.Tribuddharat, C., and M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]