Abstract
In a previous study, ecs-3, a sequence from avian pathogenic Escherichia coli (APEC) O78:K80 strain χ7122, was found to be expressed in vivo in infected chicken tissues. The region encompassing ecs-3 carries a fimbrial gene cluster that is a putative ortholog of the stg fimbrial gene cluster of Salmonella enterica serovar Typhi. This APEC fimbrial gene cluster, which we have termed stg, is a member of a distinct group of related fimbriae that are located in the glmS-pstS intergenic region of certain E. coli and S. enterica strains. Under the control of the pBAD promoter, the production of Stg fimbriae was demonstrated by Western blotting and immunogold electron microscopy with E. coli K-12. Transcriptional fusions suggest that stg expression is influenced by the carbohydrate source and decreased by the addition of iron and that Fur plays a role in the regulation of stg expression. stg sequences were associated with APEC O78 isolates, and stg was phylogenetically distributed among E. coli reference strains and clinical isolates from human urinary tract infections. Stg fimbriae contributed to the adherence of a nonfimbriated E. coli K-12 strain to avian lung sections and human epithelial cells in vitro. Coinfection experiments with APEC strain χ7122 and an isogenic Δstg mutant demonstrated that compared to the wild-type parent, the Δstg mutant was less able to colonize air sacs, equally able to colonize lungs, and able to more effectively colonize tracheas of infected chickens. Stg fimbriae, together with other adhesins, may therefore contribute to the colonization of avian respiratory tissues by certain APEC strains.
Avian pathogenic Escherichia coli (APEC) is associated mainly with extraintestinal diseases in chickens, turkeys, and other avian species and causes severe economic losses to the poultry industry (12). The most common form of these diseases is avian colibacillosis, which starts as a respiratory infection (airsacculitis) and is frequently followed by generalized infections such as perihepatitis, pericarditis, and septicemia (12). APEC belongs to a limited number of serogroups, of which O1, O2, and O78 are the most common (12). Phylogenetic analyses have indicated that most APEC strains belong to a few distinct clonal groups (19, 59). Several virulence factors, such as adhesins, iron sequestering systems, capsular and lipopolysaccharide antigens, and toxins, have been reported for APEC (27).
Although many virulence factors are required for bacterial pathogenicity, adhesin-mediated colonization of different tissues is one of the earliest events occurring during an infection. Adhesins are either assembled into hair-like appendages (fimbriae or pili) or are directly associated with the bacterial cell surface (afimbrial adhesins) (55). Several adhesins have been described so far for APEC, including type 1 (F1A), P (F11), and AC/I (avian E. coli I) fimbriae, curli, and the temperature-sensitive hemagglutinin (Tsh) (12, 27). P fimbriae have been reported mostly for human uropathogenic E. coli and are present in only a minority of APEC isolates (12, 27). The expression of P fimbriae in the air sacs and other internal organs but not in the trachea of inoculated chickens supports the idea that they are important only in later stages of infection (47). Type 1 fimbriae are characterized by the ability to adhere to d-mannose residues of epithelial cells and mucosa (24). They were shown to be expressed by bacteria colonizing the respiratory tracts of experimentally infected chickens (14) and may contribute to the initial colonization of the avian respiratory tract (12, 47). AC/I fimbriae, which belong to the S fimbrial adhesin family, mediate adherence to avian epithelial tissues in vitro and in vivo and have been found on a limited number of O78 APEC isolates (3, 61). Curli are thin, coiled aggregative structures found on the surfaces of most E. coli isolates (27, 41). Curli promote binding to extracellular matrix proteins and major histocompatibility complex class I molecules (42, 53) and also agglutinate chicken erythrocytes (9). Curli cloned from an APEC O78 strain mediated the internalization of E. coli K-12 by eukaryotic cells (20). Tsh, a temperature-sensitive hemagglutinin identified from APEC strain χ7122 (O78:K80:H9) (48), was shown to be associated with pathogenic isolates of high lethality for day-old chicks (16). Experimental infection studies showed that although Tsh contributes to the development of lesions in the air sacs, other virulence factors must contribute to the pathogenicity of strain χ7122 in the lower respiratory tract and extrarespiratory tissues (16).
A number of putative adhesin-encoding genes of strain χ7122 that were expressed in vivo in chicken tissues were previously identified by the selective capture of transcribed sequences (15). One of the pathogen-specific fragments identified (ecs-3) shared homology at the peptide level with putative fimbrial ushers present in E. coli O157:H7 and Salmonella enterica serovar Typhi. The related fimbrial gene clusters are located between the conserved genes glmS and pstS. These fimbriae also share some similarities with the long polar (LP) fimbriae of S. enterica serovar Typhimurium and E. coli strains EDL933 O157:H7 and 83/39 (6, 37, 56), each of which is encoded by operons located between the conserved genes yhjX and yhjW. This report describes the identification and characterization of the fimbrial gene cluster corresponding to fragment ecs-3 of APEC strain χ7122. We have termed the genes encoding these fimbriae stg, as the predicted fimbrial system that they encode is most similar to Stg of serovar Typhi and is distinct from LP fimbriae encoded by gene clusters within the yhjW-yhjX regions in serovar Typhimurium and certain E. coli strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. In addition, selected clinical or environmental isolates from various sources were used to screen for the presence of stgC. The 72 members of the Escherichia coli reference (ECOR) collection represent a diversity of E. coli strains that have been phylogenetically grouped by multilocus enzyme electrophoresis (MLEE) (39). The 298 APEC clinical isolates were previously described elsewhere (16). Thirty-two E. coli fecal isolates from healthy poultry were kindly provided by John M. Fairbrother (University of Montréal). APEC and avian E. coli environmental isolates were previously classified for virulence on the basis of lethality for 1-day-old chicks following subcutaneous inoculation, where lethality class 1 (LC1) corresponds to the high-lethality class, LC2 to the low-lethality class, and LC3 to the nonlethal class (16). Among the total number of APEC isolates tested, the serogroup of 187 isolates was determined. Human extraintestinal pathogenic E. coli (ExPEC) isolates included a diversity of strains from urosepsis and other extraintestinal infections in the United States. These strains were grouped phylogenetically by either MLEE or multiplex PCR (10). Bacteria were routinely grown in Luria-Bertani (LB) broth (Gibco) or on tryptic soy agar (TSA) (Gibco) at 37°C. Additional media used were MacConkey agar (Gibco) and M9 medium (35). When required, antibiotics were added at the following concentrations: 50 μg/ml for kanamycin (Km), 100 μg/ml for ampicillin (Ap), 50 μg/ml for chloramphenicol (Cm), and 15 μg/ml for nalidixic acid. For plasmid pBAD18-Cm and its derivative, chloramphenicol was used at a concentration of 10 μg/ml. The transformation of E. coli strains was routinely carried out by using calcium/manganese-based or electroshock methods as described previously (23).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| DH5α | F− λ− φ80 Δ(lacZYA-argF)endA1 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | Invitrogen |
| MG1655 | F−λ−rph-1 | 8 |
| ORN103 | thr-1 leu-6 thi-1 Δ(argF-lac)U169 xyl-7 ara-13 mtl-2 gal-6 rspL tonA2 minA minB Δ(fimEACDFGH) | 43 |
| ORN172 | thr-1 leuB thi-1 Δ(argF-lac)U169 xyl-7 ara-13 mtl-2 gal-6 rspL tonA2 supE44 Δ(fimBEACDFGH)::kan pilG1 | 60 |
| QC2517 | MG1655 recD1901::Tn10 Δfur::cat | 17 |
| E. coli clinical strains and derivatives | ||
| CFT073 | Uropathogenic E. coli strain O6:K2:H1 | 58 |
| χ7122 | Wild-type APEC O78:K80:H9, gyrA Nalr | 49 |
| EDL933 | E. coli O157:H7, EHEC strain | 45 |
| QT51 | Strain χ7122 ΔlacZYA | This study |
| QT302 | χ7122 ΔstgABCD::kan | This study |
| QT826 | QT51 Δfur::cat | This study |
| QT865 | QT826::pIJ79, Pstg-lacZ single-copy integrant | This study |
| QT891 | QT51::pIJ79, Pstg-lacZ single-copy integrant | This study |
| Plasmids | ||
| pBAD18-Cm | Arabinose-inducible vector, Cmr | 22 |
| pCP20 | FLP helper plasmid, temperature-sensitive replication, Cmr and Apr | 11 |
| pCR-XL-TOPO | High-copy cloning vector, Kmr | Invitrogen |
| pIJ2 | 6.0-kb PCR fragment of stgABCD cloned into pCR-XL-TOPO; Kmr | This study |
| pIJ14 | XbaI and HindIII fragment of pIJ2 containing the stg operon cloned into pACYC184, Cmr | This study |
| pIJ32 | PCR fragment containing the stgA promoter cloned into pRS415, Apr | This study |
| pIJ39 | PCR fragment of stgABCD cloned into pBAD18-Cm, Cmr | This study |
| pIJ79 | pST76-K::Pstg-lacZ | This study |
| pKD3 | FRT-flanked kanamycin cassette template | 11 |
| pKD46 | Red recombinase expression plasmid, Apr | 11 |
| pRS415 | Operon fusion vector, Apr | 52 |
| pST76-K | Suicide vector pSC101, temperature-sensitive replicon, Kmr | 46 |
EHEC, enterohemorrhagic E. coli; Nal, naladixic acid.
DNA and genetic manipulations.
Standard methods were used for the isolation of bacterial genomic DNA, DNA manipulation, and cloning (51). Restriction enzymes and DNA ligase used in this study were purchased from New England Biolabs (NEB), Invitrogen, or Amersham-Pharmacia and used according to the suppliers' recommendations. Recombinant plasmids, PCR products, and restriction fragments were purified using plasmid mini-prep, PCR cleanup, and gel extraction kits (QIAGEN) as recommended by the supplier.
Identification, cloning, and sequencing of the APEC stg gene cluster.
The stg gene cluster was amplified from genomic DNA of strain χ7122 using Elongase enzyme mix (Invitrogen Life Technologies) with the primers glmS-F (5′-GATCTTCTACACCGTTCCGC-3′) and pstS-R (5′-TTACGCCACCGGAAGAACCG-3′) (Fig. 1A). The 5.8-kb PCR product was purified and cloned into the vector pCR-XL-TOPO using the TOPO XL PCR cloning kit (Invitrogen), resulting in plasmid pIJ2. The cloned fragment was subcloned from pIJ2 into vector pACYC184 at the XbaI and HindIII restriction sites, resulting in vector pIJ14, which was then transformed into strain MG1655. Sequencing was achieved by the generation of transposon mutants by transduction using phage λ::Tn5seq1 (36), a derivative of Tn5 that contains universal T7 and SP6 sequencing primers at its extreme ends. Total plasmid DNA of the transductants was isolated and transformed into calcium/manganese-based competent DH5α cells. The locations of the Tn5seq1 insertions were determined by PCR using the primer Tn5seq1-left (5′-AAGCTCGGATCTAATACGAC-3′) located within the transposon and primers glmS-F and pstS-R. Restriction enzyme digestions with NotI were also performed to determine the locations of Tn5seq1 insertions. Selected pIJ14::Tn5seq1 clones were sequenced by DNA LandMarks, Inc. (St. Jean sur Richelieu, Québec, Canada) using universal primers T7 and SP6.
FIG. 1.
(A) The glmS-pstS intergenic region of APEC strain χ7122. Black arrows indicate the four ORFs of the stg gene cluster. Numbers indicate the predicted number of amino acids. White arrows indicate corresponding genes present in E. coli K-12. Gray arrows indicate positions of primers used for cloning the stg encoding region (glmS-F and pstS-R) and for screening the presence of stg sequences among isolates (stgC-F and stgC-R). The small white rectangle corresponds to the region amplified for screening the presence of stg sequences among isolates. The dashed lines indicate the 5′ and 3′ products amplified to assess the presence of a full-length stg gene cluster among isolates. (B) Phylogram based on Clustal analysis of proteins sharing highest identities/similarities with the predicted stgA gene product. The APEC StgA protein is highlighted within a box. Fimbrial proteins from gene clusters inserted in the yhjW-yhjX region, which includes LpfA of serovar Typhimurium, cluster together and belong to the LP fimbrial group. Fimbrial proteins from gene clusters inserted in the glmS-pstS region, including StgA from E. coli and serovar Typhi, comprise a distinct cluster and belong to the Stg fimbrial group. Entries from top to bottom correspond to GenBank accession numbers P43660, C86029, AAO22843, AAO52822, P22595, AAL18161, AAN45247, CAD03135, AAG58930, and CAE14452. The scale indicates percent difference in similarity. E. tarda, Edwardsiella tarda; P. luminescens, Photorhabus luminescens; S. flexneri, Shigella flexneri; S. marcescens, Serratia marcescens.
Preparation of StgA-specific antisera.
NDSAYTAIDAEGKAE, a purified peptide corresponding to the C-terminal portion of StgA, was coupled to keyhole limpet hemocyanin and used to immunize two New Zealand White rabbits. Peptide synthesis and anti-StgA (α-StgA) antiserum production were provided by New England Peptide, Inc. Antiserum was absorbed consecutively using whole cells of E. coli K-12 strain ORN172 grown at 37°C.
Immunogold labeling and TEM.
The production of stg-encoded fimbriae in an E. coli clone was examined by transmission electron microscopy (TEM). The stg operon was placed under the control of the tightly regulated arabinose promoter of pBAD18-Cm. A 5,142-bp PCR product from plasmid pIJ2 was amplified using Elongase enzyme mix (Invitrogen Life Technologies) and the primers NheI-F (5′-ATGCTAGCAAGTGATGATTCATGGTAAAGG-3′) and HindIII-R (5′-TGTGAAGCTTAAAATCCCACATGTC-3′). The PCR purified product was then digested with HindIII and NheI and cloned into plasmid pBAD18-Cm. The resulting plasmid pIJ39 and the control plasmid were transformed into the nonfimbriated E. coli K-12 Δfim mutant strain ORN172. Static bacterial cultures were grown overnight at 37°C on TSA plates with 10 μg/ml chloramphenicol and 0.05% arabinose. The bacteria were recovered by centrifugation and allowed to adhere to a carbon-Formvar-coated copper grid. For immunogold labeling, bacteria were mixed with α-StgA antiserum (1:1,000) and gold-labeled anti-rabbit immunoglobulin G (12-nm diameter) and then negatively stained with uranyl acetate. The grids were allowed to dry before being analyzed with a Philips EM300 electron microscope.
Preparation of fimbrial extracts.
Bacteria were grown on large plates of tryptic soy agar medium supplemented with 10 μg/ml chloramphenicol and 0.05% arabinose for the induction of fimbrial synthesis. Following growth at 37°C for 16 to 18 h, the bacteria were harvested and resuspended in 5 ml of 75 mM NaCl-0.5 mM Tris-HCl (pH 7.0). They were then incubated at 60°C for 30 min and pelleted by centrifugation (3,000 × g for 15 min). The supernatant was then transferred to another tube, and an aliquot was taken for precipitation with concentrated trichloroacetic acid to obtain a final ratio of 1:10. The tube was incubated for 15 to 20 min on ice, followed by centrifugation at 20,000 × g for 15 min at 4°C. The protein pellet was washed twice with 0.5 M Tris-HCl-0.5 M EDTA (pH 12.0) and resuspended in 0.5 M Tris-EDTA at 1/10 of the supernatant initial volume.
Western blotting.
Fimbrial extracts were separated by sodium dodecyl sulfate (SDS)-15% polyacrylamide gel electrophoresis minigels as previously described by Laemmli. Proteins were either stained with Coomassie brilliant blue or transferred to nitrocellulose membranes (Bio-Rad) using a Mini Trans-Blot electrophoretic cell (Bio-Rad) for 60 min at 100 V. The membrane was blocked with StartingBlock supplemented with 0.05% Tween 20 (Pierce). Incubations with primary (1:5,000) and secondary (1:25,000) antibodies were carried out for 1 h at room temperature. SuperSignal West Pico chemiluminescent substrate (Pierce) was used for detection.
Construction of mutants of APEC strain χ7122.
The deletion/inactivation of lacZYA, stgABCD, or fur genes in strain χ7122 or derivatives was obtained using the lambda red recombinase system as described previously by Datsenko and Wanner (11). For the generation of the ΔlacZYA mutant of strain χ7122, the primers LacIKO-F (5′GCAGCGTATCAGGCAATTTTTATAATTTAAACTGACGTGTAGGCTGGAGCTGCTTC-3′) and LacZKO-R (5′-GGTTTCCCGACTGGAAAGCGGGCAGTGAGCGCAACGCCATATGAATATCCTCCTTAG-3′), containing regions homologous to terminal portions of lacI or lacZ, were used to amplify the cat gene flanked by FRT (FLP recognition target) sites from the template plasmid pKD3. Gene disruption was carried out by electroshock transformation of strain χ7122 carrying plasmid pKD46 with the PCR fragment containing the chloramphenicol resistance cassette. Mutants were colony purified once at 37°C and then tested for the loss of pKD46 by selecting for ampicillin sensitivity. To eliminate the FRT-flanked cat gene encoding chloramphenicol resistance, strains were transformed with the helper plasmid pCP20 that encodes FLP recombinase and mediates excision of FRT-flanked sequences. Ampicillin-resistant transformants were selected at 30°C and then colony purified nonselectively at 43°C. The strains were then tested for a loss of resistance to both chloramphenicol (loss of the FRT-flanked cat gene) and ampicillin (encoded on plasmid pCP20), a loss of the pCP20 plasmid, and the presence of all four native plasmids of strain χ7122. The ΔlacZYA derivative of strain χ7122 was named QT51. The same technique was used for the construction of the ΔstgABCD::kan mutant of strain χ7122. The primers used were stgKO-F (5′-TATAAGTGATGATTCATGGTAAAGGATATATTATATCAATGTGTAGGCTGAGCTGCTTC-3′) and stgKO-R (5′-TAGCGCACAATCTCAACAGTTATCGTCGCTGTTGCAGTAACATATGAATATCCTCCTTAG-3′). The resulting strain was named QT302. A fur mutant of strain QT51 was created as follows. Briefly, a Δfur::cat allele derived from E. coli strain QC2517 (17) was amplified with primers CMD18 (5′-ATTCTAGACTGCTGCTGGGCATCCC-3′) and CMD19 (5′-ACTCTAGACACTCCGACATCCCAAGC-3′). The Δfur::cat-containing amplicon was transferred to strain QT51 by homologous recombination using the lambda red recombinase method described above. A fur mutant of strain QT51 was designated QT826.
Generation of single-copy stg-lacZ transcriptional fusions and β-galactosidase assay.
The stgA promoter region was amplified from plasmid pIJ2 by using Elongase enzyme mix (Invitrogen Life Technologies) and the primers CMD56-R (5′-TAGGATCCAGCATTAGAGATGCCAGAG-3′) and CMD57-F (5′-CGGAATTCAAAGGCACCGACGTTGAC-3′). The PCR product was digested with EcoRI and BamHI and cloned into pRS415 (52), resulting in plasmid pIJ32. A segment of pIJ32 containing the Pstg-lacZ fusion was amplified by PCR using Elongase and primers CMD57-F (see above) and CMD173-R (5′-TAGCATGCGGAAGTAGGCTCCCATGAT-3′). The PCR product containing Pstg fused to the lacZ gene was digested with EcoRI and SphI and cloned into suicide vector pST-76K (46), generating pIJ79. To generate single-copy Pstg-lacZ fusions in the APEC strains QT51 and fur derivative QT826, pIJ79 was integrated into the genomes of these strains by homologous recombination as described previously (46). A strain carrying a single integrated copy of Pstg-lacZ in QT826 was designated QT865. A strain carrying a single integrated copy of Pstg-lacZ in QT51 was designated QT891.
The β-galactosidase assays were performed as previously described elsewhere (35). For the assays, E. coli strains were grown overnight (18 h) at 37°C on M9 agar plates supplemented with 0.2% (wt/vol) of the following carbohydrates: glycerol, mannitol, sorbitol, arabinose, ribose, xylose, glucose, mannose, and maltose or other carbon sources (pyruvate, acetate, and succinate) at 0.3% (wt/vol). To investigate the influence of iron on expression, cultures were grown on M9 plates supplemented with glucose and either 150 μM of the chelator dipyridyl (iron-limiting conditions) or 10 μM of FeCl3 (iron-replete conditions). Bacterial suspensions were prepared by resuspending cultures in 2 ml of Z buffer (60 mM NaHPO4·7H2O, NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7H2O, 50 mM β-mercaptoethanol), and samples were adjusted with Z buffer to obtain an optical density at 600 nm of 0.6. A 100-μl volume of each sample was then transferred into 900 μl of Z buffer to which 40 μl of chloroform and 20 μl of 0.1% SDS solution were added, and tubes were then vortexed for 10 s. Following 5 min of incubation at 28°C, O-nitrophenyl-β-d-galactopyranoside was then added and samples were incubated for another 3 min. The reaction was stopped by the addition of 500 μl of a 1 M Na2CO3 solution (35).
Presence of stg sequences among E. coli strains.
The presence of stg sequences was investigated among different E. coli strains by PCR amplification of a 208-bp segment of the stgC gene. Crude DNA extracts of strains were prepared by alkaline lysis. Primers specific to the putative usher encoding gene stgC, stgC-F (5′-TCTGGTTCACATACACTACG-3′) and stgC-R (5′-CCAATCATAATCTGGCTTCT-3′) (Fig. 1A), were used for screening, as the coding sequences of usher genes are typically conserved within a fimbrial type that may comprise a number of variant major subunit or adhesin alleles (24, 28). PCR conditions were as follows: 95°C for 1 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and then an extension period of 72°C for 1 min. The reactions were carried out using Taq DNA polymerase (NEB). Specificity of the primers was confirmed by the amplification of the fragment in strain χ7122 and no amplification in strains known not to contain the stg operon on the basis of their genome sequences (MG1655, CFT073, and EDL933). For 25 of the stgC-positive strains, the glmS-pstS intergenomic regions were also analyzed by PCR amplification of the 5′ and 3′ regions by using the glmS-F/stgC-R and stgC-F/pstS-R primer pairs, respectively (Fig. 1A).
Bacterial association with human epithelial cells.
The ability of strains ORN103 and ORN103(pIJ2) to adhere to epithelial cell monolayers was assessed using human bladder-derived (UM-UC-3; ATCC no. CRL-1749) and human intestine-derived (INT407; ATCC no. CCL-6) epithelial cell lines. For both quantitative and qualitative assays, epithelial cells were grown to semiconfluence (about 3.5 × 105 cells/ml) in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum and rinsed three times before the adherence assays. Bacteria were grown overnight on TSA plates containing kanamycin and resuspended in phosphate-buffered saline (PBS), pH 7.4, to an optical density at 600 nm of 0.6 (∼3 × 108 CFU). Approximately 106 CFU was added to each well. The 24-well plates were then centrifuged at 800 rpm for 5 min, incubated at 37°C in 5% CO2 for 90 min, and rinsed three times. PBS-0.1% deoxycholic acid sodium salt was added to each well, and samples were diluted and spread on TSA plates containing kanamycin for enumeration by viable colony counts. Data are expressed as the percentage of the initial inoculum introduced. Assays were carried out in triplicate wells in two separate experiments.
Bacterial adherence in vitro to chicken lung sections.
Adherence assays were performed by using a method adapted from Nowicki et al. (38). Lung tissues from two chickens were removed and frozen in Tissue-Tek OCT compound (Sakura) at −80°C. Tissues were cut in 5-μm sections on glass slides using a cryostat (Miles 4553) and fixed with methanol. For the adherence assay, bacteria were grown overnight on TSA plates with appropriate antibiotics and suspended in PBS, pH 7.4, containing 1% (wt/vol) bovine serum albumin. Fifty microliters of a 5 × 109 bacterial suspension was added to the tissue sections, which were incubated in a moist chamber at 4°C for 45 min. Slides were rinsed twice in PBS, pH 7.4, for 5 min with gentle agitation and then fixed and stained using a Diff-Quick stain set (Dade Behring, Inc.). Adherence was observed using a Nikon Eclipse E800 microscope equipped with a Nikon Coolpix 990 digital camera.
Chicken infection model.
An E. coli intratracheal infection model using 2-week-old White Leghorn chickens preinfected with infectious bronchitis virus was used as described previously (2). Briefly, 14 axenic chickens (12 days old) were infected with a 104 mean embryo infective dose of infectious bronchitis virus serotype Massachusetts strain M41 by the oculonasal route. Two days later, each bird was inoculated via the trachea with a 100-μl mixed suspension of 1.6 × 108 CFU containing APEC strain χ7122 (56%) and its isogenic ΔstgABCD::kan derivative, QT302 (44%), which were cultured overnight in LB broth at 37°C without agitation. Six days following coinfection with the E. coli strains, chickens were euthanized and necropsied. Bacterial counts were determined from the whole homogenized trachea and the left lung and from swabbings of the caudal thoracic air sacs as previously described (14). The numbers of wild-type and mutant bacteria were obtained by determining the numbers of colonies that were sensitive (wild-type strain χ7122) and resistant (ΔstgABCD::kan mutant strain QT302) to kanamycin.
Statistical analyses.
Statistical analyses were performed using the Prism 4.0b software package (GraphPad Software). The chi-square test was used to compare frequency distribution of stg sequences among E. coli isolates. For coinfections, the Mann-Whitney test was used. The t test was used for a comparison of adherence to tissues and for β-galactosidase assays.
Nucleotide sequence accession number.
The nucleotide sequence of the stg encoding region of strain χ7122 was assigned GenBank accession no. AY530785.
RESULTS
Identification of the stgABCD gene cluster and phylogenetic relationship with other fimbrial gene clusters.
Pathogen-specific transcripts of strain χ7122 were previously identified in a chicken infection model by selective capture of transcribed sequences (15). Transcript ecs-3 shares sequence identity at the peptide level with the putative fimbrial ushers StgC of serovar Typhi strain CT18 (44) and Z5222 of E. coli O157:H7 strain EDL933 (45). In serovar Typhi strain CT18 and E. coli O157:H7 strain EDL933, stgC and gene Z5222, respectively, are parts of fimbrial operons located between the conserved genes glmS and pstS. By using primers corresponding to glmS and pstS, a 5,842-bp DNA fragment was amplified from strain χ7122 genomic DNA and cloned (Fig. 1A). In strain χ7122, a region spanning 5,423 bp and containing four open reading frames (ORFs) that comprise a fimbrial gene cluster is inserted 1 nucleotide 3′ of the glmS stop codon, resulting in a replacement of a 205-bp span within the corresponding glmS-pstS intergenic region in E. coli K-12 strains. The glmS-pstS intergenic region from strain χ7122 exhibits a G+C content of 43%, which is considerably less than the average 50.8% G+C content of E. coli K-12 (8).
Thus far, a number of fimbrial gene clusters inserted into either the glmS-pstS region or the yhjW-yhjX intergenic region have been identified in different E. coli strains (13, 25, 37, 56) and all of the gene clusters encoding these fimbriae have been termed lpf, based largely on amino acid identity to LP fimbrial proteins of serovar Typhimurium. Sequencing of the glmS-pstS intergenic region from strain χ7122 led to the identification of a fimbrial gene cluster that shares high similarity to lpf gene clusters from E. coli O113:H21 (13) and E. coli 789 (25). Analyses of the current DNA database demonstrate that there are two main groups of putative or confirmed fimbrial systems present in either E. coli or S. enterica strains that demonstrate more similarity to either LP fimbriae of serovar Typhimurium or Stg fimbriae of serovar Typhi. This is exemplified by a phylogenetic tree based on Clustal analysis of predicted fimbrial gene products sharing the highest identity-similarity scores with APEC StgA (Fig. 1B). Phylograms for APEC StgB, StgC, and StgD and related gene products similarly resulted in the generation of the same two distinct groupings (data not shown). On the basis of a comparison of predicted gene products, the gene clusters located within the glmS-pstS intergenic region in E. coli strains are all more closely related to one another than to those inserted in the yhjX-yhjW intergenic region, which is the insertional location of the lpf gene cluster in serovar Typhimurium. On the basis of these results, we have termed the fimbrial gene cluster present in the glmS-pstS intergenic region of strain χ7122 stg, as the fimbrial gene products it encodes are orthologous to those of Stg fimbriae of serovar Typhi and are similarly encoded in the glmS-pstS intergenic region. The stgABCD gene cluster of strain χ7122 contains four ORFs (Fig. 1A). The predicted sizes of the APEC stgABC gene products are identical to those of the predicted products of the lpfO113 and lpf789 gene clusters, whereas the lpfDO113 gene product is predicted to be 265 amino acids long (13) compared to a predicted 357-amino-acid precursor for the stgD and lpfD789 gene products.
Immunodetection of Stg fimbriae.
As the environmental conditions that are favorable for the expression of stg from its native promoter are unknown, we used the tightly regulated expression of stg genes from the arabinose-inducible promoter of plasmid pBAD18-Cm to induce the expression of Stg fimbriae. stg genes were cloned into plasmid pBAD18-Cm, generating plasmid pIJ39, and transformed into the fimbria-negative E. coli K-12 strain ORN172. Strain ORN172(pIJ39) was grown overnight at 37°C on TSA plates containing 0.05% arabinose, and cells were then analyzed by TEM following immunogold labeling with α-StgA antiserum. In the absence of arabinose, no fimbriae were visualized and very few gold particles were observed (Fig. 2A). When ORN172(pIJ39) was grown in the presence of arabinose, peritrichous filamentous structures were observed and immunogold labeling was localized primarily to these structures (Fig. 2B). Fimbrial extracts of strain ORN172(pIJ39), grown in the presence or absence of arabinose, were examined by Western blotting to confirm the specificity of StgA detection. Following Western blotting of the extracts, a specific band, with an apparent molecular mass of 23 kDa reacting with the α-StgA antiserum, was present in the extract of strain ORN172(pIJ39) grown in the presence of arabinose. Liquid chromatography-mass spectrophotometry analysis of the trypsin-treated product that reacted specifically with α-StgA antiserum confirmed that this band corresponded to the predicted StgA protein (data not shown). The StgA-specific band was absent when ORN172(pIJ39) was grown without arabinose (Fig. 2C). In addition, no fimbriae or StgA-specific bands were observed with either immunogold TEM or Western blotting of fimbrial extracts of the vector control strain ORN172(pBAD18-Cm) in either the absence or presence of arabinose (not shown).
FIG. 2.
(A and B) Immunodetection of Stg fimbriae and immunogold labeling with anti-StgA antiserum and observation by transmission electron microscopy. Bars = 200 nm. Strain ORN172(pIJ39) was grown overnight on TSA plates at 37°C without arabinose (A) or with 0.05% arabinose (B). Arrows indicate some of the StgA-labeled gold particles associated with fimbrial structures. (C) Immunoblot of fimbrial extracts obtained from ORN172(pIJ39) cells grown under the same conditions as those described above. Samples were migrated on an SDS-15% polyacrylamide gel electrophoresis gel and incubated with anti-StgA-specific antiserum. Lane 1, extract from cells grown without arabinose; lane 2, extract from cells grown with 0.05% arabinose. Molecular mass markers are indicated to the right. A band reacting with StgA-antiserum, at an apparent molecular mass of 23 kDa, was detected only in cells induced with arabinose.
Transcription of the stg operon is regulated by carbon source and iron.
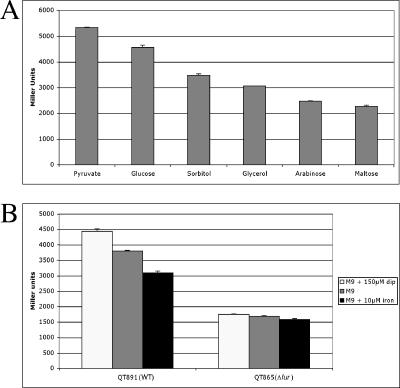
To investigate the regulation of stg gene expression in the native APEC strain, a stg::lacZ fusion was inserted into the chromosome of strain QT51, a ΔlacZYA derivative of strain χ7122, generating strain QT891. Strain QT891 was used to determine the influence of a number of carbon sources on stg expression. The expression of the promoter fusion following overnight growth on M9 plates with pyruvate, glucose, sorbitol, glycerol, arabinose, or maltose as a carbon source is presented (Fig. 3A). β-Galactosidase was expressed at higher levels in M9 medium with either pyruvate or glucose as the carbon source (Fig. 3A). Among carbon sources tested, maltose and arabinose gave the lowest expression values. Promoter expression in strain QT891 was 1.17-fold higher in M9 medium with pyruvate, the carbohydrate with the highest promoter expression, than in M9 medium with glucose (M9-glucose). The lowest promoter expression was in M9 medium containing maltose, in which the mean expression was 0.43 times that observed when cells were grown in M9-pyruvate (P < 0.0001). In M9 medium containing sorbitol, glycerol, or arabinose, promoter expression levels were intermediate. Of the several other carbohydrates that were tested, ribose, xylose, mannose, acetate, succinate, and mannitol showed intermediate expression levels similar to that of sorbitol (results not shown).
FIG. 3.
(A) β-Galactosidase activity expressed from the PstgA::lacZ fusion in APEC strain QT891 grown overnight at 37°C on M9 minimal medium plates supplemented with different carbohydrates or other carbon sources. (B) Effect of iron on β-galactosidase activity expressed from the PstgA::lacZ fusion in APEC strain QT891 and isogenic Δfur strain QT865. M9, M9-glucose. Error bars indicate standard deviations.
In order to determine whether iron availability or Fur, the global iron-dependent regulator of E. coli, had a regulatory effect on the stg promoter in an APEC strain, a Pstg-lacZ single-copy integrant of strain QT826 was constructed and termed QT865. The effects of various iron concentrations on β-galactosidase expression by strains QT891 and QT865 were then assessed. As shown in Fig. 3B, there was a significant decrease in β-galactosidase expression in strain QT891 with increasing amounts of iron (P < 0.005). Specifically, the expression decreased by 14% from M9-glucose containing 150 μM dipyridyl (M9-dipyridyl) to M9-glucose alone. A further decrease, 30% lower than that in M9-dipyridyl, was observed in M9 supplemented with 10 μM iron. Compared to that for strain QT891, stg promoter expression under all conditions was decreased for the isogenic Δfur derivative QT865 (P < 0.0001) (Fig. 3B). For M9-dipyridyl, expression was 2.5-fold higher in QT891 than in QT865. Furthermore, in M9-glucose, promoter expression was 2.2-fold higher in QT891, and similarly, in M9 medium with 10 μM of added iron, expression was 1.96-fold higher. In addition, iron availability also had a less marked effect on β-galactosidase expression levels in the Δfur strain QT865 (P < 0.02). These results indicate that increased iron concentrations decreased PstgA activity in the APEC wild-type strain background and that Fur appears to contribute to increased expression of stg regardless of iron availability in the medium.
Distribution of stgC among the ECOR collection, E. coli isolates from urinary tract infections, and avian E. coli.
To investigate whether the stg gene cluster demonstrates a phylogenetic distribution among the ECOR collection, PCR analysis was performed using the stgC-F and stgC-R primers, which are specific for a 208-bp fragment of stgC (Fig. 1A). Among the 72 members of the ECOR collection, which were arranged into five phylogenetic groups (A, B1, B2, D, and E) on the basis of multilocus enzyme electrophoresis (39), the presence of stg sequences was significantly associated with phylogenetic groups B1 and D relative to the other groups (P < 0.001) (Table 2). stg sequences were present in all of the strains belonging to group B1 and 75% of the strains belonging to group D, whereas only 8% of group A strains and none of the B2 or E group strains contained stg sequences.
TABLE 2.
Distribution of stg-positive strains within phylogenetic groups among the ECOR collection and ExPEC isolates
| Phylogenetic group | Value for E. coli from:
|
|||
|---|---|---|---|---|
| ECOR collection
|
Human extraintestinal infections
|
|||
| Total no. of strains | No. of stg-positive strainsa (%) | Total no. of isolates | No. of stg-positive isolatesa (%) | |
| A | 25 | 2 (8) | 15 | 7 (47) |
| B1 | 16 | 16 (100)c | 11 | 8 (72)c |
| B2 | 15 | 0 (0) | 24 | 0 (0) |
| D | 12 | 9 (75)c | 41 | 37 (90)c |
| E | 4 | 0 (0) | NAb | NA |
| Total | 72 | 27 (38) | 91 | 52 (57) |
Strains positive by PCR amplification using the stgC-F and stgC-R primers.
NA, not applicable.
For both the ECOR collection and ExPEC isolates, stg sequences were significantly associated with phylogenetic groups B1 and D relative to groups A and B2 (P < 0.001).
To complement the ECOR strains, 91 ExPEC isolates from human disease were also investigated for the presence of stg sequences. The ExPEC isolates tested belonged to phylogenetic group A, B1, B2, or D, as determined either by MLEE or by a triplex PCR method (10, 32). Analysis demonstrated that, as with the ECOR collection, stg sequences were highly associated with ExPEC isolates belonging to phylogenetic groups B1 and D relative to groups A and B2 (P < 0.001). No stgC-positive isolates belonging to phylogenetic group B2 were identified in ExPEC (Table 2). As the stg fimbrial gene cluster was initially identified in the APEC O78 strain χ7122, the presence of stg sequences was also investigated in APEC isolates (Table 3). stg sequences were present in 133 of 298 (44.6%) of APEC isolates. stg was significantly associated (P < 0.05) with APEC isolates relative to those from the feces of healthy poultry, since only 8 of 32 (25%) of environmental isolates contained stg sequences (Table 3). There was no significant association with the presence of stg sequences and the lethality class of APEC or environmental isolates (Table 3). Among the most common serogroups associated with avian disease (O1, O2, and O78), stg was significantly associated with APEC isolates belonging to serogroup O78 (Table 4). That is, stg sequences were present in 90.7% of the APEC isolates belonging to serogroup O78, compared to 14.3% of serogroup O1 isolates, and none of strains belonging to serogroup O2 were stgC positive (Table 4). The determination of the phylogenetic grouping of APEC isolates by triplex PCR demonstrated that strains from serogroups O1 and O2 belonged to phylogenetic group B2, whereas O78 strains never belonged to this phylogenetic group. This finding correlates with the low prevalence of stg sequences observed in ECOR and ExPEC strains belonging to group B2 (Table 2).
TABLE 3.
Distribution of stg among APEC and environmental isolates according to lethality class
| Lethality classb | Value for E. coli isolates from:
|
|||
|---|---|---|---|---|
| APEC
|
Environment
|
|||
| Total no. of isolates | No. of stg-positive isolatesa (%) | Total no. of isolates | No. of stg-positive isolates (%) | |
| LC1b | 222 | 101 (46) | 1 | 1 (100) |
| LC2 | 38 | 18 (47) | 12 | 3 (25) |
| LC3 | 38 | 14 (37) | 19 | 4 (21) |
| Total | 298 | 133 (45)c | 32 | 8 (25) |
Positive PCR amplification using the stgC-F and stgC-R primers.
Lethality classes were defined as follows: LC1, 50% lethal dose of <108 CFU; LC2, 50% lethal dose of ≥108 CFU; LC3, not lethal at ≥108 CFU (16).
stg sequences were significantly associated with APEC isolates relative to environmental isolates (P < 0.05).
TABLE 4.
Association between the O serogroups of APEC strains and the presence of stg sequences
| Serogroup | No. of stg-positive strainsa (%) | No. of strains tested |
|---|---|---|
| O1 | 4 (14.3) | 28 |
| O2 | 0 (0) | 46 |
| O78 | 78 (90.7)c | 86 |
| Otherb | 12 (44.4)c | 27 |
Positive PCR amplification using the stgC-F and stgC-R primers.
Other serogroups included in this grouping are O6, O8, O11, O15, O18, O21, O22, O35, O45, O54, O55, O71, O83, O115, and O131.
stg sequences were significantly associated with strains belonging to serogroup O78 or “other serogroups” relative to strains from serogroups O1 and O2 (P < 0.001).
For 25 of the stgC-positive strains, the conserved size and location of stg genes were assessed by PCR amplification of overlapping regions: the 5′ region using the glmS-F/stgC-R primers and the 3′ region using the stgC-F/pstS-R primers (Fig. 1A). Twenty-four of 25 stgC-positive isolates amplified DNA fragments of the same size as those from strain χ7122. These results suggest that the genomic location of stg at the glmS-pstS intergenic region is conserved and that most E. coli stgC-positive isolates are likely to contain complete full-length copies of stg gene clusters.
Contribution of stg genes to adherence of E. coli to human epithelial cells and chicken respiratory tissues.
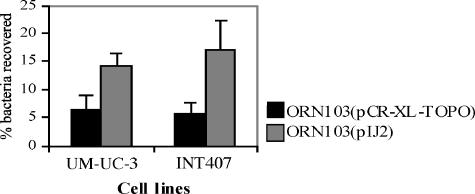
As stg genes were identified in 57% of the 91 ExPEC isolates tested from humans, we determined whether the stg gene cluster could confer adherence of E. coli to human bladder epithelial cells (UM-UC-3) and intestinal epithelial cells (INT407). Plasmid pIJ2 (stgABCD) was transformed into strain ORN103, and adherence assays were performed using ORN103(pCR-XL-TOPO) as a negative control. Strain ORN103 is an E. coli K-12 derivative that lacks type 1 fimbriae and does not produce any other known adhesins (43). ORN103(pIJ2) adhered to both epithelial cell lines to a significantly greater extent than did the negative control strain ORN103(pCR-XL-TOPO) (Fig. 4). Compared with control strain ORN103(pCR-XL-TOPO), ORN103(pIJ2) was at least twice as adherent to UM-UC-3 cells (14.2% of initial bacteria recovered versus 6.5% for ORN103; P < 0.001) and at least three times more adherent to INT407 cells (17.2% of initial bacteria recovered versus 5.6% for ORN103; P < 0.001). To examine the capacity of Stg fimbriae to mediate adherence to avian respiratory tissues, ORN103(pIJ2) and a control strain were tested for adherence to chicken lung sections in a qualitative assay. ORN103 cells containing stg demonstrated adherence in a clustered fashion to both lung and tracheal tissues, whereas the control strain adhered to a much lesser degree (Fig. 5). Although the adhered clusters of ORN103(pIJ2) cells were not uniformly distributed, the clustered adherence pattern was not observed for the control strain, with very few bacteria adhering to tissues. Adherence tests of wild-type strain χ7122 compared to that of Δstg mutant strain QT302 to either human epithelial cells or chicken lung sections did not demonstrate any discernible differences in adherence, even when 0.5% mannose was added to inhibit adherence mediated by type 1 fimbriae (data not shown).
FIG. 4.
Bacterial adherence assays using human UM-UC-3 kidney and INT407 intestinal epithelial cell lines. The percentage of the initial bacterial inoculum associated with epithelial cells after 90 min of incubation is indicated. Bacteria containing stg genes (pIJ2) were significantly more adherent (P < 0.001) to both cell lines than was the negative control strain ORN103(pCR-XL-TOPO). Error bars indicate standard deviations.
FIG. 5.
Adherence of Stg-positive strain ORN103(pIJ2) (A) and Stg-negative control strain ORN103(pCR-XL-TOPO) (B) to chicken lung sections. Bacteria were grown on TSA, and 50 μl of a 5 × 109 bacterial suspension was added to the tissue sections. Slides were stained using a Diff-Quick stain set.
Contribution of stg to colonization of chicken respiratory tissues in vivo.
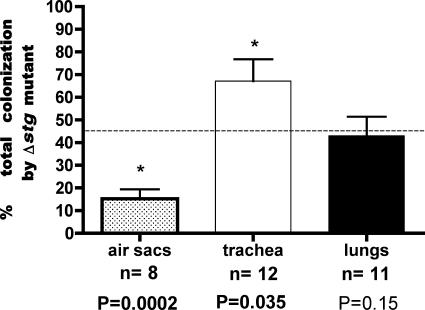
To assess whether stg contributes to respiratory colonization of APEC strain χ7122, competitive coinfections with APEC strain χ7122 and its isogenic ΔstgABCD mutant derivative strain QT302 were tested in a chicken intratracheal infection model in 14 White Leghorn chickens (2 weeks old). Two of the 14 inoculated chicks died 4 days postinoculation and were not included in the experiment. Six days postinfection with E. coli, bacterial numbers in the trachea, air sacs, and lungs of chickens were determined. Figure 6 presents the mean proportion of the ΔstgABCD mutant relative to the total bacterial numbers recovered in each of the tissues of infected chickens. In the air sacs, the wild-type strain significantly outcompeted the ΔstgABCD mutant. By contrast, in the trachea, the ΔstgABCD mutant significantly outcompeted the wild-type parent. Levels of colonization in the lungs by the wild-type parent and the ΔstgABCD mutant were similar.
FIG. 6.
Comparative colonization of respiratory tissues 6 days following coinfection of chickens with APEC strain χ7122 and isogenic ΔstgABCD::kan derivative QT302. Bars represent the mean proportion (as a percentage) of the Δstg mutant compared to the total CFU of bacteria recovered. Error bars indicate standard errors of the means. Asterisks indicate significant differences in the proportions of Δstg mutant and wild-type APEC χ7122 recovered from tissues compared to the input ratio. P values obtained using the Mann-Whitney test are indicated. The dashed line indicates the initial input ratio of the inoculum of the Δstg mutant. n = number of tissue samples from which either strain could be recovered from 12 analyzed tissues.
DISCUSSION
Sequencing of the glmS-pstS intergenomic region of APEC strain χ7122 identified a four-gene operon that we have named the E. coli ortholog of stg from serovar Typhi (Fig. 1A). The stg gene cluster is most similar to other putative fimbrial operons in E. coli (O island 154) and serovar Typhi (stg) that are also located between glmS and pstS (Stg fimbrial group) (Fig. 1B). Database and alignment analyses demonstrate that APEC Stg fimbriae are more closely related to this distinct group of fimbriae than to other related fimbriae located in the yhjW-yhjX intergenic region, such as LP fimbriae of serovar Typhimurium (6) and LP fimbriae of E. coli (37, 56) (Fig. 1B).
The stg gene cluster from strain χ7122 is highly similar to fimbrial gene clusters that were named lpf in E. coli O113:H21, rabbit enteropathogenic E. coli O15:H7 (13), and E. coli 789 (25). The conserved locations of the stg and closely related gene clusters within a number of different E. coli strains and its absence from E. coli K-12 and pathogenic strains CFT073 and EDL933 prompted us to investigate whether this fimbrial gene cluster was phylogenetically distributed. Screening of the ECOR collection for stg sequences demonstrated that stg is associated with phylogenetic groups B1 and D and absent from phylogenetic group B2 (Table 2). The screening of a number of ExPEC clinical isolates similarly indicated the presence of stgC in most E. coli isolates from phylogenetic groups B1 and D, whereas it was absent from all B2 ExPEC isolates tested (Table 3). Further, the screening of APEC isolates demonstrated that stg is common among isolates of serogroup O78, which do not belong to phylogenetic group B2, but rare among those from serogroups O1 and O2, which commonly belong to group B2 (Table 4).
These results indicate that the stg gene cluster is phylogenetically distributed in E. coli and largely absent from E. coli strains belonging to phylogenetic group B2. Some molecular phylogeny studies suggest that the B2 group represents the most ancestral lineage of E. coli (18, 29). The stg gene cluster may have been acquired relatively recently by horizontal gene transfer since it exhibits an overall G+C content of 43%, which is considerably lower than the E. coli K-12 mean genomic DNA G+C content of 50.8%. Further evidence supporting the probable horizontal acquisition of the stg gene cluster is its genomic location. The 3′ terminal end of the glmS gene and its terminator are a preferential insertion site for transposon Tn7, which has been termed attTn7 (21). Taken together, these results suggest that the acquisition of stg genes likely occurred after the branching of the B2 lineage and that stg genes may have been acquired by certain E. coli strains belonging to other lineages. In a similar way, the related stg and lpf operons may have been acquired or lost by certain clonal lineages or serovars of Salmonella enterica during its evolution (5, 57).
Several environmental factors, such as carbon source, aliphatic amino acids, iron, temperature, and electron acceptors other than oxygen, can regulate fimbrial expression (30). lacZ fusion assays using the stgA promoter indicate that this operon is transcribed in a wild-type strain background and that carbohydrate source can affect expression levels (Fig. 3A). The carbohydrate source is known to affect the expression of many bacterial genes, including those encoding fimbriae. Glucose may inhibit fimbrial expression by catabolite repression (30). However, following growth on M9 glucose medium, cells expressed the second highest level of stg promoter activity after pyruvate. Overall, expression results from the stg-lacZ fusions suggest that Stg fimbrial expression differed depending on the carbon source but was not greatly inhibited by any of the sources tested.
The ecs-3 sequence corresponding to a fragment of the stgC gene was initially identified as an APEC-specific gene expressed in the pericardium of infected chickens (15). In systemic tissues such as the pericardium, it is likely that iron availability to the bacterial cells is limited due to iron sequestration by host ferroproteins. This is supported by the identification of aerobactin and salmochelin siderophore-encoding genes that were also expressed in the pericardial tissues of infected chickens (15). Siderophore-encoding genes are typically repressed in iron-replete conditions as their transcription is reduced by the Fur-Fe+2 complex. The expression of the stgA promoter decreased as the amount of available iron in the medium increased, suggesting that stg expression is iron regulated (Fig. 3B). However, in the fur mutant strain, this decrease was much less striking, and overall promoter expression was about twofold lower in the fur mutant compared to that in the wild-type APEC strain. Fur acts mainly as a repressor of the E. coli iron acquisition genes under iron sufficiency, as the Fur-Fe+2 complex actively binds to regulatory regions (known as Fur boxes) to inhibit the transcription of these genes. Analysis of the region 5′ of stg demonstrated a number of putative Fur-binding sites; however, matches to Fur consensus sequences were low. In addition, by using a Fur titration assay, the stg promoter encoded on a high-copy plasmid did not deregulate Fur-mediated repression of an iron-regulated iucC::lacZ fusion under iron-replete conditions, suggesting that Fur is unlikely to interact with the stg promoter (data not shown). Several genes are positively regulated by Fur and apparently do not possess a Fur box (1, 33). Using transcriptional profiling, McHugh et al. (34) recently identified 101 genes in E. coli K-12 that are regulated by the Fe2+-Fur complex. Forty-eight of these genes were induced by Fur (34). Hence, in some cases and as we observed with stg, Fur may enhance the expression of certain genes. However, the involvement of Fur may be indirect and implicate other regulators (1).
The introduction of the stg genes into a nonfimbriated E. coli K-12 strain conferred the production of fimbrial structures. Stg fimbriae were peritrichously distributed, and anti-StgA antibodies were associated with fimbrial structures following immunogold labeling and electron microscopy. In addition, the antiserum specifically reacted with a band corresponding to the major fimbrial subunit following Western blotting (Fig. 2). Stg fimbriae and other fimbriae belonging to this group of adhesins mediate bacterial attachment to a number of cell/tissue types. The expression of Stg fimbriae increased the adherence of nonfimbriated E. coli K-12 cells to both human epithelial cell lines and avian respiratory tissues. Specifically, Stg fimbriae mediated increased adherence to human bladder (UM-UC-3) and intestinal (INT407) cell lines. Similarly, in nonfimbriated E. coli K-12 strains, lpfO113 conferred increased adherence to CHO-K1 cells (13) and lpf of E. coli EDL933 conferred increased adherence to HeLa and MDBK cells (56). Moreover, LP fimbriae of E. coli EDL933 demonstrated a clustered adherence pattern to cell lines. A similar clustered adherence pattern to INT407 or UM-UC-3 cells as well as chicken lung sections was observed from E. coli K-12 cells expressing APEC Stg fimbriae. In seeming contrast, for in vitro adherence assays, we observed no difference in the adherence of the wild-type APEC strain compared to that of its isogenic Δstg mutant to epithelial cells or chicken respiratory tissues. However, this actually is not surprising, as APEC strain χ7122 produces other known adhesins (e.g., type 1 fimbriae, Tsh, and curli) that may mediate adherence to cells. Similarly, Bäumler et al. (7) demonstrated that LP fimbriae of serovar Typhimurium mediated the adherence and invasion of Hep-2 cells but were not essential for adhesion to T-84, INT407, or HeLa cell lines. However, the lpf operon of serovar Typhimurium was also shown to be involved in the colonization of murine ileal Peyer's patches of mice in vivo (7). Recently, a direct role for Lpf789 in the adherence of E. coli strain 789 to human kidney epithelial cells in vitro was demonstrated (25).
A number of adhesins that mediate adherence of APEC to avian tissues or cells have been identified. These include type 1 fimbriae (12, 27), P fimbriae (26, 47), AC/I fimbriae (3, 61), curli (9, 20), and Tsh (16, 48). Although several adherence factors were identified in different APEC strains, none were demonstrated to be essential for the initial stages of infection in chickens (2, 15, 27, 31). Other adhesins may therefore be involved in the initial stages of colonization of the chicken respiratory tract. Evidence that Stg fimbriae contribute to adherence in avian respiratory tissues was obtained by performing adhesion assays on chicken lung tissue sections using E. coli strain ORN103(pIJ2). The strain harboring the stg operon adhered to chicken lung tissues to a greater extent than did the control strain (Fig. 5). The observation that Stg fimbriae mediated adherence of nonfimbriated E. coli K-12 to avian frozen tissue sections suggested a possible role for this adhesin in the colonization of the avian respiratory tract by certain APEC strains. Coinfection studies with the APEC wild-type strain and a ΔstgABCD derivative, QT302, demonstrated that the loss of Stg fimbriae resulted in a decreased capacity to colonize the air sacs (Fig. 6). These results are the first obtained that demonstrate an individual role for an adhesin that promotes the colonization of APEC to avian respiratory tissues. Further, since the air sacs are an initial port of entry for APEC and are commonly colonized by APEC during infection, Stg fimbriae, in concert with other adhesins, such as curli, type 1 fimbriae, or Tsh, may contribute to initial steps of APEC pathogenesis. By contrast, the loss of Stg fimbriae resulted in an improved capacity to colonize the trachea (Fig. 6), which is analogous to the enhanced tracheal colonization ability associated with the loss of the type 1 adhesin by APEC O2 strain MT78 (2). It is not immediately clear how the inactivation of a fimbrial adhesin can result in improved colonization of certain tissues. As has been noted for type 1 fimbriae, it is possible that the expression of a fimbrial adhesin in certain tissues may be a disadvantage by promoting adherence to mucus glycoproteins or specific receptors on phagocytic cells, which could result in increased bacterial clearance or killing (40, 47). In addition, it is possible that the inactivation of one fimbrial adhesin may result in greater expression of another adhesin, which can result in increased adherence to cells or tissues. The expression and phase variation of different adhesins involves cross talk between fimbrial systems, and recent evidence with ExPEC strains causing urinary tract infections demonstrated that the inactivation or constitutive expression of a fimbrial system can alter the expression of other adhesins (54). In human ExPEC strains, type 1 and P fimbriae have been shown to contribute to virulence in urinary tract infection models (4, 50). Since stg was associated with certain ExPEC and APEC strains and Stg fimbriae mediated adherence to both avian tissues and bladder epithelial cells, it will be of interest to investigate the contribution of Stg fimbriae in concert with other known adhesins to the pathogenesis of APEC as well as ExPEC associated with urinary tract infections.
Acknowledgments
Kind thanks to K. Hantke, P. Orndorff, B. Wanner, and J. M. Fairbrother for the gifts of strains or plasmids.
Funding for this project was provided by Natural Sciences and Engineering Research Council of Canada (NSERC) individual research grants to C.M.D. and F.D., the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, and National Research Initiative (NRI) Competitive Grants Program/United States Department of Agriculture grant 00-35212-9408 (J.R.J.). M.H.L. was the recipient of a Fondation Armand-Frappier Scholarship, and S.L. received a summer studentship from NSERC.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Arné, P., D. Marc, A. Brée, C. Schouler, and M. Dho-Moulin. 2000. Increased tracheal colonization in chickens without impairing pathogenic properties of avian pathogenic Escherichia coli MT78 with a fimH deletion. Avian Dis. 44:343-355. [PubMed] [Google Scholar]
- 3.Babai, R., G. Blum-Oehler, B. E. Stern, J. Hacker, and E. Z. Ron. 1997. Virulence patterns from septicemic Escherichia coli O78 strains. FEMS Microbiol. Lett. 149:99-105. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 5.Bäumler, A. J., A. J. Gilde, R. M. Tsolis, A. W. van der Velden, B. M. Ahmer, and F. Heffron. 1997. Contribution of horizontal gene transfer and deletion events to development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J. Bacteriol. 179:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bäumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 9.Brown, P. K., C. M. Dozois, C. A. Nickerson, A. Zuppardo, J. Terlonge, and R. Curtiss III. 2001. MlrA, a novel regulator of curli (Agf) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349-363. [DOI] [PubMed] [Google Scholar]
- 10.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 13.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dozois, C. M., N. Chanteloup, M. Dho-Moulin, A. Brée, C. Desautels, and J. M. Fairbrother. 1994. Bacterial colonization and in vivo expression of F1 (type 1) fimbrial antigens in chickens experimentally infected with pathogenic Escherichia coli. Avian Dis. 38:231-239. [PubMed] [Google Scholar]
- 15.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dozois, C. M., M. Dho-Moulin, A. Brée, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escobar-Paramo, P., O. Clermont, A. B. Blanc-Potard, H. Bui, C. Le Bouguenec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 19.Ewers, C., T. Janssen, S. Kiessling, H. C. Philipp, and L. H. Wieler. 2004. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet. Microbiol. 104:91-101. [DOI] [PubMed] [Google Scholar]
- 20.Gophna, U., M. Barlev, R. Seijffers, T. A. Oelschlager, J. Hacker, and E. Z. Ron. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gringauz, E., K. A. Orle, C. S. Waddell, and N. L. Craig. 1988. Recognition of Escherichia coli attTn7 by transposon Tn7: lack of specific sequence requirements at the point of Tn7 insertion. J. Bacteriol. 170:2832-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan, D., J. Jessee, and F. R. Bloom. 1995. Techniques for transformation of E. coli, p. 1-35. In D. M. Glover and B. D. Hames (ed.), DNA cloning I: core techniques, 2nd ed. Oxford University Press, New York, N.Y.
- 24.Hultgren, S. J., C. Hal Jones, and S. Normark. 1996. Bacterial adhesins and their assembly, p. 2730-2756. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 25.Ideses, D., D. Biran, U. Gophna, O. Levy-Nissenbaum, and E. Z. Ron. 2005. The lpf operon of invasive Escherichia coli. Int. J. Med. Microbiol. 295:227-236. [DOI] [PubMed] [Google Scholar]
- 26.Kariyawasam, S., T. J. Johnson, and L. K. Nolan. 2006. The pap operon of avian pathogenic Escherichia coli strain O1:K1 is located on a novel pathogenicity island. Infect. Immun. 74:744-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Ragione, R. M., and M. J. Woodward. 2002. Virulence factors of Escherichia coli serotypes associated with avian colisepticaemia. Res. Vet. Sci. 73:27-35. [DOI] [PubMed] [Google Scholar]
- 28.Le Bouguenec, C., M. Archambaud, and A. Labigne. 1992. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 30:1189-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15:1685-1695. [DOI] [PubMed] [Google Scholar]
- 30.Low, D., B. Braaten, and M. Van Der Woude. 1996. Fimbriae, p. 146-157. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 31.Marc, D., P. Arné, A. Brée, and M. Dho-Moulin. 1998. Colonization ability and pathogenic properties of a fim− mutant of an avian strain of Escherichia coli. Res. Microbiol. 149:473-485. [DOI] [PubMed] [Google Scholar]
- 32.Maslow, J. N., T. S. Whittam, C. F. Gilks, R. A. Wilson, M. E. Mulligan, K. S. Adams, and R. D. Arbeit. 1995. Clonal relationships among bloodstream isolates of Escherichia coli. Infect. Immun. 63:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 36.Nag, D. K., H. V. Huang, and D. E. Berg. 1988. Bidirectional chain-termination nucleotide sequencing: transposon Tn5seq1 as a mobile source of primer sites. Gene 64:135-145. [DOI] [PubMed] [Google Scholar]
- 37.Newton, H. J., J. Sloan, V. Bennett-Wood, L. M. Adams, R. M. Robins-Browne, and E. L. Hartland. 2004. Contribution of long polar fimbriae to the virulence of rabbit-specific enteropathogenic Escherichia coli. Infect. Immun. 72:1230-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowicki, B., H. Holthofer, T. Saraneva, M. Rhen, V. Vaisanen-Rhen, and T. K. Korhonen. 1986. Location of adhesion sites for P-fimbriated and for 075X-positive Escherichia coli in the human kidney. Microb. Pathog. 1:169-180. [DOI] [PubMed] [Google Scholar]
- 39.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ofek, I., J. Goldhar, Y. Keisari, and N. Sharon. 1995. Nonopsonic phagocytosis of microorganisms. Annu. Rev. Microbiol. 49:239-276. [DOI] [PubMed] [Google Scholar]
- 41.Olsen, A., A. Jonsson, and S. Normark. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652-655. [DOI] [PubMed] [Google Scholar]
- 42.Olsen, A., M. J. Wick, M. Morgelin, and L. Bjorck. 1998. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 66:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orndorff, P. E., and S. Falkow. 1985. Nucleotide sequence of pilA, the gene encoding the structural component of type 1 pili in Escherichia coli. J. Bacteriol. 162:454-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 45.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 46.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pourbakhsh, S. A., M. Dho-Moulin, A. Bree, C. Desautels, B. Martineau-Doize, and J. M. Fairbrother. 1997. Localization of the in vivo expression of P and F1 fimbriae in chickens experimentally inoculated with pathogenic Escherichia coli. Microb. Pathog. 22:331-341. [DOI] [PubMed] [Google Scholar]
- 48.Provence, D. L., and R. Curtiss III. 1994. Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect. Immun. 62:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Provence, D. L., and R. Curtiss III. 1992. Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or curli production. Infect. Immun. 60:4460-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts, J. A., B. I. Marklund, D. Ilver, D. Haslam, M. B. Kaack, G. Baskin, M. Louis, R. Mollby, J. Winberg, and S. Normark. 1994. The Gal(α1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. USA 91:11889-11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 53.Sjobring, U., G. Pohl, and A. Olsen. 1994. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA). Mol. Microbiol. 14:443-452. [DOI] [PubMed] [Google Scholar]
- 54.Snyder, J. A., B. J. Haugen, C. V. Lockatell, N. Maroncle, E. C. Hagan, D. E. Johnson, R. A. Welch, and H. L. Mobley. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Townsend, S. M., N. E. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and A. J. Bäumler. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White, D. G., M. Dho-Moulin, R. A. Wilson, and T. S. Whittam. 1993. Clonal relationships and variation in virulence among Escherichia coli strains of avian origin. Microb. Pathog. 14:399-409. [DOI] [PubMed] [Google Scholar]
- 60.Woodall, L. D., P. W. Russell, S. L. Harris, and P. E. Orndorff. 1993. Rapid, synchronous, and stable induction of type 1 piliation in Escherichia coli by using a chromosomal lacUV5 promoter. J. Bacteriol. 175:2770-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yerushalmi, Z., N. I. Smorodinsky, M. W. Naveh, and E. Z. Ron. 1990. Adherence pili of avian strains of Escherichia coli O78. Infect. Immun. 58:1129-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]