Abstract
Isovaleryl-coenzyme A (IV-CoA) is the starting unit for some secondary metabolites and iso-odd fatty acids in several bacteria. According to textbook biochemistry, IV-CoA is derived from leucine degradation, but recently an alternative pathway that branches from the well-known mevalonate-dependent isoprenoid biosynthesis has been described for myxobacteria. A double mutant was constructed in Myxococcus xanthus by deletion of genes involved in leucine degradation and disruption of mvaS encoding the 3-hydroxy-3-methylglutaryl-coenzyme A synthase. A dramatic decrease of IV-CoA-derived iso-odd fatty acids was observed for the mutant, confirming mvaS to be involved in the alternative pathway. Additional quantitative real-time reverse transcription-PCR experiments indicated that mvaS is transcriptionally regulated by isovalerate. Furthermore, feeding studies employing an intermediate specific for the alternative pathway revealed that this pathway is induced during fruiting body formation, which presumably increases the amount of IV-CoA available when leucine is limited.
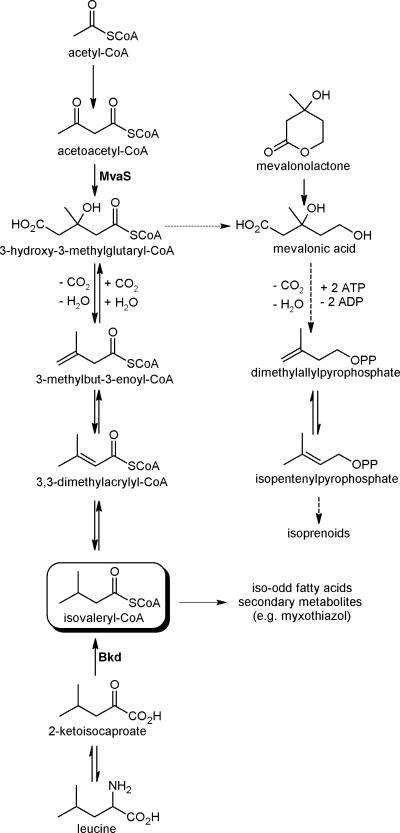
Coenzyme A (CoA) thioesters of short branched-chain carboxylic acids like isovaleryl-CoA (IV-CoA), isobutyryl-CoA (IB-CoA), or 2-methylbutyryl-CoA (2MB-CoA) are thought to be derived from the degradation of the branched-chain amino acids leucine, valine, and isoleucine via the branched-chain keto acid dehydrogenase complex, respectively. Besides being important intermediates in the degradation of these abundant amino acids, the thioesters are important starting units in the biosynthesis of secondary metabolites and fatty acids (FAs). Well-known examples for IB-CoA- or 2MB-CoA-primed secondary metabolites are the avermectins (8), but there are also several secondary metabolites that use IV-CoA as a starting unit (e.g., myxothiazol [24] and aurafuron [15]). In FA biosynthesis, IV-CoA, IB-CoA, and 2MB-CoA are the starting units of iso-odd, iso-even, and ante-iso-odd FAs, respectively, which can be found in various bacteria and can substitute for unsaturated fatty acids to maintain membrane fluidity at different temperatures (12). In all myxobacteria investigated so far, iso-odd fatty acids are the dominant fatty acid family and can make up to 75% of all fatty acids, as shown for Myxococcus xanthus (2, 3, 7). Analysis of bkd mutants of the myxobacteria M. xanthus and Stigmatella aurantiaca showed that iso-odd fatty acids and IV-CoA-derived secondary metabolites are still produced in these mutants although at a reduced level (16, 23). Further feeding experiments with 13C-labeled acetate revealed that the IV-CoA in the residual iso-odd FAs or secondary metabolites is derived from three acetate units by following an incorporation pattern also found in the mevalonate-dependent biosynthesis of isoprenoids (16, 17). The alternative pathway to IV-CoA in myxobacteria was confirmed by incorporation of pathway intermediates into the compounds of interest, and 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) was proposed as the central branching point between isoprenoid and IV-CoA biosynthesis (Fig. 1) (16, 17). The key step in the proposed new pathway is the decarboxylation/dehydration of HMG-CoA to 3,3-dimethylacrylyl-CoA or 3-methylbut-3-enoyl-CoA, which we were able to show previously in vitro (17). This reaction requires ATP and can be regarded as analogous to the reaction of mevalonate diphosphate to isopentenyl diphosphate in isoprenoid biosynthesis (18).
FIG. 1.
Biosynthesis of IV-CoA and isoprenoids in myxobacteria. Dashed arrows indicate multistep reactions. MvaS, 3-hydroxy-3-methylglutaryl-CoA synthase; Bkd, branched-chain keto acid dehydrogenase complex.
Here we present data showing that the HMG-CoA synthase (MvaS) is indeed involved in the biosynthesis of IV-CoA in bkd mutants. Additionally, we show that the alternative pathway to IV-CoA is active in the wild type during fruiting body formation and that transcription of the corresponding mvaS gene is regulated by isovaleric acid.
MATERIALS AND METHODS
General information.
Myxococcus xanthus strains were grown in CTT medium (14) supplemented with kanamycin (40 μg/ml), mevalonolactone (MV) (1 mM; Sigma-Aldrich, Taufkirchen, Germany), and isovalerate (IVA) (1 mM) when appropriate. Fruiting body formation for the isolation of FAs was performed on a large scale on TPM agar (6) with a cell density of 6 × 108 cells per cm2 of agar surface, similar to published procedures (9). For feeding experiments, [dimethyl-D6]3,3-dimethylacrylate ([D6]DMAA) (16) or l-[5,5,5-D3]leucine ([D3]leucine) (Deutero GmbH, Kastellaun, Germany) was dissolved in methanol or water and added to liquid or solid media prior to inoculation (1 mM final concentration).
Construction of mutants.
Strain DK5643 (Δbkd) was generated by PCR amplification of fragments upstream and downstream of the genomic region encoding the E1α and E1β subunits of the branched-chain keto acid dehydrogenase, which were then fused together in a third PCR using the outer primers and both previous fragments as templates in one reaction (10). Briefly, the upstream and downstream regions of the bkd locus were amplified with the primer pairs HBesgdel-1 (5′-CGGAATTCGAGGAGCGCCTCATCGAAATG-3′) and HBesgdel-2 (5′-GTGGGTGCCGCTCTTAGTCGTTGCCGTTGATG-3′) and HBesgdel-3 (5′-CATCAACGGCAACGACTAAGAGCGGCACCCAC-3′) and HBesgdel-5 (5′-CGGGATCCTATCGACGTGGCGGAACACG-3′), respectively. HBesgdel-1 and -5 contain EcoRI and BamHI sites (underlined), whereas HBesgdel-2 and -3 are complementary to each other but with half of the primer originating from the upstream region (italics) and the other half from the downstream region. Both fragments (∼620 bp) were purified and used as the template in a second PCR with HBesgdel-1 and -5 as primers, leading to a 1.2-kbp fragment. This fragment was digested with EcoRI and BamHI and cloned into pBJ114 digested with the same enzymes. The resulting plasmid, pΔesg, carrying an in-frame deletion of the C-terminal part of the E1α subunit-encoding gene and the complete E1β subunit-encoding gene, was electroporated into M. xanthus DK1622 to construct a markerless Δbkd mutant via galK counterselection with pBJ114 as described previously (11). To construct an MXAN_4267 knockout, an internal 766-bp fragment of MXAN_4267 was amplified by PCR using the primer pair Mx619f (5′-GGAATTCGCAAGGTGGCCGTGGTGGTGTG-3′) and Mx619r (5′-GGAATTCGCATCCGCCGGCAGCTTCATCAG-3′) and cloned into plasmid pCR2.1-TOPO (Invitrogen). The resulting plasmid was purified from Escherichia coli TOP10 (Invitrogen) and introduced into M. xanthus DK1622 and DK5643 by electroporation as described previously (9), resulting in strains DK5645 (mvaS::kan) and DK5624 (Δbkd mvaS::kan), respectively. Because the plasmid cannot replicate in M. xanthus, kanamycin-resistant electroporants result from homologous recombination. These incorporate the plasmid into the chromosome and thereby disrupt MXAN_4267, which was verified as described previously by use of a PCR protocol based on a plasmid-specific and a gene-specific primer pair (9). Due to the loss of the ability to produce HMG-CoA for isoprenoid biosynthesis in the double mutant, DK5624 had to be supplemented with mevalonolactone.
Quantitative RT-PCR.
Cultures of DK1622 and DK5643 were grown in CTT medium overnight with or without addition of isovalerate (1 mM) prior to dilution in fresh medium supplemented with IVA where appropriate. For RNA extraction, samples were taken during vegetative growth from wild-type or mutant cultures at different time points and stored at −20°C until the last sample was taken. Then, all samples were resuspended with TRIzol (1 ml/108 cells for 5 min at 25°C), 200 μl of chloroform/ml TRIzol was added, and the mixtures were shaken thoroughly, incubated for 3 min at 25°C, and centrifuged for 15 min at 12,000 rpm (4°C) in a tabletop centrifuge. The supernatant was transferred into a new tube, and 500 μl isopropanol/ml TRIzol was added and incubated for 10 min at 25°C after mixing. After repetition of the centrifugation step, the RNA in the pellet was washed with 75% ethanol, dried at 25°C, and resuspended in 20 to 50 μl of diethyl pyrocarbonate-treated water for 10 min at 55°C. Real-time reverse transcription-PCR (RT-PCR) (using the primers RT619-f [5′-GGGCATCGACCACTCGAAAC-3′] and RT619-r [5′-GCGTGCTGCGTGTCATAGGT-3′], leading to a amplicon 90 bp in size), standardization, and quantification were performed as described recently (13). Briefly, all real-time PCR measurements were run on a Rotor-Gene 3000 machine (Corbett Research, Australia) and data analysis was conducted with Rotor-Gene software 6.0.23 in the dynamic tube normalization mode. As a normalization signal, 16S RNA was used, and standard curves were made in (at least) duplicate, using 10-fold serial dilutions of plasmids carrying the 90-bp amplicon over 4 orders of magnitude. For every single run, each raw copy number per reaction was normalized against the 16S RNA signal. The mean relative value of the 16S RNA copy number was calculated for each growth kinetic experiment, and each single 16S RNA copy number was divided by the mean value. The resulting relative number was then taken to divide the raw copy number of each run, which led to the results shown in Fig. 2.
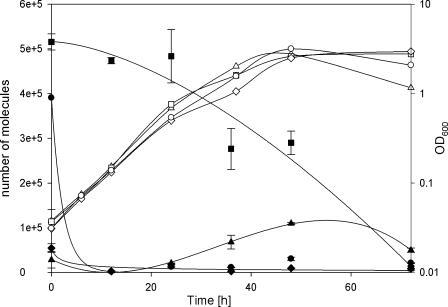
FIG. 2.
HMG-CoA transcript numbers (filled symbols) during vegetative growth (open symbols) of M. xanthus DK1622 (wild type; ▴), DK5643 (Δbkd; ▪), DK5643 plus 1 mM IVA (•), and DK5643 plus 1 mM IVA (IVA-supplemented preculture; ⧫). Mean values of two to three independent experiments are shown, with error bars representing the data ranges.
Fatty acid analysis.
Identification and relative quantification of FAs were conducted as described previously by use of a Sherlock microbial identification system (Microbial ID, Newark, DE) after transforming all bound and free FAs into their methyl esters (16). However, due to several unknown FAs in M. xanthus which were not annotated or wrongly annotated by the MIDI system, we performed gas chromatography-mass spectrometry (GC-MS) analysis of all samples after methylation and trimethylsilylation of hydroxy-FAs similar to a published procedure (19). After harvesting, the cell pellet was resuspended in an aqueous NaCl solution (1%, vol/vol). An aliquot of this suspension was transferred to a glass vial and centrifuged, and the pellet was dried in a vacuum concentrator. Five hundred microliters of a mixture of methanol, toluene, and sulfuric acid (50:50:2, vol/vol/vol) was added, and the vial was capped with a Teflon-lined screw cap and incubated at 55°C overnight. After the mixture was cooled to room temperature, 400 μl of an aqueous NH4HCO3 solution (0.5 M) was added, and phase separation was achieved by centrifugation. To 75 μl of the upper organic phase, 25 μl of N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA) (Macherey-Nagel, Düren, Germany) was added, and the mixture was allowed to react for 30 min at 37°C. This step was required to improve detection of hydroxy-FAs.
GC-MS was carried out on an Agilent 6890N gas chromatograph with a 5973 electron impact mass selective detector (Agilent, Waldbronn, Germany) using a dimethyl-(5% phenyl)-polysiloxane capillary column (Agilent HP-5ms, 0.25 mm by 30 m by 0.25 μm) and helium as the carrier gas at a flow rate of 1 ml/min. One microliter of the sample was injected in split mode (split ratio, 10:1). The column temperature was kept at 150°C for 5 min, increased to 260°C at a rate of 5°C/min, and then ramped to 300°C at 30°C/min and held at 300°C for 5 min. Other temperatures were as follows: inlet, 250°C; GC-MS transfer line, 280°C; ion source, 230°C; and quadrupole, 150°C. The mass selective detector was operated in scan mode, scanning the mass range from m/z 40 to 700. Data analysis was carried out with AMDIS software (22), version 2.64 (NIST, Gaithersburg, MD), using the value “integrated signal” for quantification. Amounts were calculated in percentages relative to the sum of integrals of a predefined subset of FA methyl esters, including their labeled forms when labeled precursors had been fed.
RESULTS AND DISCUSSION
The HMG-CoA synthase is involved in the biosynthesis of IV-CoA.
In order to enable the construction of double mutants in both pathways leading to IV-CoA, a markerless bkd mutant (DK5643) was constructed by following published procedures (11). This strain was the starting point to knock out genes which might be involved in the alternative pathway. To prove the postulated central role of the HMG-CoA synthase for isovaleryl-CoA-derived compounds, we constructed a knockout of mvaS encoding the HMG-CoA synthase in DK5643 and DK1622 (wild type). We expected Mx_0619 (also called MXAN_4267; www.tigr.org) to represent mvaS because it turned out to be the closest homologue to HMG-CoA synthases from Streptomyces griseolosporeus (NCBI accession number BAB07822) and was identified in the M1 genome (9) of M. xanthus DK1622 by a BLASTP search (1). mvaS was therefore disrupted by plasmid integration, resulting in strains DK5624 (Δbkd mvaS::kan) and DK5645 (mvaS::kan). Myxobacteria employ IV-CoA as a precursor for isoprenoid biosynthesis in addition to the standard mevalonate pathway (4, 5). Therefore, a disruption of both pathways leading to IV-CoA was expected to be lethal due to the requirement of isoprenoids for several essential processes (e.g., as membrane anchors during cell wall biosynthesis or as part of prenylated quinones in the respiratory chain) (18). Following this hypothesis, DK5624 was grown in CTT medium with or without MV supplementation. No growth could be observed without addition of MV. Furthermore, growth rate and final cell density correlated with increasing MV concentrations and showed a maximum at 1 mM MV (data not shown). Accordingly, construction and maintenance of DK5624 were performed on MV-supplemented medium (1 mM) to complement the lack of HMG-CoA biosynthesis and enable isoprenoid biosynthesis.
GC-MS analysis of the constructed strains showed that no change in the fatty acid profile could be observed for DK5645 (mvaS::kan) (data not shown) whereas a strong reduction in the amount of iso-FAs was recorded for DK5624 (Δbkd mvaS::kan) in comparison to the wild type and the DK5643 (Δbkd) mutant. This is exemplified by the amount of the major FA iso15:0, which is decreased from 41.9% in the wild type to 16.8% or 3.8% in the bkd mutant or bkd mvaS double mutant, respectively (Table 1). Furthermore, several iso-FAs present only in minor amounts in the wild type have not been detected in the mutants. The residual amount of iso-FAs in the double mutant results from a second but minor Bkd activity in M. xanthus, which was described previously (16). Moreover, feeding experiments with labeled leucine and acetate confirmed that the starting unit of the residual iso-FAs is derived from leucine exclusively (data not shown). As a consequence of the strong decrease of iso-FAs, the relative amount of straight-chain FAs is increased (exemplified by up to 40% of total FAs for 16:1ω5c in the double mutant). The wild-type FA profile could be restored in the double mutant by the addition of IVA as described previously for bkd mutants (2, 16, 23). Furthermore, when IVA was present, no MV was required for vegetative growth, which is in good agreement with previous results indicating that leucine is a major source of isoprenoids in myxobacteria (4, 5).
TABLE 1.
Fatty acid composition of cells of M. xanthus strains grown with or without IVA
| Fatty acida | % Total fatty acidsb
|
||||
|---|---|---|---|---|---|
| DK1622 | DK5643
|
DK5624
|
|||
| −IVA | +IVA | −IVA | +IVA | ||
| iso11:0 | 0.3 | 0.3 | |||
| iso13:0 | 0.9 | 0.4 | 0.4 | ||
| iso15:1ω9c | 0.7 | 0.9 | 0.7 | ||
| iso15:0 | 41.9 | 16.0 | 48.1 | 3.8 | 47.9 |
| iso15:0 3OH | 2.1 | 0.9 | 1.8 | 0.6 | 1.8 |
| iso17:2ω5c ω11c | 2.9 | 0.5 | 2.5 | 2.4 | |
| iso17:1ω11c | 1.4 | 0.7 | 1.6 | 1.5 | |
| iso17:1ω5c | 2.9 | 1.3 | 2.6 | 3.0 | |
| iso17:0 | 6.2 | 5.3 | 5.6 | 1.2 | 5.8 |
| iso17:0 2OH | 2.7 | 1.3 | 2.0 | 2.0 | |
| iso17:0 3OH | 3.3 | 1.6 | 2.9 | 0.7 | 3.1 |
| iso16:1 | 0.8 | 0.4 | 1.4 | 0.4 | |
| iso16:0 | 2.9 | 3.5 | |||
| 9:0 | 0.3 | 0.6 | |||
| 10:0 | 0.4 | 0.6 | 0.6 | 0.6 | 0.6 |
| 13:0 | 0.5 | ||||
| 14:1ω9c | 1.5 | 0.9 | 2.1 | 0.5 | 1.6 |
| 14:1ω5c | 0.4 | 0.4 | 0.4 | ||
| 14:0 | 4.4 | 7.0 | 4.2 | 7.7 | 3.9 |
| 15:1ω10c | 0.9 | 1.9 | 0.7 | 2.8 | 1.0 |
| 15:1ω4c | 0.7 | 1.6 | 0.3 | 3.3 | 0.8 |
| 15:0 | 0.5 | 4.8 | 11.5 | ||
| 16:2ω5c ω11c | 4.5 | 4.9 | 6.4 | 3.5 | 6.0 |
| 16:1ω11c | 1.0 | 2.9 | 1.1 | 2.5 | 1.1 |
| 16:1ω5c | 11.1 | 31.0 | 8.9 | 40.0 | 10.9 |
| 16:0 | 1.9 | 8.3 | 1.6 | 9.1 | 1.0 |
| 16:0 2OH | 0.6 | 0.5 | |||
| 16:0 3OH | 0.7 | 1.8 | 0.6 | 2.9 | 0.8 |
| 17:0 | 0.8 | ||||
| 9.912 | 0.6 | ||||
| 9.923 | 0.8 | 0.4 | 0.6 | ||
| 11.024 | 0.5 | 0.5 | |||
| 7.072 | 6.1 | 0.8 | 3.2 | 2.1 | |
| 16.575 | 1.0 | 0.7 | 0.5 | ||
For nonidentified fatty acids (last five rows), retention times (in minutes) are given.
Fatty acid composition is given in percentages of total fatty acids of cells of M. xanthus DK1622 (wild type), DK5643 (Δbkd), and DK5624 (Δbkd mvaS::kan) with (+IVA) or without (−IVA) the addition of 1 mM IVA.
Transcription of mvaS is strongly induced in bkd mutants but decreased after addition of isovalerate.
Previous feeding studies showed that the activity of the alternative pathway can be detected only in bkd mutants (16, 17). Therefore, quantitative RT-PCR analysis of mvaS (MXAN_4267) was performed to investigate if the assumed induction of the novel pathway results from transcriptional activation of the involved genes. In the wild type, maximum transcription of mvaS was observed at the beginning of the stationary phase. However, the absolute number of mvaS transcripts was five- to sevenfold higher in the bkd mutant than in the wild type and seemed to decrease during growth (Fig. 2). Growth of the bkd mutant in IVA-supplemented medium resulted in a decrease of transcription to approximately 3% of that of the nonsupplemented bkd mutant. Even more interesting, dilution of the bkd mutant into IVA-supplemented medium resulted in a dramatic drop of mvaS transcription to only about 1% of the initial value during the first 12 h, resulting in the transcription level observed for the IVA-supplemented bkd strain. Altogether, these results indicate a transcriptional regulation of mvaS by IVA or a compound derived thereof. Whether this regulation is direct and similar to the well-known feedback inhibition of early biosynthetic pathway enzymes by the end products of these pathways (18) remains to be elucidated.
The alternative pathway to isovaleryl-CoA is induced during fruiting body formation.
In order to identify a biological function of the alternative pathway to IV-CoA, feeding experiments with [D6]DMAA were performed under various growth conditions in DK1622. This compound was shown to be a specific intermediate of the pathway in recent feeding experiments (16, 17). No incorporation of deuterium label into iso-FAs could be observed during vegetative growth in rich medium during any stage of the growth phase (samples were taken over a period of 3 days where the culture grew from lag to late stationary phase). However, feeding of [D6]DMAA during fruiting body formation resulted in labeling of 3.8%, 15.7%, and 3.2% of the three most abundant FAs, iso15:0, iso17:0, and iso17:0 2OH, respectively, after 48 h of development. As a control experiment for the activity of the leucine degradation pathway, labeling of these fatty acids by [D3]leucine was also investigated during fruiting body formation. These experiments resulted in incorporation rates of 58.1%, 71.8%, and 64.2% into iso15:0, iso17:0, and iso17:0 2OH, respectively. Thus, leucine seems to be a much better precursor of iso-FAs than DMAA, which was additionally confirmed by feeding [D6]DMAA and [D3]leucine to the same culture. No incorporation of [D6]DMAA could be observed in this experiment, while [D3]leucine was incorporated almost as efficiently as without addition of [D6]DMAA.
The results of these feeding experiments show that during fruiting body formation the alternative pathway is active and that leucine is preferred over HMG-CoA as the precursor for iso-FAs in M. xanthus. Additionally, it was shown that de novo FA biosynthesis takes place during development. This is surprising since FAs are expected to be present in excess during development because 20 to 80% of all cells die during this process (depending on the experimental conditions) (20, 25) and therefore set free their cellular components, including FAs. Moreover, by generating a round myxospore from a rod-shaped vegetative cell, the overall cellular volume is maintained while the surface area including the membranes is significantly reduced (21).
Leucine is an essential amino acid for M. xanthus DK1622 (6). This also holds true for the two other branched-chain amino acids valine and isoleucine. However, the important function of leucine as a precursor for the most abundant FA family, the iso-odd FAs, indicates an additional explanation for the importance of leucine besides its function as a protein building block. We therefore postulate the alternative pathway to function as an additional IV-CoA supply route under leucine-limiting conditions.
Acknowledgments
We thank Taifo Mahmud for providing [D6]DMAA and R. Welch, J. Jakobsen, B. Goldman, TIGR, and the Monsanto Company for providing access to the M. xanthus genome sequence prior to publication.
We also thank the Deutsche Forschungsgemeinschaft for financial support (grants to H.B.B. and R.M.).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bode, H. B., J. S. Dickschat, R. M. Kroppenstedt, S. Schulz, and R. Müller. 2005. Biosynthesis of iso-fatty acids in myxobacteria: iso-even fatty acids are derived from (alpha)-oxidation of iso-odd fatty acids. J. Am. Chem. Soc. 127:532-533. [DOI] [PubMed] [Google Scholar]
- 3.Bode, H. B., M. W. Ring, D. Kaiser, A. C. David, R. M. Kroppenstedt, and G. Schwär. 2006. Straight-chain fatty acids are dispensable in the myxobacterium Myxococcus xanthus for vegetative growth and fruiting body formation. J. Bacteriol. 188:5632-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode, H. B., S. C. Wenzel, H. Irschik, G. Höfle, and R. Müller. 2004. Unusual biosynthesis of leupyrrins in the myxobacterium Sorangium cellulosum. Angew. Chem. Int. Ed. Engl. 43:4163-4167. [DOI] [PubMed] [Google Scholar]
- 5.Bode, H. B., B. Zeggel, B. Silakowski, S. C. Wenzel, H. Reichenbach, and R. Müller. 2003. Steroid biosynthesis in prokaryotes: identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol. Microbiol. 47:471-481. [DOI] [PubMed] [Google Scholar]
- 6.Bretscher, A. P., and D. Kaiser. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickschat, J. S., H. B. Bode, R. M. Kroppenstedt, R. Müller, and S. Schulz. 2005. Biosynthesis of iso-fatty acids in myxobacteria. Org. Biomol. Chem. 3:2824-2831. [DOI] [PubMed] [Google Scholar]
- 8.Dutton, C. J., S. P. Gibson, A. C. Goudie, K. S. Holdom, M. S. Pacey, J. C. Ruddock, J. D. Bu'Lock, and M. K. Richards. 1991. Novel avermectins produced by mutational biosynthesis. J. Antibiot. (Tokyo) 44:357-365. [DOI] [PubMed] [Google Scholar]
- 9.Jakobsen, J. S., L. Jelsbak, R. D. Welch, C. Cummings, B. Goldman, E. Stark, S. Slater, and D. Kaiser. 2004. σ54 enhancer binding proteins and Myxococcus xanthus fruiting body development. J. Bacteriol. 186:4361-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jelsbak, L., M. Givskov, and D. Kaiser. 2005. Enhancer-binding proteins with a forkhead-associated domain and the sigma54 regulon in Myxococcus xanthus fruiting body development. Proc. Natl. Acad. Sci. USA 102:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneda, T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kegler, C., K. Gerth, and R. Müller. 2006. Establishment of a real-time PCR protocol for expression studies of secondary metabolite biosynthetic gene clusters in the G/C-rich myxobacterium Sorangium cellulosum So ce56. J. Biotechnol. 121:201-212. [DOI] [PubMed] [Google Scholar]
- 14.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 15.Kunze, B., H. Reichenbach, R. Müller, and G. Höfle. 2005. Aurafuron A and B, new bioactive polyketides from Stigmatella aurantiaca and Archangium gephyra (myxobacteria). J. Antibiot. (Tokyo) 58:244-251. [DOI] [PubMed] [Google Scholar]
- 16.Mahmud, T., H. B. Bode, B. Silakowski, R. M. Kroppenstedt, M. Xu, S. Nordhoff, G. Höfle, and R. Müller. 2002. A novel biosynthetic pathway providing precursors for fatty acid biosynthesis and secondary metabolite formation in myxobacteria. J. Biol. Chem. 277:32768-32774. [DOI] [PubMed] [Google Scholar]
- 17.Mahmud, T., S. C. Wenzel, E. Wan, K. W. Wen, H. B. Bode, N. Gaitatzis, and R. Müller. 2005. A novel biosynthetic pathway to isovaleryl-CoA in myxobacteria: the involvement of the mevalonate pathway. Chembiochem 6:322-330. [DOI] [PubMed] [Google Scholar]
- 18.Michal, G. 1999. Biochemical pathways, vol. 1. Spektrum Akademischer Verlag, Heidelberg, Germany.
- 19.Minnikin, D. E., L. Alshamaony, and M. Goodfellow. 1975. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole-organism methanolysates. J. Gen. Microbiol. 88:200-204. [DOI] [PubMed] [Google Scholar]
- 20.O'Connor, K. A., and D. R. Zusman. 1988. Reexamination of the role of autolysis in the development of Myxococcus xanthus. J. Bacteriol. 170:4103-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimkets, L., and T. W. Seale. 1975. Fruiting-body formation and myxospore differentiation and germination in Myxococcus xanthus viewed by scanning electron microscopy. J. Bacteriol. 121:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein, S. E. 1999. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10:770-781. [Google Scholar]
- 23.Toal, D. R., S. W. Clifton, B. A. Roe, and J. Downard. 1995. The esg locus of Myxococcus xanthus encodes the E1 alpha and E1 beta subunits of a branched-chain keto acid dehydrogenase. Mol. Microbiol. 16:177-189. [DOI] [PubMed] [Google Scholar]
- 24.Trowitzsch-Kienast, W., V. Wray, K. Gerth, H. Reichenbach, and G. Höfle. 1986. Antibiotika aus Gleitenden Bakterien. XXVIII. Biosynthese des Myxothiazols in Myxococcus fulvus Mx f16. Liebigs Ann. Chem. 10:93-98. [Google Scholar]
- 25.Wireman, J. W., and M. Dworkin. 1977. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J. Bacteriol. 129:796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]