Abstract
Buchnera aphidicola is the endosymbiotic bacterium of the pea aphid. Due to its small genome size, Buchnera lacks many essential genes for autogenous life but obtains nutrients from the host. Although the Buchnera cell is nonmotile, it retains clusters of flagellar genes that lack the late genes necessary for motility, including the flagellin gene. In this study, we show that the flagellar genes are actually transcribed and translated and that the Buchnera cell surface is covered with hundreds of hook-basal-body (HBB) complexes. The abundance of HBB complexes suggests a role other than motility. We discuss the possibility that the HBB complex may serve as a protein transporter not only for the flagellar proteins but also for other proteins to maintain the symbiotic system.
Buchnera aphidicola sp. strain APS belongs to the gamma subdivision of the Proteobacteria, which are closely related to Escherichia coli. It is the primary endosymbiotic bacterium of the pea aphid, Acyrthosiphone pisum (Harris), and the bacterial cells are harbored by large, differentiated cells called bacteriocytes in the fat body (13). The symbiotic relationship between Buchnera and the pea aphid dates back approximately 200 million years and is so tight that neither partner can survive without the other; the aposymbiotic aphid grows poorly and produces few or no offspring, and Buchnera isolated from bacteriocytes of the host aphids is nonculturable (5, 7, 8).
As a result of an evolutionary reduction of genome size, the Buchnera genome contains only 600 kbp, which is about one-seventh of the Escherichia coli genome and which suggests that Buchnera lacks many of the essential genes for the autogenous life. In fact, Buchnera lacks most of the genes required not only for the biosynthesis of cell surface components, substrate-specific receptors, and membrane transporters but also for two-component regulatory systems. Compare the genes in Buchnera to those in E. coli; 3 versus 190 for ABC transporters, 5 versus 52 phosphotransferase system genes, and 0 versus 88 genes for two-component response regulators (14). Clearly, Buchnera needs ways of obtaining nutrients from the host, but little is known about the import system.
The Buchnera genome contains flagellar genes, although the cells are nonmotile. Why does the nonmotile Buchnera retain a large set of flagellar genes? Are they being coopted for some other use? In the present study we show that the flagellar genes are expressed, and the proteins are integrated into the flagellar hook-basal body (HBB). The HBB seems to be an evolutionally degenerated form of the flagellum that has lost motility function. However, to our surprise, the number of HBBs is on the order of hundreds per cell, indicating that the structures are unlikely to be a nonfunctional remnant but rather form an actively functioning apparatus. We discuss the possibility that the flagellar apparatus may be involved in the transport of proteins between the symbionts.
MATERIALS AND METHODS
Materials.
A long-established parthenogenetic clone of the pea aphid, Acyrthosiphon pisum (Harris) was maintained on young broad bean plants, Vicia faba (L.) under a long-day regime with 16 h of light and 8 h of dark at 15°C. B. aphidicola sp. strain APS is surgically isolated from bacteriocytes of the aphid.
Reverse transcription-PCR (RT-PCR) and sequencing.
Twenty-six sets of primers were constructed, according to the genome sequence of Buchnera, to generate specific fragments for flagellar genes. The genes (forward primer/reverse primer/predicted length of the transcript, in base pairs) were as follows: fliE (TAATTTGTTAGACGCACAAAAAAAAGACAG/TGTTGACTCATAATTTCTTGATAAGCTGAT/246), fliF (AATGGGGGAAAGTAAGAATCTACATGATGA/ATCACCCTGGCCTCCAAATAAAATATCTAT/306), fliG (GCGTTGTTATTAATGGCAATAGGTTCTGAT/TTTCTAAAGCCTCTTTTAAAAGAGACGTGC/271), fliH (AAGAGGGTGTTTTCTTAAAAAAACCACAGT/AAATAAGACGACATAATTGCTGCCATCTAG/213), fliI (GAGGTGGTAGGATTGAACACATCTATTGGT/TTAGGCAATTGATCTAATGGTTGACCTCAG/288), fliJ (ATTAATATTAGGTGTGTCGGTACATCAATG/GAATATGACTATCATTGATAATTGCGTCCT/249), fliK (TAAAATCAATCCCATGATCAGTAAAAGACA/AGGCATATAACTGTCTAAAAATGTTTTGAC/307), fliM (AATGGGGGAAAGTAAGAATCTACATGATGA/ATCACCCTGGCCTCCAAATAAAATATCTAT/289), fliN (AGATGTTGACAAAAATTTATTACCCCAAGA/ACTACAATTTCTCCAGATGCAATTAAATGA/257), fliP (AGTCATTTGCATACGTTTCCGTCTAATTCA/AGAAATGCTGGAAGAAAAGTCAGAGATGTC/299), fliQ (TAATGCTATGAAAGTTGCCTTGATTATTGC/TGCATATAATCCAGCATAACACCTAACATC/201), fliR (TCTGTTGCACCTATTTTTAAGGAAAAACTG/AACGAGATATTATAGAGGTGCCAATTTGAC/301), flhB (TGCGGATTCGTCAAGCAATGAAAGCTGTTA/CCAAACCCATGCTAAAACTTCTGCAACAGC/321), flhA (AAACAACTCAAT GGCAAATACTTGCTGGTC/GACCAAACGATTCAATTACTCTTCCTG CTG/303), flgN (CTTTAGAACGAATATTAAAACAAGAATGTC/ACAGATTTTTTATATGATGAAGACAATTCT/325), flgA (TACAATCAATTCGGTCAGTGCAACTGTTCA/GTTCCTCGGGGAATTTTTCTATTCGCTACA/302),flgB (TTGTTCTCAAGACAAGAAATATTATCTGCT/TCAATTCTTTCTCTATCCATATTTACGGTG/260), flgC (ATTGCAGGTTCAGCTATGATTGCACAATCG/CCTGGTAGCTTCTTGCTGCTGCGATATTAT/322), flgD (GGAATGCAAAAACTAAACAATACCGTGGAT/ACCATCCCAAAAGAA ATTATGTCTACCAGC/273), flgE (TGAAACTGGACGAGATTTGGATTTAGGAAT/TAGTTTGTTGTCAGAATTATCCACACTGCT/328), flgF (TGCGTTGTTATTAATGGCAATAGGTTCTGA/CGTGCCTTTTTTTTCTCCTAATGCTTTAGT/246), flgG (ATGGATTTCTAAAACTGGTCTTGATGCTCA/AGCATCTGTTTTTGAAAGATTACCCTGAGT/227), flgH (TTCGGCTTAAAAATCGCACCTCGACAATAA/CAATACGTGCATCAGCTATTTC AGTGGATG/319), flgI (AAAAAATGTAGCAGCAGTAATTGTAACGGA/TATCTATTTCTCGTTCAATTGTTGCACCGT/290), flgJ (GAACTTAAAT ACCAAGTTCGTATTAATCCA/TTACTTATTTCTTGAGATAATTGTTGG TCA/194), and flgK (CGCTATTTCTGGTATGAATGCAATGAAGAT/AGCTGCTTCGATTTTAGTTGTTCATCTTGA/239). Using total RNA extracted from aphids as a template, we synthesized cDNA with random hexamers by using the SuperScript II reverse transcriptase (Gibco-BRL). The cycling parameters were as follows: 96°C for 3 min, followed by 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min for 25 cycles, and finally 72°C for 10 min. The resultant PCR products were analyzed on 2% agarose gels, and their sequences were determined by the direct cycle sequencing method.
Electron microscopic observation of negatively stained Buchnera cells.
Buchnera cells were put onto a grid, negatively stained with 1% sodium phosphotungstate (pH 6.5), and observed with a transmission electron microscope (JEM-1010; JEOL, Japan). Micrographs were taken at an accelerating voltage of 80 kV.
Proteome analysis of Buchnera.
Buchnera cells were purified from about 500 bacteriocytes and lysed in 250 μl rehydration buffer consisting of 6 M urea, 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 0.5% IPG buffer (pH 4 to 7), 0.01% bromophenol blue, and 0.28% dithiothreitol. A total of 200 μl of lysate was applied to an Immobiline DryStrip (11 cm long, pH 4 to 7; Amersham Biotech). After first-dimensional isoelectric focusing, the gel was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 12.5% polyacrylamide separating gel. The gel was stained with Coomassie brilliant blue (CBB) R-250, and about 50 of the strongly staining protein spots were cut out and in-gel digested with trypsin. The masses of the peptide fragments were analyzed on a matrix-assisted laser desorption ionization-time of flight/mass spectrometry (MALDI-TOF/MS). Proteins were identified by using the MASCOT program against the Buchnera genome database (http://www.buchnera.gsc.riken.go.jp).
RESULTS
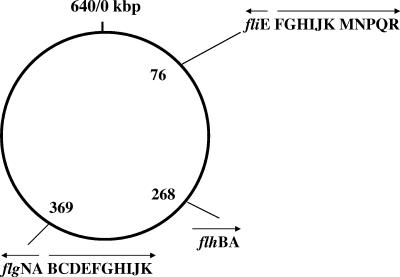
The B. aphidicola chromosome contains 26 flagellar genes even though the cells are nonmotile. The flagellar genes are arranged in five operons, which are clustered in three regions of the genome. Compared to the well-studied flagellar system of Salmonella, the order of Buchnera flagellar genes in operons is highly conserved, as can be seen in the alphabetical order of gene alignment (Fig. 1). The sequence homology of the flagellar proteins between the two species varies from as high as 75% for FlgG to as low as 20.8% for FliJ (Table 1). Note that the six core proteins of the type III secretion system (T3SS) (FlhA, FlhB, FliI, FliP, FliQ, and FliR) are highly conserved, with 40% sequence homology compared to those of Salmonella (2). All of these genes found in Buchnera are required for assembly of the so-called HBB complex (3, 10, 12). The genes missing from the corresponding set of Salmonella flagellar genes include fliA, fliC, fliD, fliL, fliO, fliS, fliT, flgM, and flgL, which are required for filament formation and which are expressed only during the last stage of flagellar assembly (1). In addition, Buchnera lacks both flhDC of the master operon and motAB required for motor function. Before we determined what function, if any, the flagellar genes might serve, we sought to determine whether the Buchnera flagellar genes are expressed.
FIG. 1.
Organization of the flagellar genes on the chromosome of Buchnera aphidicola sp. strain APS. The genome size is 640,681 bp and holds 583 genes. Flagellar genes are densely packed in three clusters on the genome. Arrows indicate the directions of transcription.
TABLE 1.
Comparison of Buchnera and Salmonella flagellar gene productsa
| Gene product | Buchnera sequence homology vs Salmonella (%) | pI
|
|
|---|---|---|---|
| Buchnera | Salmonella | ||
| FliE | 40.6 | 6.5 | 6.0 |
| FliF | 38.2 | 10.2 | 6.3 |
| FliG | 40.4 | 9.7 | 4.7 |
| FliH | 24.3 | 9.9 | 4.6 |
| FliI | 55.6 | 9.8 | 6.1 |
| FliJ | 20.8 | 10.9 | 7.8 |
| FliK | 34.8 | 9.9 | 4.9 |
| FliM | 22.1 | 8.6 | 5.5 |
| FliN | 33.6 | 9.0 | 4.4 |
| FliP | 45.9 | 10.5 | 9.5 |
| FliQ | 63.6 | 5.7 | 4.9 |
| FliR | 42.6 | 10.4 | 6.5 |
| FlhB | 41.2 | 10.6 | 9.0 |
| FlhA | 62.1 | 9.9 | 5.5 |
| FlgN | 21.1 | 10.4 | 5.3 |
| FlgA | 29.0 | 10.8 | 10.4 |
| FlgB | 38.1 | 10.6 | 5.3 |
| FlgC | 55.9 | 10.3 | 5.3 |
| FlgD | 34.7 | 8.9 | 4.7 |
| FlgE | 37.3 | 4.8 | 4.8 |
| FlgF | 33.6 | 10.4 | 4.6 |
| FlgG | 75.0 | 4.5 | 4.7 |
| FlgH | 53.8 | 10.3 | 8.7 |
| FlgI | 48.4 | 10.5 | 9.0 |
| FlgJ | 30.9 | 9.1 | 8.8 |
| FlgK | 27.9 | 5.3 | 4.8 |
The amino acid sequence homology of the flagellar proteins between the two species and their isoelectric points are listed for comparison.
To examine transcription of flagellar genes, reverse transcription-PCR (RT-PCR) was performed using the primer pairs constructed according to the genome sequences (see Materials and Methods). As shown in Fig. 2, amplification was detected for all 26 genes. No bands were detected in samples lacking the reverse transcriptase. The sizes of the RT-PCR products agreed with those predicted from the genome sequences. By direct cycle sequencing analysis, the nucleotide sequence of each gene product was shown to match exactly that expected from the genome sequence, thus validating the active transcription of all flagellar genes in Buchnera cells.
FIG. 2.
Detection of flagellar gene expression by RT-PCR. Transcripts of 26 flagellar genes were amplified by RT-PCR with the specific primers listed in the text and electrophoresed on 2% agarose gel. +, PCR after RT; −, PCR without RT. M, molecular markers of 100-bp ladder. OmpF is the positive control.
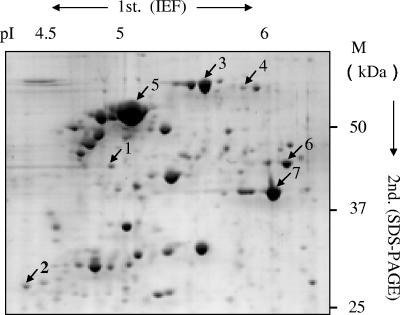
To examine the translation of the flagellar genes, proteomic analysis was performed. Whole lysate of Buchnera cells surgically isolated from aphids was analyzed by SDS-PAGE. This was followed by peptide mass fingerprinting analysis on the MALDI-TOF/MS). By systematic analysis of the bands from SDS gels, we could identify nine flagellar proteins: FliH, FlgF, FlhA, FlgE, FlgH, FlgI, FliG, FliM, and FliI (data not shown). Bands in the region, which overlapped with many different proteins, were separated by two-dimensional PAGE. Among more than 200 proteins separated in the acidic region of the two-dimensional gel, about 50 major spots were analyzed. The largest spot was MopA, a GroEL-like chaperonin 60, which is also called symbionin (13). The other major spots included DnaK, RpsA (30S ribosomal protein S1), TufB (elongation factor EF-Tu), and OmpF-like protein. Two flagellar proteins, FlgE for the hook and Flg G for the distal rod, were further identified among minor spots (Fig. 3). It should be noted that the copy numbers of FlgE and FlgG are too small (120 and 26 subunits per flagellum, respectively [9]) to detect in the crude preparation by CBB staining, and thus the immunoblotting is routinely used for detection. The CBB-stained spots of these proteins in Fig. 3 indicate that FlgE and FlgG are abundant in this crude preparation.
FIG. 3.
Two-dimensional electrophoretic profile of Buchnera proteins. Buchnera cells isolated from bacteriocytes were lysed in rehydration buffer, and the aliquot was applied onto an Immobiline DryStrip (pI 4 to 7). After isoelectric focusing, followed by SDS-12.5% PAGE, peptide mass fingerprinting analysis by using MALDI-TOF/MS was performed. The largest spot was MopA, a GroEL-like 60-kDa chaperonin. Numbers: 1, FlgE; 2, FlgG; 3, DnaK; 4, RpsA; 5, MopA, 6, TufB; 7, OmpF-like protein.
From our results, we conclude that the Buchnera flagellar genes are not pseudogenes but are actually transcribed and translated in cells. It is well known that the flagellar gene expression is directly controlled by the master genes, flhDC in Salmonella, which are in turn under the regulation of other genes that sense environmental changes. Since Buchnera cells are confined in a seemingly stable milieu of the bacteriocyte and buffered against environmental changes, the absence of these regulatory genes in Buchnera suggests that the flagellar genes may be constitutively expressed.
Knowing that the flagellar genes are expressed, we expect to find flagellar structures assembled on the cell surface, and hence we examined intact Buchnera cells isolated from bacteriocytes by transmission electron microscopy. Cells appeared partially translucent (Fig. 4) when stained with 1% sodium phosphotungustate. Either membranes were breached by a slight osmotic shock into the staining solution and/or are simply the Buchnera since the outer membrane lacks the porins. There were no filamentous structures observed on the cell surface. Instead, we observed hundreds of structures that span between the inner and outer membranes (Fig. 4). The structure resembles the flagellar basal body, with MS- and PL-ring complexes and rods penetrating them. Some particles have a hook-like structure, albeit too short to curve, that extends from the PL-ring complex (Fig. 4, insets). The dimensions of these features are similar to those of Salmonella (3, 10). Our images are evidence that the expressed flagellar genes are assembled into flagellar HBBs. It should be noted that the Buchnera HBB is peritrichously distributed and that our count of a few hundred particles on the periphery of the cell in Fig. 4 provides an estimate of at most a thousand HBBs per cell.
FIG. 4.
Electron micrograph of the negatively stained Buchnera. A Buchnera whole cell shows that the periphery is covered with HBBs. Insets: A and B, enlarged views of the peripheral area of the cell surface; C and D, typical HBB-like particles are further enlarged. Buchnera organisms isolated from the bacteriocyte were stained with 1% sodium phosphotungstate. The specimen was observed by transmission electron microscopy. The scale bar for the main image is 500 nm; the scale bars for insets A and B is 100 nm and for insets C and D is 20 nm.
In conclusion, the flagellar genes on the Buchnera genome are transcribed and translated, and the flagellar proteins assembled into hundreds of HBBs, which cover the cell surface of Buchnera.
DISCUSSION
The genome analysis of B. aphidicola sp. strain APS has revealed that Buchnera retains flagellar genes coding proteins for the hook and the basal body but not for the filament. The genomic information has also suggested that the partial structure of the flagellum, if it exists, might be used for purposes other than motility to enrich the symbiosis life (15). We report here the first experimental evidence of the flagellar structure existing in Buchnera.
We have found hundreds of HBBs on the cell surface of Buchnera. The HBBs are peritrichously distributed over the surface. Since there are no master genes that control the flagellar number in peritrichously flagellated species such as E. coli and Salmonella, Buchnera flagellar proteins might be constitutively expressed, distributed, and assembled in the membrane without control. Why are so many HBBs necessary for Buchnera? Buchnera is nonmotile, and the flagella lack the essential components of motility such as a filament (the propeller) and the Mot complex (the stator of the motor). These facts suggest that the Buchnera HBBs may have functions distinct from motility. The most plausible function is protein transporter, as the flagellar apparatus belongs to the family of T3SSs (1, 4, 6, 11, 16).
The T3SS proteins that are secreted do not have signal sequences and are thus secreted without cleavage. In other bacterial species such as Salmonella, the flagellar HBB complex exports the flagellar proteins that form the rod, hook and filament; the other T3SS structures secrete effectors of virulence in order to invade the host. The appearance of the rod and part of the hook in Buchnera is evidence that the HBB complex can function to export proteins. Considering the reduced genome size of Buchnera, which lacks the full set of genes encoding the membrane transporters, we suggest that the Buchnera HBB is a plausible candidate for an export apparatus to transfers Buchnera proteins to the host. Can the HBBs export proteins other than flagellar proteins? There is evidence that some virulence factors are also secreted through the flagellar secretion apparatus in Yersinia (16), but more examples are necessary to support this hypothesis.
There is an ATPase at or near the gate of the flagellar export apparatus; it is thought to unfold proteins to be secreted or to insert them into the axial export channel. However, the energy of transport itself has not been identified. One candidate of the energy source is the membrane potential. The pI values of the HBB components are mostly basic (9.0 to 10.0), whereas those of Salmonella are acidic (4.0 to 5.0) (15; see also Table 1). This is also true for the whole Buchnera proteins, suggesting that the electrostatic environment, especially the membrane potential, of Buchnera may be quite different from that of other bacteria. It will be interesting to determine the polarity of the electric potential in Buchnera. We believe that the reason for the abundance of the HBB on the surface of Buchnera will be apparent once we determine the function they perform.
Acknowledgments
We thank Yoshiyuki Sakaki and the technical staff of RIKEN GSC for proteome analysis, and Masaomi Kanbe for transmission electron microscopic analysis. We thank Kazuhiro Kutsukake and Ritsu Kamiya for invaluable comments, the late Hajime Isikawa for his encouragement, and David DeRosier for critical reading of the manuscript.
This study was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Technology of Japan (M.M.) and by the Softnano-machine project, Japan Science and Technology Agency (S.A.).
REFERENCES
- 1.Aizawa, S.-I. 1996. Flagellar assembly in Salmonella typhimurium. Mol. Microbiol. 19:1-5. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, S.-I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 3.Aizawa, S.-I., E. D. Gay, J. J. Christopher, M. M. Robert, and S. Yamaguchi. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aizawa, S.-I., I. B. Zhulin, L. Marquez-Magana, and G. W. Ordal. 2002. Chemotaxis and motility, p. 437-452. In A. L. Sonenshein, (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 5.Baumann, P., L. Baumann, C.-Y. Lai, D. Rouhbakhsh, N. A. Moran, and M. A. Clark. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu. Rev. Microbiol. 49:55-94. [DOI] [PubMed] [Google Scholar]
- 6.Blocker, A., K. Komoriya, and S.-I. Aizawa. 2003. Type III secretion system and bacterial flagella: insight into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria, Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa, H. 1989. Biochemical and molecular aspects of endosymbiosis in insects. Int. Rev. Cytol. 116:1-45. [DOI] [PubMed] [Google Scholar]
- 9.Jones, C. J., R. M. Macnab, H. Okino, and S.-I. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 10.Kubori, T., N. Shimomura, S. Yamaguchi, K. Namba, and S.-I. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 11.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S.-I. Aizawa. 1998. Supermolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 12.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morioka, M., and H. Ishikawa. 1998. Insect chaperonin 60: symbionin. Methods Enzymol. 290:181-193. [DOI] [PubMed] [Google Scholar]
- 14.Munson, M. A., P. Bauman, and M. G. Kinsey. 1991. Buchnera, a new genus and Buchnera aphidicola, new species, a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int. J. Syst. Bacteriol. 41:566-568. [Google Scholar]
- 15.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 16.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]