Abstract
StpA has functional similarity to its homologue, the nucleoid structuring protein H-NS. It binds to AT-rich, planar, bent DNA and constrains DNA supercoils. In addition, StpA acts as an RNA chaperone. StpA and H-NS also form heterodimers. However, cellular levels of StpA are low due to repression of stpA by H-NS and negative autoregulation. Here we show that effective (30-fold) repression of stpA transcription requires a downstream regulator element located within the stpA coding region. In addition, we show that StpA represses stpA threefold in an hns null mutant. In contrast, repression of the bgl operon, another H-NS-repressed system, is not achieved by StpA alone. It becomes StpA dependent in the presence of a fusion protein encompassing the N-terminal 37 amino acids of H-NS, which comprise the core of the dimerization domain. StpA also effectively complements H-NS-I119T, a mutant defective in specific DNA binding, in repression of the bgl operon. Thus, StpA complements H-NS proteins defective in DNA binding to repress bgl, while in autoregulation of stpA it acts autonomously, indicating a difference in the mechanisms of repression.
The 15.3-kDa StpA protein of Escherichia coli is 58% identical to H-NS, a pleiotropic repressor and nucleoid structuring protein (14, 44). StpA binds to AT-rich, planar, bent DNA and constrains supercoils, as does its homologue H-NS, but with a higher affinity (24, 38). In addition to these features which are functionally similar to those of H-NS, StpA is an RNA chaperone. StpA was initially identified as a suppressor of a T4 td intron mutant defective in splicing, where stpA is carried on a multicopy plasmid (44, 45). StpA catalyzes annealing of the mutant td intron RNA and promotes its self-splicing (45). In addition, StpA binds to MicF, a small regulatory RNA, resulting in destabilization of MicF (7). MicF is known to interact with the ompF RNA, triggering its degradation and thus resulting in downregulation of the OmpF outer membrane porin (8).
Genetic and biochemical data demonstrated that StpA and H-NS form heterodimers (21, 22, 40). Heterodimer formation might be important in the modulation of H-NS activity by StpA, and heterodimers may have specific functions. Furthermore, StpA was found to be unstable in hns mutants, while it is stable in the wild type, which demonstrated that heterodimer formation with H-NS protects StpA from degradation by Lon (21, 22). The cellular level of StpA is significantly lower than that of its abundant homologue, H-NS, most likely due to repression of stpA by H-NS (37, 46). In hns mutants, StpA negatively autoregulates transcription of stpA (37, 46). Furthermore, stpA expression is temperature regulated, activated during exponential growth, regulated by Lrp, and induced by osmotic stress (16, 37).
Overexpression of StpA in hns mutants can restore the repression of several H-NS-repressed systems, such as proU, bgl, the synthetic 5A6A galP1 promoter, and hns (21, 40, 46). Likewise, chromosomally encoded StpA partially compensates for positive regulation of translation of malT by H-NS (20). Therefore, StpA is considered a molecular backup of H-NS (46). However, StpA may have distinct functions from those of H-NS, since StpA has unique features, mutation of stpA has no effect on repression by H-NS, and complementation of hns mutants occurs only partially and often requires artificially high levels of StpA (16). Furthermore, in several studies of complementation of hns mutants by StpA, hns mutants with insertions or replacements of the hns coding sequence were used, which may express truncated H-NS proteins (for example, see references 20 and 46). For example, H-NS proteins with truncation of the C-terminal DNA-binding domain are sufficient to render repression of bgl StpA dependently, indicating that StpA acts as an adapter to target H-NS mutants defective in DNA binding to bgl (17, 18, 29, 39).
Here we addressed the regulation of stpA and bgl by H-NS and by StpA. We found that repression of stpA by H-NS is significantly more effective when a downstream sequence encompassing the stpA coding region is present. In addition, StpA autoregulates stpA, with threefold repression, in an hns null mutant, i.e., independent of H-NS. In contrast, repression of bgl by StpA occurs only in the presence of the N-terminal H-NS protein domain or an H-NS-I119T mutant, i.e., in the presence of mutant H-NS proteins defective in specific DNA binding. This supports the model that StpA complements such H-NS mutants to repress bgl and may indicate that dimerization and/or oligomerization by H-NS is required for repression of bgl by StpA or that StpA nucleates (or targets) the defective H-NS proteins to bgl.
MATERIALS AND METHODS
Strains and plasmids.
The genotypes of the E. coli strains and the relevant structures of the plasmids used for this study are listed in Table 1. All experiments were performed using isogenic E. coli K-12 CSH50 (28) derivatives (Table 1). Transductions were performed using phage T4GT7 (41). Integration of lacZ reporter constructs into the chromosomal phage lambda attachment site attB was performed as described previously (10, 12). Briefly, strain S541 or mutant derivatives of this strain harboring the temperature-sensitive, integrase-expressing plasmid pLDR8 (10) were transformed with religated originless BamHI fragments carrying attP, a spectinomycin resistance cassette, and the respective lacZ reporter gene fusion (Table 1). Integrants were selected on LB-spectinomycin plates at 42°C. Independent colonies were tested by PCR to verify the integration of a monomer into attB and the integrity of the lacZ fusion construct. Two independent clones were used in the enzyme assays. Strain S3215 (S2509 Δhns::kanKD4), which has an exact deletion of hns (including the start codon and last codon), was constructed as described previously (6), using primers S665 (TCTATTATTACCTCAACAAACCACCCCAATATAAGTTTGAGATTACTACAgtgtaggctggagctgcttcg) and S672 (AAATCCCGCCGCTGGCGGGATTTTAAGCAAGTGCAATCTACAAAAGATTAcatatgaatatcctccttagttcctattcc). The sequence that is complementary to the template pKD3 or pKD4 for PCR is in lowercase letters.
TABLE 1.
E. coli K-12 strains and plasmids used for this study
| Strain or plasmid | Genotype or structurea | Constructionb or reference |
|---|---|---|
| Strains | ||
| CSH50 | bgl° Δ(lac-pro) ara thi | 27 |
| M182 | Δ(lacIPOZYA)74 galU galK strA | 9 |
| PD32 | MC4100 hns-206::amp | 46 |
| S159 | M182 stpA::tet | 46 |
| S160 | M182 hns::kan | 46 |
| S541 | Δbgl-AC11 ΔlacZ-Y217 ara thi (derived from CSH50) | 12 |
| S614 | S541 hns::amp | × T4GT7 (PD32) |
| S2303 | S541 hns::kan | × T4GT7 (S160) |
| S1195 | S541 attB::[SpecrPlacUV5 bglG(orf) (+95 to 971) lacZ] | × pKESD49 (13) |
| S1258 | S1195 hns::amp | 13 |
| S2323 | S1195 hns::kan | × T4GT7 (S160) |
| S1371 | S1195 stpA::tet | × T4GT7 (S159) |
| S1767 | S1371 hns::amp | × T4GT7 (PD32) |
| S2476 | S1371 hns::kan | × T4GT7 (S160) |
| S1213 | S541 attB::[SpecrPbgl (+25) lacZ] | × pKEKB30 (13) |
| S1471 | S1213 hns::amp | × T4GT7 (PD32) (13) |
| S2442 | S1213 hns::kan | × T4GT7 (S160) |
| S1437 | S1213 stpA::tet | × T4GT7 (S159) |
| S3158 | S1437 hns::kan | × T4GT7 (S160) |
| S2217 | S541 attB::[SpecrstpA (−540 to +466) lacZ] | × pKES121 |
| S2262 | S2217 hns::amp | × T4GT7 (PD32) |
| S2519 | S2217 hns::kan | × T4GT7 (S160) |
| S2221 | S541 attB::[SpecrPstpA (−540 to +41) lacZ] | × pKES123 |
| S2266 | S2221 hns::amp | × T4GT7 (PD32) |
| S2523 | S2221 hns::kan | × T4GT7 (S160) |
| S2258 | S2221 stpA::tet | × T4GT7 (S159) |
| S2951 | S2258 hns::amp | × T4GT7 (PD32) |
| S2604 | S2258 hns::kan | × T4GT7 (S160) |
| S2509 | S541 attB::[SpecrPstpA stpA(orf) (−540 to +466) lacZ] | × pKETW3 |
| S2844 | S2509 hns::amp | × T4GT7 (PD32) |
| S2554 | S2509 hns::kan | × T4GT7 (S160) |
| S2561 | S2509 stpA::tet | × T4GT7 (S159) |
| S2953 | S2561 hns::amp | × T4GT7 (PD32) |
| S2633 | S2561 hns::kan | × T4GT7 (S160) |
| S3215 | S2509 Δhns::kanKD4 | This work |
| S2513 | S541 attB::[SpecrPstpA (−540 to +98) lacZ] | × pKETW5 |
| S2848 | S2513 hns::amp | × T4GT7 (PD32) |
| S2557 | S2513 hns::kan | × T4GT7 (S160) |
| S2565 | S2513 stpA::tet | × T4GT7 (S159) |
| S2955 | S2565 hns::amp | × T4GT7 (PD32) |
| S2637 | S2565 hns::kan | × T4GT7 (S160) |
| S2741 | S541 attB::[SpecrPlacUV5 stpA(orf) (+1 to +466) lacZ] | × pKETW17 |
| S2850 | S2741 hns::amp | × T4GT7 (PD32) |
| S2770 | S2741 hns::kan | × T4GT7 (S160) |
| S2766 | S2741 stpA::tet | × T4GT7 (S159) |
| S2957 | S2766 hns::amp | × T4GT7 (PD32) |
| S2774 | S2766 hns::kan | × T4GT7 (S160) |
| Plasmids | ||
| pLG | pSC101 neo | 40 |
| pLG-HNS | hns in pLG | 40 |
| pLG-HNS | hns-L26P in pLG | 40 |
| pLG-HNS | hns-E53G-T55P in pLG | 40 |
| pLG-HNS | hns-I119T in pLG | 40 |
| pKETW18 | pSC101 cat | This work |
| pKETW20 | hns wt with hns promoter in pKETW18 | This work |
| pKETW21 | hns-L26P with hns promoter in pKETW18 | This work |
| pKETW22 | hns-E53G-T55P with hns promoter in pKETW18 | This work |
| pKETW23 | hns-I119T with hns promoter in pKETW18 | This work |
| pKETW15 | lacIqlacO3-Ptac-lacO, multiple cloning site in pACYC, cat | This work |
| pKETW13 | hns-His6 in pKETW15 | This work |
| pKETW14 | hns::amp-His6 in pKETW15 | This work |
| pKETW7 | hnsKN-His6 in pKETW15 | This work |
| pKETW8 | hns::kan-His6 in pKETW15 | This work |
| pKEM30 | lacIqlacO3-Ptac-lacO, multiple cloning site, HA tag in pACYC, neo | This work |
| pKETW10 | hnsKN-HA under control of lacIqtac in pKEM30 | This work |
| pKEM51 | lacIqtac hns-HA in pSC101 Cm (pKETW18) | This work |
For the lacZ reporter constructs integrated into attB, the relevant structure is given. Positions of cloned fragments and fusions are given in brackets, e.g., Pbgl (+25) indicates that the lacZ gene was fused downstream of base pair +25 relative to the transcription start of the bgl promoter, and stpA(orf) (+1 to +466) indicates that a stpA fragment encompassing positions +1 to +466 relative to the transcription start and carrying an ATG-to-ATA mutation of the translation start was cloned. The lacUV5 promoter encompasses a fragment from positions −40 to +1 relative to the transcription start and thus lacks lacO. Compiled sequences of all plasmids are available upon request. For other details, see references 11-13, 26, and 35. Δhns::kanKD4 was constructed as described in Material and Methods.
Transductions and integrations into attB were performed as described in Materials and Methods.
Plasmids were constructed according to standard techniques (1, 34). All lacZ reporter fusions were derived from a plasmid that carries a pACYC177 origin, the neo gene (4), attP, and an omegon-spectinomycin resistance cassette (33) as described previously (12). Site-specific mutations and fusions of stpA or bgl to lacZ were constructed by PCR. Plasmids pKETW20 to pKETW23 for expression of the dominant-negative hns mutants were constructed by PCR amplification of fragments encompassing hns and hns mutants, using the pLG plasmid series (40) as the template and primers S601 (GCGTCGACTTATTGCTTGATCAGGAAATCGTC) and S602 (ACTCTAGATCCTTACATTCCTGGCTATTGCA). The PCR fragments were subjected to restriction digestion with XbaI and SalI (restriction sites are indicated by underlining), and the fragments were cloned into XbaI- and SalI-digested pKETW20, which is a pSC101-derived vector conferring chloramphenicol resistance. Plasmids pKETW13, pKETW14, pKETW8, and pKETW7 were constructed by PCR amplification of hns, hns-206::amp, hns::kan, and hnsKN (i.e., the 5′ open reading frame of hns::kan). The PCR fragments encompass 26 bp upstream of the ATG codon. At the 3′ end of the coding sequence, six histidine codons (CAT) and a stop codon (TGA) were added. The PCR fragments were digested with the restriction enzyme BglII or BamHI and cloned into plasmid pKETW15. Plasmid pKETW15 is a pACYC-derived vector that confers chloramphenicol resistance and carries a lacIq tacOP cassette followed by a multiple cloning site. Plasmids pKEM51 and pKETW10 were constructed similarly by PCR amplification of hns or hnsKN and cloning of the PCR fragments. In these plasmids, expression is driven by tacOP, and the hns or hnsKN open reading frame is fused to the lacZ Shine-Dalgarno sequence. At the C terminus of the open reading frame, a sequence (TACCCATACGATGTTCCAGATTACGCTtaa) coding for a hemagglutinin (HA) tag followed by a stop codon (lowercase) was added in frame. All regions of plasmids that were derived from PCR fragments were sequenced. The relevant structures of the plasmids are shown schematically in the figures and in Table 1. Details of constructions and compiled sequences of the plasmids are available upon request. Media and plates were used as described previously (12). Antibiotics were added to final concentrations of 25 μg/ml (kanamycin), 50 μg/ml (ampicillin), 15 μg/ml (chloramphenicol), 50 μg/ml (spectinomycin), and 12.5 μg/ml (tetracycline) where necessary.
Determination of β-galactosidase activity.
Enzyme activities were determined for cultures grown to exponential phase (optical density at 600 nm [OD600] of 0.5) in LB medium (Difco). LB medium was used because hns mutants grow poorly in minimal medium. The cultures were inoculated from fresh overnight cultures and harvested after growth at 37°C to an OD600 of 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a concentration of 1 mM, where indicated. β-Galactosidase assays were performed as described previously (27). The enzyme activities were determined from at least three independent cultures, and standard deviations were <10%.
Western blotting and immunodetection of H-NS and StpA.
Cultures were grown in LB medium at 37°C to an OD600 of 0.5. For expression of HA-tagged H-NS, IPTG was added to a final concentration of 1 mM, and the growth of cultures was stopped on ice after 2 h of induction. For StpA quantification, chloramphenicol was added to stop translation. Samples were taken just prior to chloramphenicol addition as well as 40, 80, and 120 min after its addition. The cells were harvested by centrifugation and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (25) at a concentration of 0.1 OD600 unit per 40 μl sample buffer. Five microliters (for detection of H-NS-HA) or 2 μl (for quantification of StpA) of cell suspension was separated by SDS-PAGE (18%), using an SE600 16-cm gel electrophoresis unit (GE Healthcare). The gel was blotted onto a 0.45-μm-pore-size polyvinylidene difluoride transfer membrane, using a TE70 semidry blotting apparatus (GE Healthcare). The blot was handled using a standard Western blotting protocol (22). For H-NS detection, monoclonal rat antiserum directed against the HA tag (Roche Diagnostics) was used as the primary antibody, at a concentration of 0.2 μg/ml. Horseradish peroxidase-coupled anti-rat antibody and an enhanced chemiluminescence kit (GE Healthcare) were used for visualization according to the manufacturer's recommendations. For quantification of StpA, a monoclonal rabbit antiserum directed against StpA was used as the primary antibody, as described previously (5). Alexa fluor 680-conjugated goat anti-rabbit immunoglobulin G (IgG) (heavy plus light chains; Molecular Probes) was used as the secondary antibody at a concentration of 0.5 μg/ml. Visualization and quantification were performed using an Odyssey imaging system (Li-Cor Biosciences) according to the manufacturer's instructions.
RESULTS
Regulation of stpA by H-NS and StpA.
The stpA gene is known to be repressed by H-NS and by StpA, as shown by stpA promoter-lacZ fusions and Northern analyses (16, 37, 46). To examine the transcriptional regulation of stpA by H-NS and StpA in more detail, we constructed a series of stpA-lacZ reporter constructs (Fig. 1). In these reporter constructs, stpA fragments which encompass the stpA promoter and the stpA coding region (from positions −540 to +466 relative to the transcription start [16, 37]) (Fig. 1A and B) or the promoter only (from positions −540 to +41 and +540 to +98) (Fig. 1C and D) were fused to lacZ. In addition, the stpA open reading frame was inserted between a constitutive promoter (lacUV5) and the lacZ gene (Fig. 1E). In this lacUV5-stpA(orf)-lacZ construct, translation of stpA is prevented by mutation of the start codon from ATG to ATA [indicated by stpA(orf)]. The stpA-lacZ reporter constructs were integrated into the chromosome at the phage lambda attB attachment site, and their expression was tested in the wild type, in stpA and hns single mutants, and in stpA hns double mutants grown in LB medium to the mid-exponential growth phase (OD600 of 0.5).
FIG. 1.
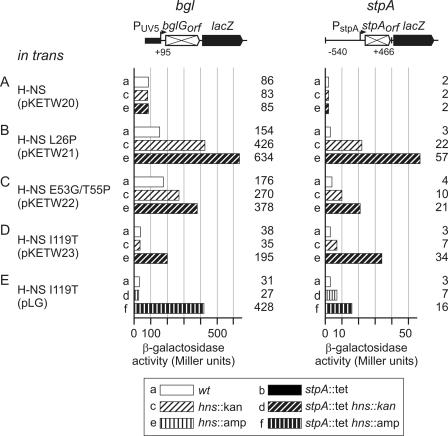
Regulation of stpA-lacZ fusions by H-NS and StpA. The expression level directed by stpA-lacZ fusions integrated into the chromosome was determined for the wild type (a) and for stpA::tet (b), hns::kan (c), hns::kan stpA::tet (d), hns::amp (e), hns::amp stpA::tet (f), and Δhns-kanKD4 (g) mutants (see legend in figure). The positions of the stpA fragments are given relative to the transcription start (16, 37). The lacZ gene carries its native ribosome binding site, and the lacUV5 promoter, which lacks lacO, encompasses positions −40 to +1 relative to the transcription start. stpAorf indicates that the stpA mRNA cannot be translated due to a mutation of the ATG start codon of stpA to ATA. β-Galactosidase activities were determined for cultures grown in LB to an OD600 of 0.5.
The stpA-lacZ reporter carrying the stpA promoter and the stpA coding region (from positions −540 to +466) fused to lacZ expressed 65 units of β-galactosidase activity in the wild type (Fig. 1A, bar a). In the hns::kan mutant, the expression level increased threefold, to 183 units (Fig. 1A, compare bars c and a). Surprisingly, the expression level directed by this construct was significantly higher (371 units) in the hns::amp mutant (Fig. 1A, compare bars e and c).
The stpA promoter-stpA-lacZ fusion encompasses the sequence from positions −540 to +466 relative to the transcription start. A fusion encompassing a stpA fragment from −300 to +466 gave similar results (data not shown). Since this stpA-lacZ fusion encodes StpA, its expression was not tested in hns stpA double mutants. To address autoregulation by StpA, we constructed a derivative in which the translation initiation codon of stpA was mutated to ATA (Fig. 1B). The expression level directed by this stpA promoter-stpA(orf)-lacZ fusion was significantly lower: in the wild type, only 3 units of β-galactosidase activity was detected (Fig. 1B, bar a). This low level of expression suggests polarity within the nontranslated stpA gene. The introduction of an stpA::tet allele into the wild-type strain had no effect, as 3 units of β-galactosidase activity was detected, as seen in the wild type (Fig. 1B, compare bars a and b). In the hns::kan mutant, expression increased ∼6-fold, to 17 units (Fig. 1B, bar c), while in the hns::amp mutant the expression level increased ∼12-fold, to 35 units (Fig. 1B, bar e). However, in the hns::kan stpA::tet and hns::amp stpA::tet double mutants, expression increased to similar values, 89 and 88 units, respectively, i.e., 30-fold higher than that of the wild type (Fig. 1B, bars d and f). These data show that H-NS is sufficient for repression of stpA and that chromosomally encoded StpA causes a threefold (in the hns::amp mutant) to sixfold (in the hns::kan mutant) repression of stpA. Maximal expression of stpA was obtained in the hns stpA double mutants only.
Difference between hns::amp and hns::kan alleles.
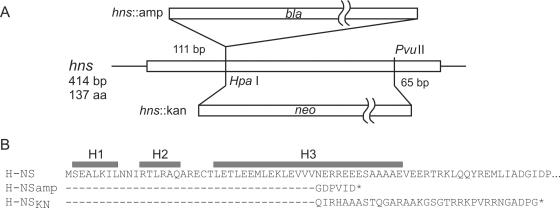
The stpA promoter-stpA-lacZ fusion (Fig. 1A) and the stpA promoter-stpA(orf)-lacZ fusion (Fig. 1B) were expressed at twofold higher levels in the hns::amp mutant than in the hns::kan mutant (Fig. 1A and B, compare bars e and c). The hns::amp allele carries an ampicillin resistance cassette inserted at the single HpaI site of H-NS (Fig. 2) (9). The hns::kan allele carries a replacement of the hns internal HpaI-PvuII fragment with a kanamycin resistance cassette (Fig. 2) (42). In both of these alleles, the 37 N-terminal codons of hns remain intact (9, 42). The N-terminal 37 amino acids (aa) of the gene product encompass the first two alpha helices and one-half of the third alpha helix of the dimerization domain (3, 15). Translation of the 5′ end of hns::amp would stop after five additional codons (Fig. 2), while translation of the 5′ end of hns::kan could result in a 70-amino-acid peptide that carries the 37 N-terminal amino acids of H-NS and 33 additional amino acids (Fig. 2). This hns::kan fusion open reading frame and its putatively encoded protein are called hnsKN and H-NSKN, respectively (also see below).
FIG. 2.
Structures and coding capacities of the hns::amp and hns::kan alleles. (A) The hns::amp allele carries an insert of an ampicillin cassette at the HpaI site (9). The hns::kan allele carries a deletion of the hns internal HpaI-PvuII fragment, which is replaced by a kanamycin cassette (42, 46). (B) Fusions of the inserts to hns were sequenced, and the sequences and coding capacities of the alleles are given. Also indicated are the extents of the three N-terminal alpha helices, which form the core of the dimerization domain (3, 15). The putative peptide encoded by the 5′ end of hns::kan is called H-NSKN.
To further analyze the difference in regulation of stpA in hns::amp and hns::kan mutant strains, we constructed a Δhns::kanpKD4 mutant, in which the hns coding region was precisely deleted and replaced by a kanamycin resistance cassette (6). The expression of the stpA promoter-stpA(orf)-lacZ fusion, which directs 3 units of β-galactosidase activity in the wild type (Fig. 1B, bar a), increased to 34 units in the Δhns::kanpKD4 mutant, i.e., to an identical level as that in the hns::amp mutant (Fig. 1B, compare bars g and e). This result demonstrates that the hns::amp allele resembles a null allele and that hns::kan is not a null allele. Thus, hns::kan mutants may express the N-terminal H-NS peptide H-NSKN, which may be active in repression of stpA in concert with StpA (addressed below).
Repression of stpA requires upstream and downstream sequences.
The level of regulation using the stpA promoter-stpA(orf)-lacZ reporter is high (30-fold). It was shown for proU (23, 31, 32) and later also for other H-NS-repressed systems, including bgl (13, 36), hilA (30), and eltAB (43), that downstream sequences are required for full repression by H-NS. To address whether downstream sequences are likewise important in the repression of stpA, we constructed two stpA promoter-lacZ fusions. One of these fusions carries the promoter up to position +98 relative to the transcription start (Fig. 1C) and is thus similar to a fusion used before (37). The second stpA promoter-lacZ fusion carries the lacZ gene fused at position +41 relative to the transcription start. This fusion does not include the stpA start codon ATG (mapping at positions +42 to +44) (Fig. 1C). The longer promoter construct directed 650 units of β-galactosidase activity in the wild type (Fig. 1C, bar a). The introduction of an stpA::tet allele again had very little effect (758 units) (Fig. 1C, bar b). In the hns::kan mutant, the activity increased to 1,950 units (Fig. 1C, bar c), and in the hns::amp mutant it increased approximately threefold, to 2,395 units (Fig. 1C, bar e). In the hns::kan stpA::tet and hns::amp stpA::tet double mutants, 4,016 and 4,330 units of β-galactosidase, respectively, were expressed (Fig. 1C, bars d and f). Thus, H-NS and StpA together repress the promoter sixfold (Fig. 1C, compare bar a to bars d and f). StpA represses the promoter twofold in the absence of H-NS (Fig. 1C, compare bars d and f to bars c and e).
The shorter (fused at position +41) stpA promoter-lacZ fusion directed an even higher level of β-galactosidase activity (2,230 units) in the wild type (Fig. 1D, bar a). The mutation of stpA again had no effect (2,340 units) (Fig. 1D, bar b). In the hns::kan and hns::amp mutants, expression increased four- and fivefold (to 8,515 and 11,040 units), respectively (Fig. 1D, bars c and e). In the hns::kan stpA::tet and hns::amp stpA::tet double mutants, the expression increased to 19,075 and 20,480 units, respectively (Fig. 1D, bars d and f). The repression of the stpA promoter (+41)-lacZ fusion by H-NS and StpA together was eightfold (Fig. 1D, compare bar a to bars d and f). Repression by StpA in the absence of H-NS was twofold (Fig. 1D, compare bars d and f to bars c and e). Interestingly, there is hardly any difference between the hns::amp and hns::kan alleles in the case of stpA promoter-lacZ fusions. In these fusions, the stpA promoter extends from positions −540 to +41 or +98. Shorter stpA promoter constructs that extend from positions −300 to +41 or +98 gave similar results (data not shown).
To analyze whether downstream sequences alone confer regulation by H-NS and/or StpA, the stpA coding region [stpA(orf)] (including positions +1 to +466 relative to the transcription start) was inserted between the constitutive lacUV5 promoter and the lacZ gene (Fig. 1E). The expression levels directed by this construct were rather low in the wild type and the stpA::tet mutant (34 units and 32 units, respectively) (Fig. 1E, bars a and b). In the hns::amp mutant, the expression level increased approximately twofold, to 53 units (Fig. 1E, bar e), while in the hns::kan mutant only the wild-type level of β-galactosidase activity (34 units) was detected (Fig. 1E, bar c). In the hns::amp stpA::tet and hns::kan stpA::tet double mutants, the expression level increased to 48 and 60 units, respectively (Fig. 1E, bars f and d). Thus, the presence of the downstream sequences alone results in a rather low, possibly significant level of repression by H-NS and StpA.
Taken together, these data show that H-NS is necessary and sufficient for maximal repression of stpA, while StpA moderately represses stpA in the absence of H-NS. The data also show that downstream sequences contribute to repression by H-NS and StpA, as repression was 30-fold when lacZ was fused downstream of stpA(orf), while it was only 6- to 8-fold in the mutants carrying stpA promoter-lacZ fusions. Furthermore, the data indicate that the stpA gene exerts a strong polar effect, since the expression levels directed by the stpA promoter-lacZ fusions were 10- to 30-fold higher than those obtained when lacZ was fused downstream of stpA. The polarity was enhanced when translation of stpA was prevented, which resulted in an additional 20-fold reduction of expression. The difference between the hns::amp and hns::kan alleles, which was more pronounced with the reporter constructs carrying the downstream regulatory element, is presumably attributable to a small fusion protein, H-NSKN, encoded by the hns::kan allele (also see below). In addition, it should be noted that the growth of the hns::amp stpA::tet double mutant was more retarded than that of the hns::kan stpA::tet double mutant.
Role of StpA in regulation of bgl.
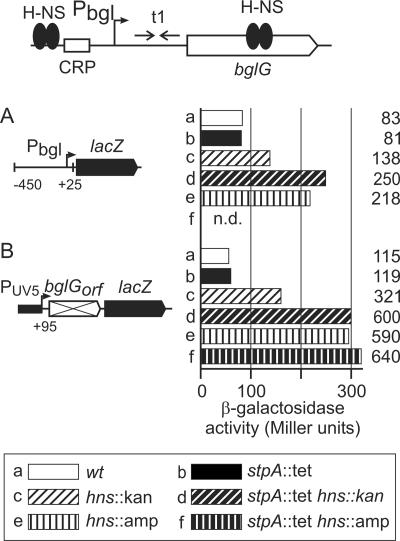
It was shown previously that StpA contributes to silencing of bgl in hns mutant backgrounds which express N-terminal fragments of the H-NS protein (17, 18). These H-NS fragments encompass amino acids 1 to 91 and 1 to 93 and include the complete dimerization and oligomerization domain (14) and a region of H-NS (aa 39 to 60) which is necessary for stabilization of StpA (3, 15, 21). It has also been shown that the requirement of these truncated H-NS mutants for complementation by StpA depends on their level of expression (17). In addition, we have shown that repression of bgl by H-NS involves two H-NS binding sites (13), as shown earlier for proU (31, 32). In bgl, one of the H-NS binding sites is located upstream of the promoter, while the second site is located approximately 600 to 700 bp downstream of the transcription start site (13) (Fig. 3, top panel).
FIG. 3.
Regulation of bgl-lacZ fusions by H-NS and StpA. At the top, a schematic view of the bgl locus is given, including the positions of H-NS binding regions upstream of the promoter, the catabolite regulator protein (CRP) binding site, and 600 to 700 bp downstream mapping within bglG. t1 indicates a transcriptional terminator in the leader of the operon, where BglG mediates antitermination depending on the availability of β-glucosides to the cell (19). The expression levels directed by chromosomal bgl-lacZ fusions specific for repression via the upstream silencer (A) or the downstream silencer (B) were determined for the wild type and for hns and stpA single and double mutants (see legend in the figure). Strains used were S1213 and derivatives (A) and S1195 and derivatives (B). n.d., not determined.
To analyze whether StpA can affect repression of bgl at both the upstream and downstream sites, we used two bgl-lacZ reporter constructs that are integrated in the chromosome at the attB site (11-13). One reporter is specific for repression via the upstream site and carries the bgl promoter fused to the lacZ gene at position +25 relative to the transcription start (Fig. 3A). A second reporter is specific for repression via the downstream site and carries a fragment downstream of bgl (from positions +95 to +971 relative to the transcription start). This downstream fragment, encompassing the nontranslatable coding region of bglG, is inserted between the constitutive lacUV5 promoter and the lacZ gene (Fig. 3B).
The bgl promoter-lacZ fusion directed 83 units of β-galactosidase activity in the wild type and 81 units in the stpA mutant (Fig. 3A, bars a and b). In the hns::kan mutant, only 138 units of activity were detected (Fig. 3A, bar c), while in the hns::amp mutant expression increased threefold (to 218 units) compared to that in the wild type (Fig. 3A, compare bars a and e), as reported previously (13). Likewise, in the hns::kan stpA::tet double mutant, expression increased threefold compared to that in the wild type (249 units) (Fig. 3A, compare bars a and d). Similar results were obtained for the bgl reporter construct specific for repression via the downstream site (Fig. 3B). Expression increased fivefold, from 115 units in the wild type to 591 units in the hns::amp mutant (Fig. 3B, compare bars a and e) (13). The expression level in the hns::kan mutant (321 units) was again lower than that in the hns::amp mutant (590 units) (Fig. 3B, compare bars e and c). The introduction of the stpA::tet allele had no effect in the wild type (Fig. 3B, compare bars a and b) or in the hns::amp mutant (Fig. 3B, compare bars f and e). However, in the hns::kan mutant, the introduction of the stpA::tet allele resulted in a twofold increase, from 321 to 600 units of β-galactosidase activity (Fig. 2B, compare bars c and d). These data show that StpA has no effect on upstream or downstream silencing of bgl in the wild type or the hns::amp mutant. In the hns::kan mutant, StpA regulates bgl approximately twofold, via both the upstream and downstream sites.
hns::kan encodes a 70-aa fusion protein, H-NSKN, comprising 37 aa of the H-NS N terminus.
The derepression level of stpA as well as of bgl was higher in the hns::amp mutant than in the hns::kan mutant. However, derepression levels were identical in the hns::amp stpA::tet and hns::kan stpA::tet double mutants (Fig. 1 and 3). This demonstrates that the hns::amp and hns::kan mutants, which are structurally different (Fig. 2), are also functionally different and that one of them may express a fragment that is functional together with StpA.
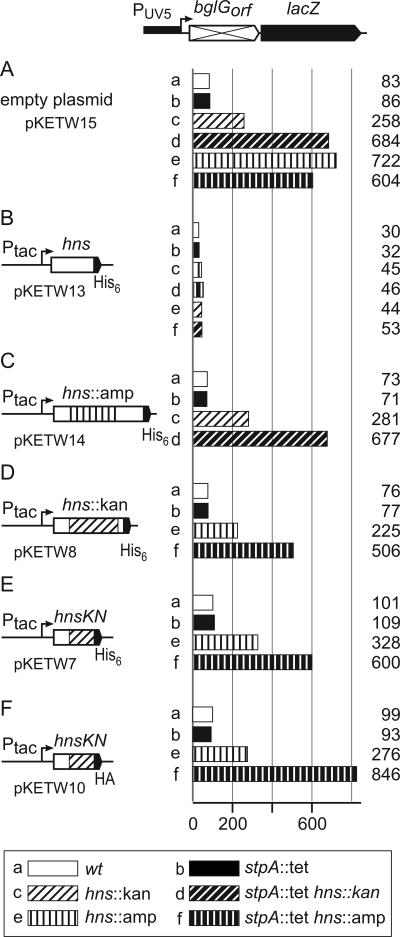
To analyze the difference between the two hns alleles, complementation analyses were performed. To this end, the wild-type hns gene, the hns::amp allele, and the hns::kan allele were cloned into pACYC-derived plasmids under the control of the tac promoter (Fig. 4). Six histidine codons were added to the 3′ end of the hns coding region in all plasmids, and all plasmids carry a lacIq gene upstream of the tac promoter. In addition, a plasmid was constructed which could express a His-tagged version of the putative H-NSKN fusion protein of 70 amino acids (Fig. 2). All of these plasmids were used for transformation of the wild type as well as of single and double hns and stpA mutants carrying the bgl-lacZ reporter specific for repression via the downstream site (Fig. 4).
FIG. 4.
Complementation shows that hns::kan is not a null allele. Strains carrying the chromosomal bgl-lacZ reporter specific for repression via the downstream site by H-NS and StpA were transformed with a control plasmid (pKETW15) (A) or with plasmids encoding wild-type hns (pKETW13) (B), hns::amp (pKETW14) (C), hns::kan (pKETW8) (D), and hnsKN (the 5′ end of hns::kan; pKETW7) (E) under the control of the tac promoter. All plasmids code for C-terminal His6-tagged H-NS variants. In addition, plasmid pKETW10, encoding an HA-tagged version of H-NSKN, (F) was used. The plasmids also carry the lacIq gene, which is present upstream of the tac promoter. Strains that carry the lacUV5 promoter-bglG(orf)-lacZ reporter include S1195 (wild type) and stpA and hns derivatives (Table 1).
Transformation of the empty vector (pKETW15) had no effect on the levels of β-galactosidase activity (compare Fig. 3B and 4A). Partial derepression was obtained in the hns::kan mutant (Fig. 4A, bar c), and complete derepression was obtained in the hns::amp single mutant (Fig. 4A, bar e) as well as in the hns::amp stpA::tet and hns::kan stpA::tet double mutants (Fig. 4A, bars d and f). In transformants carrying the plasmid (pKETW13) encoding wild-type H-NS, strong repression occurred in all strain backgrounds, with expression levels ranging from 30 to 53 units (Fig. 4B). In transformants carrying the plasmid (pKETW14) encoding the hns::amp allele, expression was not altered. Full repression occurred in the wild type and the stpA mutant (73 and 71 units, respectively) (Fig. 4C, bars a and b), partial repression occurred in the hns::kan mutant (281 units) (Fig. 4C, bar c), and complete derepression occurred in the hns::kan stpA::tet double mutant (677 units) (Fig. 4, bar d). Thus, hns::amp does not complement. However, complementation was obtained in transformants containing plasmids carrying the hns::kan allele (pKETW8) or hnsKN (pKETW7), which encodes the putative H-NSKN fusion peptide (Fig. 4D and E). As expected for the wild type and the stpA::tet mutant, full repression was measured (Fig. 4D and E, compare bars a and b). Strikingly, in the hns::amp background, complementation with hns::kan and hnsKN resulted in partial repression (225 and 328 units, respectively) (compare Fig. 4D and E, bars e, with Fig. 4A, bar e). In the hns::amp stpA::tet double mutant, hns::kan and hnsKN had no effect, as expected (compare Fig. 4D and E, bars f, with Fig. 4A, bar f). These data suggest that the hns::kan allele does indeed encode a fusion protein, H-NSKN, comprised of 37 N-terminal amino acids of H-NS and 33 additional amino acids. This fusion protein, together with StpA, causes partial repression of bgl.
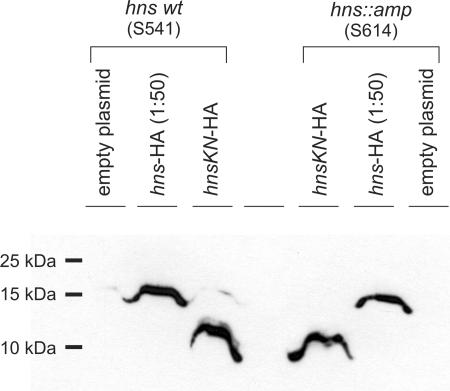
Next, the expression of the proposed N-terminal H-NSKN fusion protein was confirmed by Western blot analysis (Fig. 5). To this end, the wild type and an hns::amp mutant were transformed with a pSC101-derived plasmid encoding a C-terminally HA-tagged H-NS protein and a pACYC-derived plasmid for expression of the hnsKN allele, which codes for the H-NSKN fusion peptide with a C-terminal HA tag. In these plasmids, expression is directed by the tac promoter, and the plasmids carry the lacIq gene. The HA-tagged versions were used because a Western analysis using the pSC101-derived plasmids encoding the His-tagged H-NSKN variant, which were used in the complementation analysis, was not sensitive enough (data not shown). We confirmed that the plasmid encoding H-NSKN-HA has the same activity in complementation as the plasmid encoding the His-tagged variant of H-NSKN (Fig. 4F).
FIG. 5.
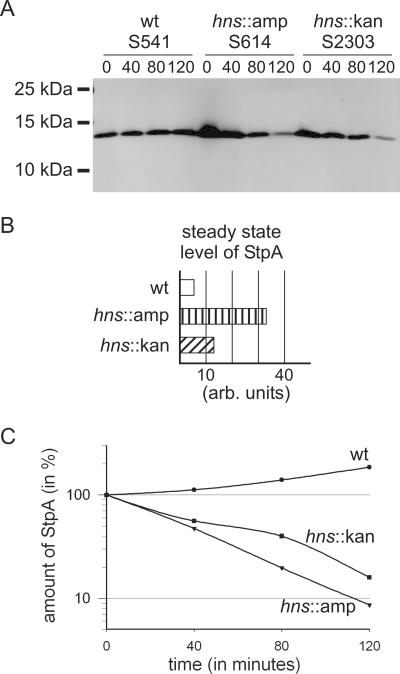
Western blot for immunodetection of the hns::kan-encoded fusion peptide H-NSKN. Transformants of strains S541 (wild type) and S614 (hns::amp) with plasmids pKEM30 (empty vector), pKEM51 (hns-HA), and pKETW10 (hnsKN-HA) were grown in LB at 37°C to an OD600 of 0.5. IPTG was added to a final concentration of 1 mM, cultures were stopped on ice after 2 h of induction, cells were harvested, and the expressed proteins were analyzed by Western blotting using an HA-specific antibody as described in Materials and Methods. To compensate for very weak expression of the H-NSKN-HA fusion peptide, a 1:50 dilution of the H-NS-HA wild-type fusion was loaded.
For Western blot analysis, the transformants were grown in LB medium with the respective antibiotics for selection of the plasmids, and expression was induced with 1 mM IPTG for 2 h prior to harvesting of the cells. Cell lysates were separated in 18% SDS-PAGE gels, blotted, and probed with HA-specific antibodies. Induction of HA-tagged hns and hnsKN revealed proteins of the expected sizes of 16.976 kDa for H-NS-HA and 9.107 kDa for H-NSKN-HA (Fig. 5). Expression of the H-NSKN-HA-tagged fusion protein was significantly lower (50 times) than that of the wild-type H-NS-HA protein, for which a 1:50 dilution was loaded. No difference was observed between the wild type and the hns::amp mutant by use of this ectopic expression system (Fig. 5). These data show that the hns::kan allele is not a null allele, and they support the conclusion that weak expression of an N-terminal H-NS 37-amino-acid fragment as part of a fusion protein is sufficient to render the expression of bgl StpA dependent.
Furthermore, we analyzed whether StpA stability is altered in the hns::kan mutant. StpA is a substrate of the protease Lon, but it is stable in wild-type cells due to the formation of heterodimers with H-NS (22). The steady-state amount and stability of StpA were determined in the wild type, in the hns::amp mutant, and in the hns::kan mutant (which expresses the N-terminal H-NSKN fusion protein). Cells were grown in LB medium to an OD600 of 0.5, and then protein synthesis was inhibited by the addition of chloramphenicol (Fig. 6) or spectinomycin (not shown). Samples were taken just prior to chloramphenicol addition and 40, 80, and 120 min after the addition of chloramphenicol. Cell lysates were separated by SDS-PAGE, blotted, and analyzed with StpA-specific antibodies that do not cross-react with H-NS (37) by quantitative Western analysis (Fig. 6A). The steady-state levels (Fig. 6B) were determined for samples taken prior to chloramphenicol addition (time zero in Fig. 6A). The steady-state level of StpA was lowest in the wild type. In the hns::amp mutant, the steady-state level was increased fourfold compared to that in the wild type, and in the hns::kan mutant it was increased twofold compared to that in the wild type (Fig. 6B). This is in agreement with the increased expression rate of stpA in the hns::amp and hns::kan mutants (Fig. 1). The stability of StpA was determined by analysis of the decrease in the amount of StpA 40, 80, and 120 min after chloramphenicol addition relative to the steady-state amount at time zero (Fig. 6A and B). In the wild type, StpA was stable, and the relative amount of StpA increased slightly after inhibition of translation, as shown before (37). In contrast, StpA was unstable in the hns::amp mutant and in the hns::kan mutant. These data show that stabilization of StpA and its functional cooperation with a fragment of the N-terminal H-NS dimerization domain can be separated.
FIG. 6.
StpA is unstable in the hns::kan mutant expressing the H-NSKN fusion protein. Wild-type E. coli and hns::amp and hns::kan mutants were grown in LB. At mid-exponential phase (OD600, 0.5), chloramphenicol was added to stop translation. Samples were taken at 0, 40, 80, and 120 min, separated by SDS-PAGE, and analyzed by Western blotting using StpA-specific antibodies. Quantification of the Western blot was done with an Odyssey imaging system (Li-Cor Biosciences). (A) Representative Western blot. (B) Steady-state amounts of StpA, determined by quantification of the signals at time zero (in arbitrary units). (C) Relative stability of StpA, as determined by signal intensities obtained at 40, 80, and 120 min relative to the intensity of the signal at time zero (normalized to 100%).
Regulation of bgl and stpA by dominant-negative H-NS mutants.
To further address the interdependence of StpA and truncated or mutated H-NS proteins in the repression of stpA and bgl, dominant-negative mutants of H-NS defective in dimerization/oligomerization or specific DNA binding were used (2, 40). Three H-NS mutants were chosen, including H-NS-L26P, H-NS-E53G/T55P, and H-NS-I119T. H-NS-L26P and H-NS-E53G/T55P carry mutations in the dimerization/oligomerization domain. These two proteins specifically bind to H-NS sites; however, they do not form extended assemblies, and no higher-order oligomers are detected in cross-links (2). In contrast, H-NS-I119T binds DNA and forms higher-order assemblies but does not specifically recognize H-NS binding sites (2).
Low-copy-number pSC101-derived plasmids encoding either wild-type H-NS or one of the H-NS mutants were used to transform the wild type, the hns mutants, and the hns stpA double mutants carrying the bgl-lacZ reporter construct specific for repression via the downstream site (Fig. 7, left column) or the stpA promoter-stpA(orf)-lacZ reporter (Fig. 7, right column). Analysis of these dominant-negative mutants in the hns::amp stpA::tet double mutant by using the pLG plasmid series pLG-HNS (wild-type H-NS), pLG-HNS-L26P, and pLG-HNS-E53G/T55P (40) turned out to be problematic due to severe growth defects and concomitant variations in the β-galactosidase assay. Therefore, we analyzed these dominant-negative H-NS mutants in the wild type, the hns::kan mutant, and the hns::kan stpA::tet double mutant by using a set of pSC101-derived plasmids encoding wild-type or mutant H-NS proteins that confer chloramphenicol resistance (Table 1). The dominant-negative mutant protein H-NS-I119T was also analyzed in the hns::amp and hns::amp stpA::tet mutants (Fig. 7E).
FIG. 7.
H-NS-I119T, a dominant-negative mutant of H-NS, specifically represses bgl together with StpA. A chromosomal bgl-lacZ fusion specific for repression of bgl by the downstream site (bgl) and a chromosomal stpA-lacZ fusion (stpA) were used to determine the dependence of dominant-negative mutants of H-NS on StpA. Strain S1195 (wild type) and its derivatives (bgl) and strain S2741 and its derivatives were transformed with pSC101-derived plasmids encoding wild-type H-NS (A), H-NS-L26P (B), HNS-E53G/T55P (C), and HNS-I119T (D and E). On these plasmids, expression of hns is directed by its own promoter. pSC101-derived plasmids pKETW20 to pKETW23 were used for analysis of hns::kan and hns::kan stpA::tet strains (A to D). The pLG-H-NS-I119T (40) plasmid was used for analysis of the hns::amp and hns::amp stpA::tet mutants (E).
In transformants carrying a plasmid encoding wild-type H-NS, the bgl and stpA reporter constructs were repressed in all strain backgrounds, as expected (Fig. 7A). When H-NS-L26P and H-NS-E53G/T55P were provided in trans, expression of bgl and the stpA reporter increased roughly 1.5-fold in the wild type, confirming the dominant-negative activity of these mutants (compare Fig. 7B and C to Fig. 7A). In the presence of H-NS-L26P, expression was rather high in the hns::kan mutant as well as in the double mutant, indicating that H-NS-L26P is inactive in repression and that StpA cannot complement this mutant (Fig. 7B). Expression of H-NS-E53G/T55P gave intermediate results, indicating that this protein has residual activity in repression of bgl and of stpA and that StpA could complement this mutant (Fig. 7C).
An unexpected result was obtained upon expression of H-NS-I119T in trans (Fig. 7D and E). This protein led to more effective repression of bgl (∼30 units) than did native H-NS (∼80 units) in the wild type and the hns::kan and hns::amp single mutants (compare the bgl results in Fig. 7A, bars a and c, with those in Fig. 7D, bars a and c, and Fig. 7E, bars a and e). Transformation of both the hns::amp stpA and hns::kan stpA double mutants with the H-NS-I119T-encoding plasmid yielded very few colonies, and cultures of these colonies grew very poorly. However, the β-galactosidase assays with these transformants were reproducible. Expression increased to 195 units in the hns::kan stpA double mutant and to 430 units in the hns::amp stpA::tet double mutant (compare the bgl results in Fig. 7D, bar e, with those in Fig. 7E, bar f). These data show that H-NS-I119T does not repress bgl in the absence of StpA. Highly effective repression by H-NS-I119T in stpA+ strains demonstrated that StpA effectively complements H-NS-I119T in repression of bgl. In contrast, the results using the stpA promoter-stpA(orf)-lacZ reporter for analysis of the dominant-negative H-NS-I119T mutant were different from those with the bgl reporter (Fig. 7D and E, compare bgl and stpA results). When H-NS-I119T was provided in trans, the expression of the stpA-lacZ reporter increased slightly (1.5-fold) in the wild type, which is in agreement with the dominant-negative activity of H-NS-I119T, as this mutant was initially identified (40). Expression increased further in the stpA mutant background with H-NS-I119T provided in trans and was highest in the stpA hns double mutants (Fig. 7D, bar d, and Fig. 7E, bar f [stpA results]). Thus, StpA effectively complements H-NS-I119T in repression of bgl but not in repression of stpA (Fig. 7D and E, compare bgl and stpA results).
DISCUSSION
We have shown that repression of the stpA gene by H-NS involves a downstream regulatory element mapping within the stpA coding region. In the presence of this element, repression of transcription, as monitored using a stpA-lacZ fusion, is 30-fold, while repression is only 6- to 8-fold in the absence of the downstream element. In addition, we found that in an hns null mutant, StpA represses its gene threefold. In contrast, StpA does not repress the bgl operon in an hns null mutant. Repression of bgl becomes StpA dependent in the presence of H-NS mutants defective in (specific) DNA binding. These data indicate a difference in the mechanisms of repression of stpA and bgl by StpA.
The specificity of regulation of stpA by H-NS is high (30-fold) in the presence of a downstream regulatory element (Fig. 1). This suggests that H-NS and StpA bind within the stpA coding region. In agreement with this, DNA structure prediction using the Bend-It web tool (http://hydra.icgeb.trieste.it/∼kristian/dna/) indicates an AT-rich planar bend within the stpA coding region centered at approximately position +130. Upstream of the promoter, a strong planar bend at position −300 relative to the transcription start was predicted. According to the present model of repression by H-NS (14), H-NS binds as a dimer to AT-rich and bent nucleation sites and then forms extended nucleoprotein complexes on the DNA. These H-NS-DNA filaments can trap RNA polymerase at a promoter by zipping the DNA strands located upstream and downstream of the promoter together (14). By consideration of this model, our data suggest that H-NS may bind upstream and downstream to the stpA promoter to form such a stable nucleoprotein complex, resulting in repression of the stpA promoter. Effective repression of other genes also requires a downstream regulatory element, as first shown for proU and later shown for other systems, including bgl, hilA, and eltAB (11, 23, 30-32, 36, 43). Results of Northern and Western analyses similarly suggest a high level of regulation of stpA by H-NS (16, 37, 46). Autorepression of stpA in Δhns or hns::amp null mutants is likewise more effective (threefold) in the presence of the stpA downstream regulatory element than in its absence (twofold). This may indicate that StpA acts similarly to H-NS. It is also possible that the RNA chaperone StpA binds to its mRNA. In addition, autorepression by StpA is enhanced in an hns::kan mutant carrying a 70-amino-acid fusion protein, H-NSKN (Fig. 4), encompassing the 37 N-terminal amino acids of H-NS, which include the first two alpha helices and half of the third alpha helix that form the core of the dimerization domain (3, 15). This H-NSKN fusion protein might stabilize the repressing nucleoprotein complex formed by StpA. These results and the finding that repression of bgl in hns::kan mutants (encoding H-NSKN) is StpA dependent demonstrate the importance of using hns null mutants in the analysis of StpA. Since in several studies hns mutants that may express H-NS fragments were used (for example, see references 20 and 46), autoregulation of stpA may be a first example of H-NS-independent regulation of transcription by chromosomally encoded StpA.
For repression of bgl, it was shown before that StpA complements truncated H-NS proteins comprising the N-terminal dimerization/oligomerization domain, including amino acids 1 to 91 (or 93) (17, 18, 21). High levels of such truncated H-NS proteins repress bgl independently of StpA (17, 29). Here we show that the H-NSKN protein encompassing the 37 N-terminal amino acids of H-NS only is sufficient to render bgl expression StpA dependent. StpA remains unstable in this background, in agreement with the result that stabilization of StpA by H-NS requires H-NS amino acids 39 to 60 (21). Furthermore, we have shown that StpA together with H-NS-I119T represses bgl more effectively than does wild-type H-NS, although H-NS-I119T is dominant negative in repression of proU (40). H-NS-I119T is also dominant negative in repression of stpA, and StpA only moderately complements H-NS-I119T in repression of stpA (Fig. 7). H-NS-I119T is defective in specific DNA binding but functional in forming extended nucleoprotein complexes (2). Taken together, these data indicate that in repression of bgl, StpA may very specifically nucleate (or target) the defective H-NS-I119T protein to the DNA, while in repression of proU or stpA the StpA protein is less specific. This difference in regulation of bgl versus stpA and proU by H-NS-I119T and StpA may further imply that growth conditions which induce stpA may allow the maintenance of strong repression of bgl, even if H-NS is less active under such conditions.
Acknowledgments
We thank Bernt Eric Uhlin, Umeå, Sweden, for a generous gift of StpA-specific antibodies and Sylvie Rimsky, France, for the pLG plasmid series encoding wild-type H-NS and dominant-negative mutants of H-NS. We gratefully acknowledge the members of the laboratory for discussion and materials, particularly S. Madhusudan, who constructed plasmid pKEM51, and V. Nagarajavel, who first detected a difference in expression of bgl in hns::kan and hns::amp mutants.
This work was funded by the Deutsche Forschungsgemeinschaft through grant Schn-371/8-1.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2005. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Badaut, C., R. Williams, V. Arluison, E. Bouffartigues, B. Robert, H. Buc, and S. Rimsky. 2002. The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 277:41657-41666. [DOI] [PubMed] [Google Scholar]
- 3.Bloch, V., Y. Yang, E. Margeat, A. Chavanieu, M. T. Auge, B. Robert, S. Arold, S. Rimsky, and M. Kochoyan. 2003. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 10:212-218. [DOI] [PubMed] [Google Scholar]
- 4.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coligan, J. E., B. M. Dunn, Hidde L.Ploegh, D. W. Speicher, and P. T. Wingfield. 2005. Current protocols in protein science. John Wiley & Sons, Inc., New York, N.Y.
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deighan, P., A. Free, and C. J. Dorman. 2000. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol. Microbiol. 38:126-139. [DOI] [PubMed] [Google Scholar]
- 8.Delihas, N., and S. Forst. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1-12. [DOI] [PubMed] [Google Scholar]
- 9.Dersch, P., K. Schmidt, and E. Bremer. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8:875-889. [DOI] [PubMed] [Google Scholar]
- 10.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration into the lambda attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 11.Dole, S., Y. Klingen, V. Nagarajavel, and K. Schnetz. 2004. The protease Lon and the RNA-binding protein Hfq reduce silencing of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 186:2708-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dole, S., S. Kühn, and K. Schnetz. 2002. Post-transcriptional enhancement of Escherichia coli bgl operon silencing by limitation of BglG-mediated antitermination at low transcription rates. Mol. Microbiol. 43:217-226. [DOI] [PubMed] [Google Scholar]
- 13.Dole, S., V. Nagarajavel, and K. Schnetz. 2004. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52:589-600. [DOI] [PubMed] [Google Scholar]
- 14.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 15.Esposito, D., A. Petrovic, R. Harris, S. Ono, J. F. Eccleston, A. Mbabaali, I. Haq, C. F. Higgins, J. C. Hinton, P. C. Driscoll, and J. E. Ladbury. 2002. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 324:841-850. [DOI] [PubMed] [Google Scholar]
- 16.Free, A., and C. J. Dorman. 1997. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 179:909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Free, A., M. E. Porter, P. Deighan, and C. J. Dorman. 2001. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 42:903-918. [DOI] [PubMed] [Google Scholar]
- 18.Free, A., R. M. Williams, and C. J. Dorman. 1998. The StpA protein functions as a molecular adapter to mediate repression of the bgl operon by truncated H-NS in Escherichia coli. J. Bacteriol. 180:994-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Görke, B. 2003. Regulation of the Escherichia coli antiterminator protein BglG by phosphorylation at multiple sites and evidence for transfer of phosphoryl groups between monomers. J. Biol. Chem. 278:46219-46229. [DOI] [PubMed] [Google Scholar]
- 20.Johansson, J., B. Dagberg, E. Richet, and B. E. Uhlin. 1998. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 180:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson, J., S. Eriksson, B. Sonden, S. N. Wai, and B. E. Uhlin. 2001. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA-stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 183:2343-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson, J., and B. E. Uhlin. 1999. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:10776-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordi, B. J. A. M., A. E. Fielder, C. M. Burns, J. C. D. Hinton, N. Dover, D. W. Ussery, and C. F. Higgins. 1997. DNA binding is not sufficient for H-NS mediated repression of proU expression. J. Biol. Chem. 272:12083-12090. [DOI] [PubMed] [Google Scholar]
- 24.Keatch, S. A., P. G. Leonard, J. E. Ladbury, and D. T. F. Dryden. 2005. StpA protein from Escherichia coli condenses supercoiled DNA in preference to linear DNA and protects it from digestion by DNase I and EcoKI. Nucleic Acids Res. 33:6540-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Madhusudan, S., A. Paukner, Y. Klingen, and K. Schnetz. 2005. Independent regulation of HNS mediated silencing of the bgl operon at two levels: upstream by BglJ and LeuO and downstream by DnaKJ. Microbiology 151:3349-3359. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Ohta, T., C. Ueguchi, and T. Mizuno. 1999. rpoS function is essential for bgl silencing caused by C-terminally truncated H-NS in Escherichia coli. J. Bacteriol. 181:6278-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olekhnovich, I. N., and R. J. Kadner. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357:373-386. [Online.] doi:org/ 10.1016/j.jmb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Overdier, D. G., and L. N. Csonka. 1992. A transcriptional silencer downstream of the promoter in the osmotically controlled proU operon of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 89:3140-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owen-Hughes, T. A., G. D. Pavitt, D. S. Santos, J. M. Sidebotham, C. S. J. Hulton, J. C. D. Hinton, and C. F. Higgins. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71:255-265. [DOI] [PubMed] [Google Scholar]
- 33.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schnetz, K. 2002. Silencing of the Escherichia coli bgl operon by RpoS requires Crl. Microbiology 148:2573-2578. [DOI] [PubMed] [Google Scholar]
- 36.Schnetz, K. 1995. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 14:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sondén, B., and B. E. Uhlin. 1996. Coordinated and differential expression of histone-like protein in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 15:4970-4980. [PMC free article] [PubMed] [Google Scholar]
- 38.Sonnenfield, J. M., C. M. Burns, C. F. Higgins, and J. C. Hinton. 2001. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie 83:243-249. [DOI] [PubMed] [Google Scholar]
- 39.Ueguchi, C., T. Suzuki, T. Yoshida, K. Tanaka, and T. Mizuno. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263:149-162. [DOI] [PubMed] [Google Scholar]
- 40.Williams, R. M., S. Rimsky, and H. Buc. 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 178:4335-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, G. G., K. Y. K. Young, G. J. Edlin, and W. Konigsberg. 1979. High-frequency generalised transduction by bacteriophage T4. Nature 280:80-82. [DOI] [PubMed] [Google Scholar]
- 42.Yamada, H., T. Yoshida, K. Tanaka, C. Sasakawa, and T. Mizuno. 1991. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 230:332-336. [DOI] [PubMed] [Google Scholar]
- 43.Yang, J., M. Tauschek, R. Strugnell, and R. M. Robins-Browne. 2005. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology 151:1199-1208. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, A., and M. Belfort. 1992. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 20:6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, A., V. Derbyshire, J. L. G. Salvo, and M. Belfort. 1995. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA 1:783-793. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, A., S. Rimsky, M. E. Reaban, H. Buc, and M. Belfort. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acids dynamics. EMBO J. 15:1340-1349. [PMC free article] [PubMed] [Google Scholar]