Abstract
The dimorphic prosthecate bacteria (DPB) are α-proteobacteria that reproduce in an asymmetric manner rather than by binary fission and are of interest as simple models of development. Prior to this work, the only member of this group for which genome sequence was available was the model freshwater organism Caulobacter crescentus. Here we describe the genome sequence of Hyphomonas neptunium, a marine member of the DPB that differs from C. crescentus in that H. neptunium uses its stalk as a reproductive structure. Genome analysis indicates that this organism shares more genes with C. crescentus than it does with Silicibacter pomeroyi (a closer relative according to 16S rRNA phylogeny), that it relies upon a heterotrophic strategy utilizing a wide range of substrates, that its cell cycle is likely to be regulated in a similar manner to that of C. crescentus, and that the outer membrane complements of H. neptunium and C. crescentus are remarkably similar. H. neptunium swarmer cells are highly motile via a single polar flagellum. With the exception of cheY and cheR, genes required for chemotaxis were absent in the H. neptunium genome. Consistent with this observation, H. neptunium swarmer cells did not respond to any chemotactic stimuli that were tested, which suggests that H. neptunium motility is a random dispersal mechanism for swarmer cells rather than a stimulus-controlled navigation system for locating specific environments. In addition to providing insights into bacterial development, the H. neptunium genome will provide an important resource for the study of other interesting biological processes including chromosome segregation, polar growth, and cell aging.
Unlike most bacteria, which reproduce by symmetric binary fission, dimorphic prosthecate bacteria (DPB) reproduce by asymmetric binary fission (e.g., Caulobacter crescentus) or budding (e.g., Hyphomonas and Hyphomicrobium species) to produce a motile swarmer cell from a nonmotile mother cell (68). The mother cell is distinguished by a presence of an appendage termed a prostheca or stalk (84), as well as generally having a holdfast that allows the cell to adhere to a surface (36). The swarmer cells, which are unable to reproduce, undergo a developmental process that results in their conversion to mother cells.
The life cycle of DPB is analogous in many ways to that of dimorphic invertebrates, and this analogy is further supported by studies that have shown that the motile offspring of DPB, as in the multicellular case, are in a “juvenile” condition in which most energy is expended on motility and little on growth (26, 67). These facts suggest that DPB are good model systems for understanding the evolution and biology of dimorphic life in general.
DPB are ubiquitous in both freshwater and marine environments but are found also to a lesser degree in soil (68). They are of considerable environmental interest since many Hyphomicrobium species can mineralize pollutants such as aromatic hydrocarbons (54), methyl chloride (34), and various alcohols, including methanol (40). In addition, various Hyphomonas species are primary colonizers of marine surfaces (6) and form biofilms necessary for the recruitment of invertebrate larvae such as those of oysters (15). DPB are members of the α-proteobacteria but currently are not considered a coherent taxonomic unit (68), and Hyphomonas and Caulobacter are even classified as members of different orders.
One member of the DPB, C. crescentus CB15, has been the subject of a genome sequencing study (62). The genus Hyphomonas was selected for sequencing as a second member of the DPB, not only for comparative purposes but also because, unlike C. crescentus, the stalk in Hyphomonas is a reproductive structure. The developing bud receives proteins, DNA, and other cellular components from the mother cell through the stalk (36). Electron micrographs suggest that these components are transported through the stalk within membranous swellings termed pseudovesicles (98). Another significant distinction in reproduction between C. crescentus and budding, prosthecate bacteria is that the reproductive ability of the latter is quite limited, with typically only about eight offspring formed during the life of the mother cell (55), while C. crescentus mother cells typically create 100 or more offspring over their lifetimes (1). This suggests that Hyphomonas is a good model system for the study of senescence.
The species of Hyphomonas, Hyphomonas neptunium, was selected for a variety of reasons. It has a faster swarmer cycle (time for new swarmer cells to mature into reproductive cells and initiate bud formation) than other Hyphomonas species (55), and its prostheca are easily distinguishable by light microscopy (12). In addition, the use of the stalk as a conduit for transfer of macromolecules from the mother cell to the developing bud in Hyphomonas in general (98) was originally discovered in H. neptunium (92). This is the first report of a whole genome sequence of a member of the family Hyphomonadaceae (47), a group of bacteria that is believed to include primary colonizers of surfaces in the ocean (6).
MATERIALS AND METHODS
Genome sequencing and assembly.
The genome of H. neptunium ATCC 15444T (the type strain of the species) was sequenced by means of the whole genome shotgun method as previously described (25). A total of 41,941 usable sequence reads were generated, of which 41,461 were incorporated into the initial assembly, yielding an average of 8.1-fold coverage across the genome. Further details of sequencing and closure are available in document 1 posted at http://www.hyphomonas.com/hnep_supp.html.
Sequence annotation.
The prediction of putative-protein coding genes and functional annotation were performed as previously described (88). The program GLIMMER (20) was used to identify the initial set of putative coding regions; from this initial list candidates consisting of fewer than 30 codons and those containing overlaps were eliminated. Frameshifts and point mutations were detected and corrected where appropriate. Additional phylogenetic analyses were performed using APIS (for automated phylogenetic inference system) (unpublished). APIS automates the process of sequence similarity, alignment, and phylogenetic inference for each protein in a genome (see below).
APIS.
APIS is a system for automatic creation and summarizing of phylogenetic trees for each protein encoded by a genome. It is implemented as a series of Ruby scripts, and the results are viewable on an internal web server which allows the user to explore the data and results in an interactive manner. The homologs used by APIS for each phylogenetic tree are obtained by comparing each query protein against a curated database of proteins from complete genomes using WU-BLAST (30). The full-length sequences of these homologs are then retrieved from the database and aligned using MUSCLE (24), and bootstrapped neighbor-joining trees are produced using QuickTree (37). As QuickTree (unlike most programs) produces bootstrapped trees with meaningful branch lengths, the inferred tree is then midpoint rooted prior to analysis, allowing automatic determination of the taxonomic classification of the organisms with proteins in the same clade as the query protein. APIS was created to address some of the weaknesses of existing automated phylogenetic systems such as PyPhy (79), in which a general-purpose protein database such as Swiss Prot (78) is used, weakening the interpretation of clades because the absence of proteins from organisms which have not had their genomes completely sequenced cannot be taken as biological evidence of the nonexistence of such proteins.
Assertions of orthology.
Putative orthologs between genomes were established by the following method. First, two BLASTP (30) analyses were run; then all proteins encoded by the first genome were compared against a database of proteins encoded by the second, and vice versa. The threshold used in these comparisons was 10−9. Orthology was asserted if two proteins were each other's best BLASTP hit (best reciprocal match).
Identification of outer membrane proteins.
A set of curated outer membrane proteins with experimental evidence were retrieved from the membrane transport protein classification database (http://www.tcdb.org) (77). The representative hidden Markov models (HMMs) for each individual family were retrieved from the Pfam protein families database (7).
The complete predicted protein sequences of H. neptunium were first searched against this outer membrane protein database (75) for similarity to known outer membrane proteins using BLASTP and for matches to the HMMs using the program hmmsearch. All of the query proteins with significant hits (cutoff of 10−3) were collected and searched against the NCBI nonredundant protein database. A web-based interface that incorporates the number of hits to the outer membrane protein database, BLASTP and HMM search E-value and score, and the description of top hits to the nonredundant protein database was implemented to facilitate the annotation processes. Up-to-date results can be viewed at the TransportDB website (http://www.membranetransport.org/) (75, 76), and the initial analysis used in the annotation of the genome is available in Table 1 posted at http://www.hyphomonas.com/hnep_supp.html.
TABLE 1.
General features of the genome of H. neptunium ATCC 15444T
| Parametera | Value |
|---|---|
| Size (bp) | 3,705,021 |
| % G+C content | 62 |
| Predicted no. of protein CDS | 3,521 |
| Avg CDS size (bp) | 953 |
| No. of unconserved hypothetical proteins | 400 (11%) |
| No. of conserved hypothetical proteins | 506 (14%) |
| % Coding of genome | 91 |
| No. of rRNA operons | 1 |
| No. of tRNAs | 43 |
CDS, coding sequences.
Chemotaxis assays.
Chemotaxis assays were performed using motile cells from an overnight culture of H. neptunium in a modified TY (2.5 g of Bacto-tryptone plus, 1.5 g of yeast extract per liter [pH 6.9], supplemented with 8 mM calcium chloride) medium washed three times in chemotaxis buffer (10 mM phosphate buffer, pH 7.0, 8 mM calcium chloride). About 50% of the cells were motile under these conditions. The swarm plate assay was performed on cells inoculated in the center of modified TY medium, LB medium, or marine broth (Gibco) solidified with 0.3% agar, an agar concentration that was found to be optimal for bacterial growth and motility under the conditions of the experiments. The chemical-in-plug assays and the temporal gradient assays were performed essentially as described previously (2, 3, 89). The following chemicals were tested in a chemical-in-plug assay and a temporal gradient assay at concentrations of 0.1 mM, 1 mM, and 10 mM: glutamate, aspartate, oxaloacetate, succinate, pyruvate, and malate. The substituted quinone, 1,4-benzoquinone, was tested as a potential repellent at concentrations of 0.1 mM, 1 mM, and 10 mM. Oxygen was tested in a temporal gradient assay as described previously (99).
Nucleotide sequence accession number.
The annotated genome sequence of H. neptunium ATCC 15444T has been deposited in the GenBank database as accession number CP000158.
RESULTS AND DISCUSSION
General characteristics.
The genome of H. neptunium ATCC 15444T is composed of a single circular chromosome. General features of the genome can be found in Table 1.
Repetitive and mobile DNA.
The H. neptunium genome was analyzed for the existence of repetitive elements. The results are summarized in Table 2. Besides repeats due to the highly conserved duplicated genes EF-Tu and DUF227, the H. neptunium genome contains transposons related to two families found in C. crescentus, ISCc2 and ISCc3. In addition, the genome contains six copies of a 370-bp intergenic repeat that may have some regulatory function. It is perhaps significant that four of these copies flank the presumed origin of replication at an approximate distance of ±100 and ±200 kbp.
TABLE 2.
Repetitive elements in the H. neptunium genomea
| Class | Avg size (bp) | No. of copies | Functionb |
|---|---|---|---|
| 1 | 1,265 | 4 | ISCc3 transposase |
| 2 | 1,019 | 8 | ISCc2 transposase |
| 3 | 608 | 2 | EF-Tu C terminus |
| 4 | 542 | 2 | EF-Tu N terminus |
| 5 | 370 | 6 | Conserved intergenic region |
| 6 | 647 | 2 | DUF227 C terminus |
| 7 | 320 | 2 | DUF227 N terminus |
Repeats listed are those over 200 bp in length as found by REPuter (42).
DUF227 is a family of proteins containing a conserved domain of unknown function.
Metabolism.
Cultured Hyphomonas species preferentially use amino acids as their carbon and energy sources (56). Consistent with this, H. neptunium ATCC 15444T did not use glucose in semisynthetic growth medium (93, 91) but was capable of using pyruvate, α-ketoglutarate, fumarate, or malate as carbon sources (22).
Despite these experimental results, it appears that the H. neptunium genome possesses all of the genes needed for both the glycolytic conversion of glucose to pyruvate and pentose phosphate biosynthetic pathways. This suggests that, while H. neptunium may not have used glucose under the conditions tested, it does have the ability to utilize glucose under some conditions. Since H. neptunium possesses genes encoding enzymes for a complete tricarboxylic acid cycle along with enzymes for the glyoxylate shunt, the ability of H. neptunium to utilize organic acids (22) is supported by genome analysis. Additionally, the genome analysis suggests that glycerol can serve as a carbon source for H. neptunium by feeding the end product of glycerol dehydrogenase into the glycolytic pathway. As expected, genes for the complete degradative pathways of the 20 standard amino acids are also present.
Aromatic and halogenated compounds also appear to be metabolized by H. neptunium since the genome contains genes that are predicted to encode enzymes involved in the degradation of various such substrates. While identifying specific substrates will require experimental evidence, it is clear that proteins encoded by certain genes (e.g., HNE_0817, HNE_0958, HNE_0987, HNE_1322, HNE_1435, HNE_1602, HNE_1663, HNE_2413, HNE_2751, and HNE_3259) are involved. It is understandable that the ability to utilize aromatic and halogenated compounds would confer adaptive advantage upon H. neptunium as it was isolated from Barcelona harbor (51), which is contaminated with such compounds (60).
The results from the Genome Properties system of The Institute for Genomic Research (33) are posted at http://www.hyphomonas.com/hnep_supp.html as Table 2. The Genome Properties system consists of a series of properties (including evidence for many metabolic pathways) that can be expressed in numerical or controlled vocabulary terms, thus easing the comparison of properties across different genomes.
Comparison with C. crescentus and Silicibacter pomeroyi.
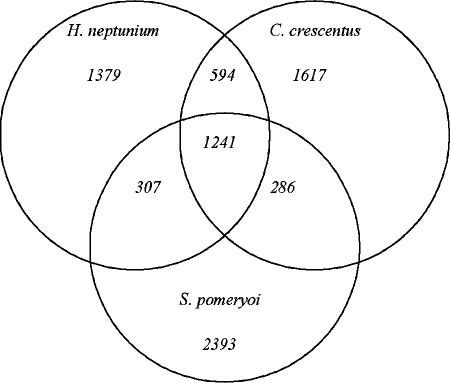
The predicted proteins of the H. neptunium genome were compared against those of C. crescentus as well as those of the only other completely sequenced member of the Rhodobacterales, S. pomeroyi (57), which is not a DPB. A Venn diagram summarizing the comparison is presented as Fig. 1. As shown, while each of the three genomes encodes numerous proteins without apparent orthologs in the other two, a core of 1,241 proteins is shared by all. As expected, this core consists primarily of proteins involved in essential processes such as transcription, translation, and basic metabolism. H. neptunium and C. crescentus share 594 proteins to the exclusion of S. pomeroyi, and many of these proteins may be related to the lifestyle of DPB (discussed in detail in subsequent sections). S. pomeroyi and H. neptunium share only 307 proteins to the exclusion of C. crescentus, among which are many flagellar proteins, transporters, and permeases.
FIG. 1.
Venn diagram showing the shared gene content between H. neptunium, C. crescentus, and S. pomeroyi. Orthology was assumed using the best reciprocal BLASTP matches, with a P value cutoff of 10−9.
There is a striking similarity between the outer membrane protein complements of C. crescentus and H. neptunium that is not shared by S. pomeroyi, namely the proliferation of predicted TonB-dependent receptors, which catalyze energy-dependent transport across the outer membrane. The C. crescentus genome was originally predicted to encode 65 TonB-dependent receptors, the highest number yet found in sequenced bacterial genomes. Subsequent studies have revealed equally high, or greater, numbers (25 to 115) in the genomes of Bacteroides species, pseudomonads, and plant-pathogenic xanthomonads. The genome of H. neptunium is predicted to encode 43 TonB-dependent receptors, in marked contrast to that of S. pomeroyi, which lacks them completely. Similarly, the genomes of both H. neptunium and C. crescentus encode predicted vitamin B12 receptors that feed directly into an ABC-type iron compound importer, while S. pomeroyi appears to lack such a construct. Proliferation of TonB-dependent receptors suggests that both C. crescentus and H. neptunium take up macromolecules (e.g., iron-siderophore complexes and vitamin B12) that are too large to be obtained via passive diffusion through the outer membrane porins. Nine of the 43 H. neptunium receptors were predicted to interact with siderophores on the basis of HMM analysis, while three appeared to bind vitamin B12; the substrate of the remainder could not be predicted with a reasonable degree of confidence. One C. crescentus TonB-dependent receptor has recently been shown to be involved in the uptake of maltodextrins (61), suggesting that these receptors in H. neptunium may similarly, actively take up nutrients in a nutrient-poor environment.
Approximately the same fraction of TonB-dependent receptors was predicted to bind siderophores in both H. neptunium and C. crescentus. Neither organism appears to encode proteins for siderophore production, suggesting that both organisms may exploit exogenous siderophores, as has been suggested for Nitrosomas europaea. Like N. europaea, both H. neptunium and C. crescentus (but not S. pomeroyi) also seem to possess a tandem pair of fecIR, genes involved in ferric citrate transport mediated by transmembrane signaling. Thus, H. neptunium and C. crescentus (and N. europaea) appear to employ similar iron-scavenging strategies in marine and freshwater environments, respectively. The lack of such strategies in S. pomeroyi may indicate that this organism is less likely to encounter iron limitation in its native environment. Coastal salt marshes, such as the marsh from which S. pomeroyi was isolated (32), have been shown to have significantly higher iron concentrations in vegetated soils than nearby unvegetated areas (23). It is possible that S. pomeroyi encounters available iron from such an iron-rich source.
To explore further the genes shared between C. crescentus, H. neptunium, and S. pomeroyi, a phylogenetic profiling (66) analysis was performed. Phylogenetic profiling is a method in which the presence or absence of presumed orthologs to a protein is examined across many complete genomes. Proteins that are only present in the genomes of organisms sharing a particular characteristic are good candidates for being involved in that feature. Consistent with the orthology study above, H. neptunium shared the highest number of proteins, 62, with C. crescentus to the exclusion of other organisms. Also consistent with the orthology study was that the second-highest number, 10, was between H. neptunium and S. pomeroyi. As expected, functions have not been ascribed to most of the 62 proteins uniquely shared between H. neptunium and C. crescentus (Table 3). This list of proteins, however, includes numerous transcriptional regulators that may be involved in the cell cycle or stalk biogenesis and several putative lipoproteins that may be involved in stalk formation or oligotrophy. Of the 10 proteins shared with S. pomeroyi (Table 4), four are flagellar proteins. Functional homologs of these flagellar proteins occur in many other bacteria, but the sequences of the S. pomeroyi and H. neptunium proteins are sufficiently divergent that they scored below the cutoff for orthology with flagellar proteins from other bacteria, suggesting that the S. pomeroyi and H. neptunium flagellar genes were derived from a common ancestor.
TABLE 3.
The 62 predicted proteins in H. neptunium sharing a putative ortholog only with C. crescentus, according to phylogenetic profiling, together with their putative orthologs
| H. neptunium protein |
C. crescentus protein
|
||
|---|---|---|---|
| ORF | Description | ORF | Description |
| HNE_0127 | Conserved hypothetical protein | CC0136 | Hypothetical protein |
| HNE_0203 | Conserved hypothetical protein | CC3602 | Hypothetical protein |
| HNE_0204 | OstA family protein | CC3601 | Hypothetical protein |
| HNE_0317 | Putative chorismate mutase, type II | CC1116 | Chorismate mutase, putative |
| HNE_0320 | Hypothetical protein | CC1485 | Hypothetical protein |
| HNE_0394 | Conserved hypothetical protein | CC0224 | Hypothetical protein |
| HNE_0430 | Ribosomal protein L35 | CC1046 | Ribosomal protein L35 |
| HNE_0439 | Phosphocarrier protein HPr | CC0241 | phosphocarrier protein HPr |
| HNE_0477 | Conserved hypothetical protein | CC2977 | Transcriptional regulator, TetR family |
| HNE_0479 | Putative membrane protein | CC1840 | RNase BN, putative |
| HNE_0570 | Transcriptional regulator, TetR family | CC2493 | Transcriptional regulator, TetR family |
| HNE_0573 | Thioesterase family protein | CC3309 | Hypothetical protein |
| HNE_0634 | Conserved hypothetical protein | CC3551 | Hypothetical protein |
| HNE_0654 | Conserved hypothetical protein | CC0356 | Hypothetical protein |
| HNE_0679 | Putative lipoprotein | CC1285 | Hypothetical protein |
| HNE_0697 | Conserved domain protein | CC1987 | Hypothetical protein |
| HNE_0728 | Conserved hypothetical protein | CC0948 | Hypothetical protein |
| HNE_0830 | Conserved hypothetical protein | CC0922 | Medium-chain fatty acid, CoA ligase |
| HNE_0851 | Putative lipoprotein | CC0699 | Hypothetical protein |
| HNE_0983 | Putative membrane protein | CC0636 | Hypothetical protein |
| HNE_1015 | Conserved hypothetical protein | CC2228 | Hypothetical protein |
| HNE_1037 | Tetratricopeptide repeat protein | CC2031 | TPR domain protein |
| HNE_1131 | Conserved domain protein | CC1589 | Hypothetical protein |
| HNE_1193 | Conserved hypothetical protein | CC0163 | Hypothetical protein |
| HNE_1388 | Conserved hypothetical protein | CC0495 | Transcriptional regulator, TetR family, putative |
| HNE_1417 | Putative lipoprotein | CC2104 | Hypothetical protein |
| HNE_1569 | Transcriptional regulator, TetR family | CC2493 | Transcriptional regulator, TetR family |
| HNE_1578 | Conserved hypothetical protein | CC1167 | Hypothetical protein |
| HNE_1665 | Conserved hypothetical protein | CC1177 | Hypothetical protein |
| HNE_1732 | Hypothetical protein | CC2341 | Hypothetical protein |
| HNE_1746 | Conserved hypothetical protein | CC1951 | Hypothetical protein |
| HNE_1840 | Exodeoxyribonuclease VII, small subunit | CC2070 | Exodeoxyribonuclease small subunit |
| HNE_1984 | Conserved hypothetical protein | CC1842 | Response regulator, putative |
| HNE_2049 | Sporulation related repeat protein | CC2007 | Hypothetical protein |
| HNE_2081 | Phosphoglycerate mutase family protein | CC1966 | Hypothetical protein |
| HNE_2096 | Conserved hypothetical protein | CC1308 | Hypothetical protein |
| HNE_2106 | Toluene tolerance protein, Ttg2 family | CC3693 | Hypothetical protein |
| HNE_2138 | Toluene tolerance protein, Ttg2 family | CC3693 | Hypothetical protein |
| HNE_2240 | Conserved hypothetical protein | CC2430 | Hypothetical protein |
| HNE_2241 | Putative chain length determinant protein | CC2431 | Hypothetical protein |
| HNE_2337 | Conserved domain protein | CC3555 | Hypothetical protein |
| HNE_2339 | Sensor histidine kinase | CC3474 | Sensor histidine kinase, putative |
| HNE_2440 | Transcriptional regulator, TetR family | CC1345 | Transcriptional regulator, TetR family |
| HNE_2607 | Transcriptional regulator, TetR family | CC1855 | Transcriptional regulator, TetR family |
| HNE_2647 | Antibiotic biosynthesis monooxygenase domain protein | CC2132 | Hypothetical protein |
| HNE_2661 | Peptidase, S54 (rhomboid) family | CC2627 | Rhomboid family protein |
| HNE_2737 | Conserved hypothetical protein | CC1453 | Hypothetical protein |
| HNE_2875 | Conserved hypothetical protein | CC2822 | Hypothetical protein |
| HNE_2914 | Conserved hypothetical protein | CC2480 | Hypothetical protein |
| HNE_2952 | RusA family protein | CC2107 | Hypothetical protein |
| HNE_2965 | Transcriptional regulator, TetR family | CC1664 | Transcriptional regulator, TetR family |
| HNE_2977 | Transcriptional regulator, TetR family | CC0772 | Transcriptional regulator, TetR family |
| HNE_3033 | Putative MraZ protein | CC2563 | Hypothetical protein |
| HNE_3111 | Putative lipoprotein | CC0125 | Hypothetical protein |
| HNE_3125 | Conserved hypothetical protein | CC3104 | Hypothetical protein |
| HNE_3206 | Conserved hypothetical protein | CC3441 | Arylesterase-related protein |
| HNE_3320 | Conserved hypothetical protein | CC0155 | Hypothetical protein |
| HNE_3401 | Conserved hypothetical protein | CC3115 | Hypothetical protein |
| HNE_3416 | S4 domain protein | CC0656 | S4 domain protein |
| HNE_3520 | Transcriptional regulator, MarR family | CC3677 | Transcriptional regulator, MarR family |
| HNE_3553 | Thiamine monophosphate synthase family protein | CC3746 | Hypothetical protein |
| HNE_3556 | Putative lipoprotein | CC3748 | Hypothetical protein |
TABLE 4.
The 10 predicted proteins in H. neptunium sharing a putative ortholog only with S. pomeroyi, according to phylogenetic profiling, together with their putative orthologs
| H. neptunium protein |
S. pomeroyi protein
|
||
|---|---|---|---|
| ORF | Description | ORF | Description |
| HNE_1542 | Conserved hypothetical protein | SPO0172 | Hypothetical protein |
| HNE_1330 | Conserved hypothetical protein | SPO3675 | Hypothetical protein |
| HNE_0593 | Radical SAM domain protein | SPO2049 | Radical SAM domain protein |
| HNE_0272 | Flagellar biosynthetic protein fliQ | SPO0179 | Flagellar biosynthetic protein FliQ |
| HNE_0269 | Putative flagellar protein | SPO0182 | Flagellar basal-body rod protein; FlgB |
| HNE_0266 | Putative flagellum biosynthesis repressor protein FlbT | SPO2036 | Sensory box sensor histidine kinase/response regulator |
| HNE_0259 | Hypothetical protein | SPO0174 | Hypothetical protein |
| HNE_0248 | Putative flagellar hook-associated protein FlgL | SPO0194 | Flagellar hook-associated protein FlgL family protein |
| HNE_3425 | Glyoxalase family protein | SPO0313 | Glyoxalase family protein |
| HNE_3078 | Conserved hypothetical protein | SPO3859 | Hypothetical protein |
Selenoproteins represent a unique feature of H. neptunium in comparison to C. crescentus and S. pomeroyi. H. neptunium encodes an l-seryl-tRNA selenium transferase (HNE_2489), a selenocysteine-specific translation elongation factor (HNE_2488), and an apparent selenoprotein of unknown function (HNE_2485).
Surface colonization.
Swarmer cells of both Caulobacter and Hyphomonas are dedicated to finding a suitable surface for colonization prior to their morphogenesis into reproductive cells. Having oligotrophic capability, these prosthecates may serve important roles as primary colonizers by initiating biofilm development (15). Caulobacter and some species of Hyphomonas synthesize a combination of retractable pili and polysaccharide holdfasts (normally polar capsules), which function in surface attachment and colonization (13), while other species of Hyphomonas use two polysaccharide structures for these functions (45). For example Hyphomonas adhaerens synthesizes both an adhesive holdfast (71) and pili (70), whereas Hyphomonas rosenbergii synthesizes a polysaccharide holdfast and a capsule that surrounds the cell (44). Interestingly, the holdfasts of 15/16 marine species and 6/10 freshwater species of Caulobacter contain GlucNac (N-acetylglucosamine) (53), as do all Hyphomonas species that have been examined to date (71). The integrity of the GlcNac is critical for the elastic properties of the holdfast (50). Thus, the presence of GlucNac is suggested to be characteristic of adhesive polysaccharides (44). It is not clear yet whether this is an example of converging or diverging evolution.
Genomic analysis suggests that H. neptunium synthesizes pili and polysaccharides. There are at least 13 open reading frames (ORFs) coding for pili biosynthesis (aside from those annotated as being involved in type IV fimbrae syntheses) and a number of ORFs potentially involved in surface polysaccharide biosynthesis. Interestingly, one of these ORFs, (HNE_3005), codes for a putative O-linked GlcNac transferase that may be involved in biosynthesis of the GlucNac residues in the holdfast. However, despite the fact that H. neptunium mother cells have holdfasts, many of the known proteins for holdfast synthesis and attachment in C. crescentus appear not to have orthologs encoded by the H. neptunium genome. For example, none of the holdfast attachment proteins encoded by the C. crescentus hfaABD gene cluster (17, 41) have apparent orthologs encoded in the H. neptunium genome, and of the proteins encoded by the hfsDAB gene cluster for holdfast synthesis in C. crescentus (82), only HfsB seems to have an ortholog in H. neptunium (HNE_2240). Therefore, it appears that holdfast synthesis and attachment in H. neptunium must be performed differently than in C. crescentus.
Motility.
The H. neptunium swarmer cell has a single, polar flagellum, which it sheds prior to differentiation into a mature mother cell (90, 92). The developing bud can elaborate a flagellum while still attached to the mother cell as cells undergoing budding are often motile, suggesting that expression of the H. neptunium flagellar genes is temporally and spatially regulated as it is in C. crescentus. Most of the genes encoding structural proteins of the flagellum and components required for flagellar assembly and function are found in a region consisting of 35 contiguous ORFs (HNE_0241 to HNE_0275) that are arranged in at least 8 operons (see Table 3 and Fig. 1 posted at http://www.hyphomonas.com/hnep_supp.html). This island of motility is similar to that in the S. pomeroyi DSS-3T genome but is in contrast to the organization of flagellar genes in C. crescentus, where most of these genes are distributed in 13 major clusters that are scattered throughout the genome to r-scan statistics (21, 39) (see Fig. 2 posted at http://www.hyphomonas.com/hnep_supp.html). Interestingly, for predicted products of most of the flagellar genes within the H. neptunium island of motility, the closest BLAST matches are with S. pomeroyi DSS-3T homologs (24 of 31) (see Fig. and Table 3 posted at http://www.hyphomonas.com/hnep_supp.html), suggesting a common ancestry for these flagellar genes. Genes outside the major H. neptunium flagellar gene cluster that have predicted roles in motility include a cheY homolog (HNE_0639), a cheR homolog (HNE_0640), and two potential flagellar regulatory genes, flhF (HNE_0942) and motR (HNE_0943; also referred to as flhG or fleN). The flhF and motR genes are not found in enteric bacteria, but they do occur in a number of flagellated bacteria, including Pseudomonas species, where they have been implicated in the polar placement of flagella or the regulation of flagellar number (18, 65).
Where it has been examined in other bacteria, a transcriptional hierarchy controls the expression of flagellar genes (52, 97). Initiation of the flagellar gene hierarchy in C. crescentus requires the response regulator CtrA (72, 74). H. neptunium ctrA (HNE_0944) appears to be in an operon with flhF and motR (see Fig. 1 posted at http://www.hyphomonas.com/hnep_supp.html), suggesting a role for ctrA in flagellar gene regulation. The activity of CtrA in C. crescentus is controlled in response to cell cycle and developmental cues by a multicomponent signal transduction network consisting of the sensor kinases CckA, PleC, DivJ, and DivL and the response regulator DivK (5). H. neptunium possesses homologs of cckA (HNE_0507), divJ (HNE_0746), and divL (HNE_0399), indicating that the earliest events in the regulation of flagellar biogenesis in H. neptunium may be similar to those in C. crescentus. Transcription of many of the flagellar genes in C. crescentus requires σ54-RNA polymerase holoenzyme in conjunction with the σ54-dependent activator FlbD and FliX, a protein that functions to both inhibit and stimulate FlbD activity (8, 14, 58, 59, 73, 95, 96). H. neptunium possesses an rpoN homolog (encodes σ54; HNE_0206) but not homologs of flbD or fliX. Inspection of sequences upstream of the flagellar genes failed to identify any good matches for potential σ54-dependent promoters [the consensus sequence is TGGCAC-N4-TTTGC(A/T)]. Taken together, these observations suggest that σ54 is not involved in H. neptunium flagellar biogenesis. Proteobacteria often employ σ28 for the expression of specific flagellar genes (43, 49, 69, 85), but H. neptunium lacks this alternative σ factor. Thus, the transcriptional control of H. neptunium flagellar genes needed in later stages of flagellar biogenesis appears to involve a mechanism that has not been described in other bacteria.
The ability of flagellated motile bacteria to seek optimum positions in gradients by chemotaxis is normally dependent on the presence of methyl-accepting chemotaxis proteins (MCPs) and a conserved set of chemotaxis genes. Bacterial chemotaxis occurs by modulation of the probability of locomotor-directional changes, brought about by reversals of the direction of flagellar rotation or modulation of flagellar rotary speed (9). CheY, which is a response regulator of a conventional two-component system, controls the probability of directional changes, and the activity of CheY is modulated by the MCPs and conserved cytoplasmic chemosensory proteins (4). An unexpected finding from the H. neptunium genome was the absence of homologs of MCPs and most of the other chemotactic proteins, including CheA, CheW, and CheB. The absence of conserved chemosensory proteins together with the presence of genes required for the biogenesis of functional flagella was also reported for Aquifex aeolicus (19), S. pomeroyi (57), and Pirellula sp. strain 1 (31), now reclassified as Rhodopirellula baltica, a member of the Planctomycetes that, like H. neptunium and C. crescentus, has a dimorphic cell cycle. In contrast to H. neptunium, C. crescentus possesses ORFs for 18 MCPs and two complete sets of chemotaxis signal transduction proteins (62). H. neptunium possesses CheY and CheR homologs, which appear to be encoded within the same operon. CheR catalyzes the methylation of specific glutamate residues within the cytoplasmic domains of MCPs (83). Since MCPs are the only known protein substrates for CheR, it is not clear which proteins, if any, H. neptunium CheR methylates or if CheR has a role in motility in H. neptunium.
We investigated motility and chemotaxis in H. neptunium using a set of standard techniques and found that the bacteria are capable of swimming motility with directional changes, resulting in a three-dimensional random walk. The bacteria swam at speeds ranging from 10 to 90 μm per s by modulating the rotary speed of a single unidirectional flagellum. Swimming cells, however, did not respond to chemotactic stimuli, including oxygen, which triggers a tactic response in all bacteria studied to date (87). In addition, in either spatial or temporal gradients of potential attractants (oxaloacetate, succinate, pyruvate, malate, aspartate, glutamate, and oxygen) and repellents (1,4-benzoquinone and oxygen), no chemotactic rings or bands were formed, and the probability of directional changes and swimming speed remained unchanged. These results are consistent with the absence of a complete set of the genes that are normally required for efficient navigation in chemical gradients. Motility that is not coupled to temporal sensing machinery may serve as a mechanism of population dispersion rather than stimulus-controlled navigation toward specific environments. A recent model for the bacterial chemotactic response (16) suggested that bacterial chemotaxis provides a competitive advantage to motile bacteria only under conditions where the cells can detect and adapt to stimuli within short distances (fewer than a few millimeters) or a short timescale (shorter than the cell division time). An H. neptunium swarmer cell, which does not replicate, may travel a relatively great distance from the attached reproductive cell that spawned it before it finds a suitable niche for attachment, differentiation, and growth. We hypothesize that H. neptunium swarmer cells are well adapted to survival in oligotrophic environments because their motility provides an efficient means for dispersion.
Signal transduction.
H. neptunium contains several genes encoding two-component signal transduction proteins: 17 sensor histidine kinase genes, 23 response regulator genes, and 3 combined sensor histidine kinase/response regulator genes were identified. Four of the histidine kinase genes were located adjacent on the chromosome to a response regulator, indicating possible functional units. Although two-component signal transduction systems were originally characterized for their role in adaptive responses to environmental changes, work in C. crescentus has revealed a major role for these signaling systems in controlling core physiological processes, including cell cycle progression and programs of morphogenesis inherent to the Caulobacter life cycle (38, 64, 80). A recent study reported the systematic deletion of each of the 106 two-component signal transduction genes encoded in the C. crescentus genome. Phenotypic analysis of these deletion strains identified 39 signaling genes required for proper cell cycle progression, cell growth, or morphogenesis, 9 of which are essential for viability (81). Since these 39 signal transduction genes are important for normal growth and cell cycle progression, we expected that many would be conserved in the closely related H. neptunium genome. In fact, H. neptunium encodes orthologs for 25 of the 39 (64%) genes required for normal growth or morphology, which includes 9 genes that are essential for viability (Table 5). In contrast, only 18 (27%) of the 67 other two-component genes in Caulobacter for which no clear phenotype was identified had orthologs in H. neptunium. Similarly, S. pomeroyi contains orthologs for 17 of the 39 (44%) Caulobacter genes required for normal growth or morphology (including 8 of the 9 essential genes), but only 9 of the 67 genes (13%) without clear phenotypes. Interestingly, 17 of the 39 Caulobacter two-component signaling genes with growth or morphology phenotypes have orthologs in both H. neptunium and S. pomeroyi (Table 5). This set of genes includes the major cell cycle regulators ctrA, cckA, divL, and divJ and includes the recently identified two-component pair cenK-cenR, which is essential for growth of Caulobacter owing to a role in regulating the cell envelope. These comparisons demonstrate that two-component signaling genes critical to core physiological processes are more highly conserved than other classes of genes. This analysis further suggests that the two-component genes with growth and morphology phenotypes in Caulobacter that are conserved in all three bacteria are crucial players in regulating cell cycle progression. A detailed investigation into the role of these genes should be productive. Similar comparisons of other classes of genes may help to identify important cell cycle regulatory genes in these organisms. Our analysis further suggests that two-component signaling genes unique to each organism or only conserved in two of the three may mediate responses or events specific to each organism. For example, in Caulobacter a phosphorelay, comprised of the kinase shkA (CC0138), the histidine phosphotransferase shpA (CC1114), and the response regulator tacA (CC3315), regulates stalk biogenesis during cell cycle progression (11). Orthologs of each gene are found in H. neptunium, which also has stalks, but are not found in S. pomeroyi, which lacks stalks. This example suggests that further comparative analysis of these genomes may unveil other genes and pathways responsible for the lifestyle of a prosthecate bacterium.
TABLE 5.
Comparison of two-component signal transduction genes shared by C. crescentus and H. neptunium
| C. crescentus ORF | Gene name | Typea | Essential? | H. neptunium ORF | S. pomeroyi ORF | Phenotypic characteristicb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell cycle | Doubling time | Swarm size | Swarm density | Motility | Stalk | Pili | Cell length | ||||||
| CC0138 | shkA | HK | HNE_0294 | Y | − | − | + | − | − | − | |||
| CC0237 | RR | HNE_1408 | SPO0710 | + | − | − | |||||||
| CC0238 | HK | HNE_1404 | SPO0711 | + | − | − | |||||||
| CC0289 | HK | HNE_3058 | SPO1947 | + | − | − | |||||||
| CC0294 | phoB | RR | HNE_2028 | SPO1953 | − | + | − | − | − | ||||
| CC0530 | cenK | HK | + | HNE_0407 | SPO0541 | ||||||||
| CC0744 | RR | HNE_0229 | Y | − | − | + | − | − | − | ||||
| CC1063 | divJ | HK | HNE_2910 | SPO0094 | Y | − | − | + | − | − | − | ||
| CC1078 | cckA | HK | + | HNE_0507 | SPO2036 | ||||||||
| CC1114 | shpA | HPT | HNE_0318 | Y | − | − | − | − | − | − | |||
| CC1740 | HK | HNE_2012 | SPO2088 | − | − | ||||||||
| CC1741 | RR | HNE_2011 | SPO2087 | − | − | ||||||||
| CC1743 | RR | + | HNE_2009 | SPO2085 | |||||||||
| CC2462 | pleD | RR | HNE_2284 | SPO2753 | Y | − | + | − | − | ||||
| CC2463 | divK | RR | + | HNE_2285 | |||||||||
| CC2482 | pleC | HK | HNE_2910 | Y | − | + | − | − | − | ||||
| CC2755 | HK | HNE_1311 | SPO1623 | − | |||||||||
| CC2931 | RR | + | HNE_3093 | SPO0251 | |||||||||
| CC2932 | HK | + | HNE_3094 | SPO2173 | |||||||||
| CC3035 | ctrA | RR | + | HNE_0944 | SPO1679 | ||||||||
| CC3315 | tacA | RR | HNE_3039 | Y | − | − | + | − | − | − | |||
| CC3471 | RR | HNE_0639 | − | + | − | − | − | ||||||
| CC3474 | HK | HNE_2339 | Y | + | − | − | |||||||
| CC3477 | RR | HNE_2344 | Y | + | − | − | |||||||
| CC3484 | divL | HK | + | HNE_0399 | SPO3868 | ||||||||
| CC3743 | cenR | RR | + | HNE_3550 | SPO3426 | ||||||||
HK, histidine kinase; RR, response regulator; HPT, histidine phosphotransferase.
Phenotypic characterization of deletions is from reference 81. Genes whose corresponding deletion strain was identified as having a cell cycle, growth, motility, or morphogenesis defect are marked with a plus sign (+). Cell cycle-regulated genes according to reference 46 are indicated by Y. For all other categories, a plus sign indicates that the phenotypic characteristic is larger than wild type and a minus (−) sign indicates that a characteristic is smaller than wild type.
Cell aging and death.
Senescence is the decrease in survival or reproduction with age and has been well documented in unicellular microorganisms with asymmetric division. For example, stalked cells of C. crescentus exhibit decreased reproductive output (number of progeny produced per cell per hour) with age (1). In this study, some C. crescentus stalked cells produced up to 130 progeny over the 300-h time course of the experiment, but many divided more slowly or stopped dividing completely with increased age. Recent studies with Escherichia coli showed that daughter cells that inherit the older poles exhibit slower growth rates, decreased reproductive output, and an increased likelihood of death, suggesting that bacteria that undergo morphologically symmetric division also exhibit senescence (86). Prosthecate, budding bacteria display an extreme form of senescence in which the mother cell gives rise to a very limited number of progeny (55, 94). H. neptunium mother cells produce up to nine progeny (R. M. Weiner, unpublished data). This pronounced senescence may be related to polar growth and the use of the stalk as a reproductive structure since the developing bud likely receives newly synthesized macromolecules (e.g., components of the cell envelope) that are not renewed in the reproductive cell. Thus, senescence of the H. neptunium mother cell may result from the accumulation of damaged cellular components or the loss of components required for reproduction following a limited number of budding events.
Alternatively, the cessation in reproductive output by H. neptunium may be mediated by a specific genetic program. Programmed cell death occurs in some bacteria with distinct developmental cycles, such as during Bacillus subtilis sporulation and fruiting body formation in Myxococcus xanthus (48). If the H. neptunium cell cycle involves a programmed cell death, candidate genes for such a program include toxin-antitoxin (TA) loci and homologs of eukaryotic apoptotic proteins. These genes are found in a variety of bacteria, but this does not preclude their potential involvement in H. neptunium senescence. TA loci have been suggested to be involved in programmed cell death in E. coli, but this idea remains controversial (27, 28, 29, 35, 63). H. neptunium has two sets of TA loci (HNE_2664/HNE_2665 and HNE_0347/HNE_0348). TA loci consist of two genes organized in an operon, one encoding a stable toxin and the other an unstable antitoxin. They were described first in plasmid and phage genomes, where they contribute to episome stability by selectively killing segregants cured of the episome, but were later found in the chromosomes of many bacteria (28, 29, 35, 63). Homologs of eukaryotic apoptotic proteins found in bacteria include caspase-like proteases, HtrA-like serine proteases, apoptotic ATPases, and NACHT family NTPases. The marine cyanobacterium Trichodesmium sp. apparently undergoes a programmed cell death in aging cultures and also in response to certain environmental and physiological stresses, and this process is correlated with an increased level and activity of caspase-like proteases (10). H. neptunium possesses three HtrA-like serine proteases (HNE_0472, HNE_1313, and HNE_2644) which could have roles in a potential cell death program.
Conclusion.
The analysis of the genome sequence of H. neptunium ATCC 15444T and its comparison to that of C. crescentus CB15 has revealed some unexpected results. For example, although the analysis is congruent with phenotypic similarities between the two organisms, the fact that H. neptunium shares twice as many proteins with C. crescentus than it does with its fellow Rhodobacter S. pomeroyi conflicts with the currently accepted 16S rRNA phylogeny. This suggests that Hyphomonas can be reclassified.
Genomic and experimental evidence indicate that H. neptunium cells are motile but not chemotactic. Furthermore, while some of the flagellar genes such as flgL and fliQ appear to be much closer to their S. pomeroyi homologs than to their homologs in C. crescentus, H. neptunium contains the potential flagellar regulatory genes flhF and flhG, which are not encoded by either of the other two genomes. The presence of additional regulatory flagellar genes that are not found in closely related organisms is consistent with the notion that motility plays an important role in the lifestyle of this organism. We speculate that motility among Hyphomonas swarmer cells is a random dispersal method. The lack of evidence for chemotaxis further supports this hypothesis.
Finally, comparative genomics has yielded valuable insights into prokaryotic diversity and evolution. The availability of the complete H. neptunium genome will be an important tool for examining unique aspects of the biology of DBP, including cell development, stalk biogenesis, and strategies for oligotrophic lifestyles. Moreover, the H. neptunium genome will provide an important resource for the study of processes for which prosthecate, budding bacteria may serve as model systems, including chromosome segregation, polar growth, and cell aging.
Acknowledgments
The sequencing and analysis of H. neptunium was funded by National Science Foundation Award 0237224 to T.R.H., Y.V.B., and N.L.W. In addition, the phylogenetic analysis was supported in part by the NSF “Assembling the Tree of Life” Award 0228651 to Jonathan Eisen, N.L.W., and K.E.N.
REFERENCES
- 1.Ackermann, M., S. C. Stearns, and U. Jenal. 2003. Senescence in a bacterium with asymmetric division. Science 300:1920. [DOI] [PubMed] [Google Scholar]
- 2.Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. [DOI] [PubMed] [Google Scholar]
- 3.Alexandre, G., S. E. Greer, and I. B. Zhulin. 2000. Energy taxis is the dominant behavior in Azospirillum brasilense. J. Bacteriol. 182:6042-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amsler, C. D., and P. Matsumura. 1995. Chemotactic signal transduction in Escherichia coli and Salmonella typhimurium, p. 89-103. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 5.Ausmees, N., and C. Jacobs-Wagner. 2003. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu. Rev. Microbiol. 57:225-247. [DOI] [PubMed] [Google Scholar]
- 6.Baier, R., A. Meyer, V. DePalma, R. Krieg, and M. Fornalik. 1983. Surface microfouling during the induction period. J. Heat Trans. 105:618-624. [Google Scholar]
- 7.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:138-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson, A. K., G. Ramakrishnan, N. Ohta, J. Feng, A. J. Ninfa, and A. Newton. 1994. The Caulobacter crescentus FlbD protein acts at ftr sequence elements both to activate and to repress transcription of cell cycle-regulated flagellar genes. Proc. Natl. Acad. Sci. USA 91:4989-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 10.Berman-Frank, I., K. D. Bidle, L. Haramaty, and P. G. Falkowski. 2004. The demise of the marine cyanobacterium Trichodesmium spp., via an autocatalyzed cell death pathway. Limnol. Oceanogr. 49:997-1005. [Google Scholar]
- 11.Biondi, E. G., J. M. Skerker, M. Arif, M. S. Prasol, B. S. Perchuk, and M. T. Laub. 2006. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 59:386-401. [DOI] [PubMed] [Google Scholar]
- 12.Blackman, M. A., and R. M. Weiner. 1975. Photomicrography of nalidixic acid treated Hyphomicrobium neptunium: inhibition of bud formation and bud separation. Can. J. Microbiol. 21:226-230. [DOI] [PubMed] [Google Scholar]
- 13.Bodenmiller, D., E. Toh, and Y. V. Brun. 2004. Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 186:1438-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brun, Y. V., and L. Shapiro. 1992. A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 6:2395-2408. [DOI] [PubMed] [Google Scholar]
- 15.Chang, Y., S. Coon, M. Walch, and R. M. Weiner. 1996. Effects of Hyphomonas biofilms on the toxicity of copper and zinc to Crassostrea gigas and C. virginica larval set. J. Shellfish Res. 15:589-595. [Google Scholar]
- 16.Clark, D. A., and L. C. Grant. 2005. The bacterial chemotactic response reflects a compromise between transient and steady-state behavior. Proc. Natl. Acad. Sci. USA 102:9150-9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole, J. L., G. G. Hardy, D. Bodenmiller, E. Toh, A. Hinz, and Y. V. Brun. 2003. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol. Microbiol. 49:1671-1683. [DOI] [PubMed] [Google Scholar]
- 18.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 20.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dembo, A., and S. Karlin. 1992. Poisson approximations for r-scan processes. Ann. Appl. Probab. 2:329-357. [Google Scholar]
- 22.Devine, R. A., and R. M. Weiner. 1990. Hyphomonas species metabolise amino acids using Krebs cycle enzymes. Microbios 62:137-153. [Google Scholar]
- 23.Doyle, M. O., and M. L. Otte. 1997. Organism-induced accumulation of iron, zinc and arsenic in wetland soils. Environ. Pollut. 96:1-11. [DOI] [PubMed] [Google Scholar]
- 24.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emala, M. A., and R. M. Weiner. 1983. Modulation of adenylate energy charge during the swarmer cycle of Hyphomicrobium neptunium. J. Bacteriol. 153:1558-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelberg-Kulka, H., B. Sat, and R. Hazan. 2001. Bacterial programmed cell death and antibiotics. ASM News 67:617-624. [Google Scholar]
- 28.Engelberg-Kulka, H., B. Sat, M. Reches, S. Amitai, and R. Hazan. 2004. Bacterial programmed cell death systems as targets of antibiotics. Trends Microbiol. 12:66-71. [DOI] [PubMed] [Google Scholar]
- 29.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 30.Gish, W. 2004. WU-BLAST. Distributed by the author. [http://blast.wustl.edu].
- 31.Glockner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot, W. Ludwig, D. Gade, A. Beck, K. Borzym, K. Heitmann, R. Rabus, H. Schlesner, R. Amann, and R. Reinhardt. 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 100:8298-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haft, D. H., J. D. Selengut, L. M. Brinkac, N. Zafar, and O. White. 2005. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics 21:293-306. [DOI] [PubMed] [Google Scholar]
- 34.Hartmans, S., A. Schmuelke, A. M. Cook, and T. Leisinger. 1986. Methyl-chloride: naturally occurring toxicant and C-1 growth substrate. J. Gen. Microbiol. 132:1139-1142. [Google Scholar]
- 35.Hayes, F. 2005. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 36.Hirsch, P. 1974. Budding bacteria. Annu. Rev. Microbiol. 28:391-444. [DOI] [PubMed] [Google Scholar]
- 37.Howe, K., A. Bateman, and R. Durbin. 2002. QuickTree: building huge neighbour-joining trees of protein sequences. Bioinformatics 18:1546-1547. [DOI] [PubMed] [Google Scholar]
- 38.Hung, D., H. McAdams, and L. Shapiro. 2000. Regulation of the Caulobacter cell cycle, P. 361-378. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington D.C.
- 39.Karlin, S., J. Mrázek, and A. M. Campbell. 1996. Frequent oligonucleotides and peptides of the Haemophilus influenzae genome. Nucleic Acids Res. 24:4263-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köhler, J., and A. C. Schwartz. 1982. Oxidation of aromatic aldehydes and aliphatic secondary alcohols by Hyphomicrobium spp. Can. J. Microbiol. 26:65-72. [Google Scholar]
- 41.Kurtz, H. D., and J. Smith. 1992. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J. Bacteriol. 174:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurtz, S., and C. Schleiermacher. 1999. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics 15:426-427. [DOI] [PubMed] [Google Scholar]
- 43.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langille, S. E., G. G. Geesey, and R. M. Weiner. 2000. Polysaccharide-specific probes inhibit adhesion of Hyphomonas rosenbergii strain VP-6 to hydrophilic strains. J. Ind. Microb. Biotechnol. 25:81-85. [Google Scholar]
- 45.Langille, S. E., and R. M. Weiner. 1998. Spatial and temporal deposition of Hyphomonas strain VP-6 capsules involved in biofilm formation. Appl. Environ. Microbiol. 64:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling the Caulobacter cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 47.Lee, K. B., C. T. Liu, Y. Anzai, H. Kim, T. Aono, and H. Oyaizu. 2005. The hierarchical system of the “Alphaproteobacteria”: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 55:1907-1919. [DOI] [PubMed] [Google Scholar]
- 48.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 6:2863-2874. [DOI] [PubMed] [Google Scholar]
- 50.Li, G., C. S. Smith, Y. V. Brun, and J. X. Tang. 2005. The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J. Bacteriol. 187:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liefson, E. 1964. Hyphomicrobium neptunium sp. nov. Antonie Leeuwenhoek 30:249-256. [DOI] [PubMed] [Google Scholar]
- 52.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 53.Merker, R. I., and J. Smit. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl. Environ. Microbiol. 54:2078-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moaledj, K. 1978. Qualitative analysis of an oligocarbophilic aquatic microflora in the Plussee. Arch. Hydrobiol. 82:98-113. [Google Scholar]
- 55.Moore, R. L. 1981. The biology of Hyphomicrobium and other prosthecate, budding bacteria. Annu. Rev. Microbiol. 35:567-594. [DOI] [PubMed] [Google Scholar]
- 56.Moore, R. L., R. M. Weiner, and R. Gebers. 1984. Genus Hyphomonas Pongratz 1957 nom. rev. emend., Hyphomonas polymorpha Pongratz 1957 nom. rev. emend., and Hyphomonas neptunium (Liefson 1964) comb. nov. emend. (Hyphomicrobium neptunium). Int. J. Syst. Bacteriol. 34:71-73. [Google Scholar]
- 57.Moran, M. A., A. Buchan, J. M. Gonzalez, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. L. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 58.Muir, R. E., and J. W. Gober. 2004. Regulation of FlbD activity by flagellum assembly is accomplished through direct interaction with the trans-acting factor, FliX. Mol. Microbiol. 54:715-730. [DOI] [PubMed] [Google Scholar]
- 59.Mullin, D. A., S. M. Van Way, C. A. Blankenship, and A. H. Mullin. 1994. FlbD has a DNA-binding activity near its carboxy terminus that recognizes ftr sequences involved in positive and negative regulation of flagellar gene transcription in Caulobacter crescentus. J. Bacteriol. 176:5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narbonne, J. F., M. Daubèze, P. Baumard, H. Budzinski, C. Clérandeau, F. Akcha, P. Mora, and P. Garrigues. 2001. Biochemical markers in mussel, Mytilus sp., and pollution monitoring in European coasts: data analysis, p. 215-236. In P. Garrigues, H. Barth, C. H. Walker, and J. F. Narbonne (ed.), Biomarkers in marine organisms: a practical approach. Elsevier Science B.V., Amsterdam, The Netherlands.
- 61.Neugebauer, H., C. Herrmann, W. Kammer, G. Schwarz, A. Nordheim, and V. Braun. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 187:8300-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, C. M. Fraser, and J. A. Eisen. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nystrom, T. 2003. Conditional senescence in bacteria: death of the immortals. Mol. Microbiol. 48:17-23. [DOI] [PubMed] [Google Scholar]
- 64.Ohta, N., T. W. Grebe, and A. Newton. 2000. Signal transduction and cell cycle checkpoints in developmental regulation of Caulobacter, p. 341-360. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 65.Pandza, S., M. Baetens, C. H. Park, T. Au, M. Keyhan, and A. Matin. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36:414-423. [DOI] [PubMed] [Google Scholar]
- 66.Pellegrini, M., E. M. Marcotte, M. J. Thompson, D. Eisenberg, and T. O. Yeates. 1999. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA 96:4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poindexter, J. S. 2004. Dimorphic prosthecate bacteria: the genera Caulobacter, Asticcacaulis, Hyphomicrobium, Pedomicrobium, Hyphomonas, Thiodendron. In The prokaryotes: an evolving electronic resource for the microbiological community, version 3.17. Springer-Verlag, New York, N.Y. [Online.] http://link.springer-ny.com/link/service/books/10125/.
- 69.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 70.Quintero, E. J., K. Busch, and R. M. Weiner. 1998. Spatial and temporal deposition of adhesive extracellular polysaccharide capsule and fimbriae by Hyphomonas strain MHS-3. Appl. Envir. Microbiol. 64:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quintero, E. J., and R. M. Weiner. 1995. Evidence for the adhesive function of the exopolysaccharide of Hyphomonas strain MHS-3 in its attachment to surfaces. Appl. Envir. Microbiol. 61:1897-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 73.Ramakrishnan, G., and A. Newton. 1990. FlbD of Caulobacter crescentus is a homologue of the NtrC (NRI) protein and activates σ54-dependent flagellar gene promoters. Proc. Natl. Acad. Sci. USA 87:2369-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reisenauer, A., K. Quon, and L. Shapiro. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol. 181:2430-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ren, Q., K. H. Kang, and I. T. Paulsen. 2004. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 32:284-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ren, Q., and I. T. Paulsen. 2005. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 1:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider, M., M. Tognolli, and A. Bairoch. 2004. The Swiss-Prot protein knowledgebase and ExPASy: providing the plant community with high quality proteomic data and tools. Plant Physiol. Biochem. 42:1013-1021. [DOI] [PubMed] [Google Scholar]
- 79.Sicheritz-Ponten, T., and S. G. Andersson. 2001. A phylogenomic approach to microbial evolution. Nucleic Acids Res. 29:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skerker, J. M., and M. T. Laub. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2:325-337. [DOI] [PubMed] [Google Scholar]
- 81.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith, C., A. Hinz, D. Bodenmiller, D. E. Larson, and Y. V. Brun. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 185:1432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Springer, W. R., and D. E. J. Koshland. 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. USA 74:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Staley, J. T. 1968. Prosthecomicrobium and Ancalomicrobium: new prosthecate freshwater bacteria. J. Bacteriol. 95:1921-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Starnbach, M. N., and S. Lory. 1992. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6:459-469. [DOI] [PubMed] [Google Scholar]
- 86.Stewart, E. J., R. Madden, G. Paul, and F. Taddei. 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 88.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 89.Tso, W.-W., and J. Adler. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118:560-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wali, T. M., G. R. Hudson, D. A. Danald, and R. M. Weiner. 1980. Timing of swarmer cell cycle morphogenesis and macromolecular synthesis by Hyphomicrobium neptuniumin synchronous culture. J. Bacteriol. 144:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weiner, R. M. 2005. Genus Hyphomonas, p. 179-187. In J. T. Staley et al. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part C. Springer, New York, N.Y. [Google Scholar]
- 92.Weiner, R. M., and M. A. Blackman. 1973. Inhibition of deoxyribonucleic acid synthesis and bud formation by nalidixic acid in Hyphomicrobium neptunium. J. Bacteriol. 116:1398-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weiner, R. M., M. Melick, K. O'Neill, and E. Quintero. 2000. Hyphomonas adhaerens sp. nov., Hyphomonas johnsonii sp. nov. and Hyphomonas rosenbergii sp. nov., marine budding and prosthecate bacteria. Int. J. Syst. Evol. Microbiol. 50:459-469. [DOI] [PubMed] [Google Scholar]
- 94.Whittenbury, R., and C. S. Dow. 1977. Morphogenesis and differentiation in Rhodomicrobium vannielii and other budding and prosthecate bacteria. Bacteriol. Rev. 41:754-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wingrove, J. A., E. K. Mangan, and J. W. Gober. 1993. Spatial and temporal phosphorylation of a transcriptional activator regulates pole-specific gene expression in Caulobacter. Genes Dev. 7:1979-1992. [DOI] [PubMed] [Google Scholar]
- 96.Wu, J., A. K. Benson, and A. Newton. 1995. Global regulation of a sigma 54-dependent flagellar gene family in Caulobacter crescentus by the transcriptional activator FlbD. J. Bacteriol. 177:3241-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu, J., and A. Newton. 1997. Regulation of the Caulobacter flagellar gene hierarchy: not just for motility. Mol. Microbiol. 24:233-239. [DOI] [PubMed] [Google Scholar]
- 98.Zerfas, P. M., M. Kessel, E. J. Quintero, and R. M. Weiner. 1997. Fine-structure evidence for cell membrane partitioning of the nucleoid and cytoplasm during bud formation in Hyphomonas species. J. Bacteriol. 179:148-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhulin, I. B., V. A. Bespalov, M. S. Johnson, and B. L. Taylor. 1996. Oxygen taxis and proton motive force in Azospirillum brasilense. J. Bacteriol. 178:5199-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]