Abstract
A growing body of evidence suggests that the WhiB-like proteins exclusive to the GC-rich actinomycete genera play significant roles in pathogenesis and cell division. Each of these proteins contains four invariant cysteine residues and a conserved helix-turn-helix motif. whmD, the Mycobacterium smegmatis homologue of Streptomyces coelicolor whiB, is essential in M. smegmatis, and the conditionally complemented mutant M. smegmatis 628-53 undergoes filamentation under nonpermissive conditions. To identify residues critical to WhmD function, we developed a cotransformation-based assay to screen for alleles that complement the filamentation phenotype of M. smegmatis 628-53 following inducer withdrawal. Mycobacterium tuberculosis whiB2 and S. coelicolor whiB complemented the defect in M. smegmatis 628-53, indicating that these genes are true functional orthologues of whmD. Deletion analysis suggested that the N-terminal 67 and C-terminal 12 amino acid residues are dispensable for activity. Site-directed mutagenesis indicated that three of the four conserved cysteine residues (C90, C93, and C99) and a conserved aspartate (D71) are essential. Mutations in a predicted loop glycine (G111) and an unstructured leucine (L116) were poorly tolerated. The region essential for WhmD activity encompasses 6 of the 10 residues conserved in all seven M. tuberculosis WhiBs, as well as in most members of the WhiB family identified thus far. WhmD structure was found to be sensitive to the presence of a reducing agent, suggesting that the cysteine residues are involved in coordinating a metal ion. Iron-specific staining strongly suggested that WhmD contains a bound iron atom. With this information, we have now begun to comprehend the functional significance of the conserved sequence and structural elements in this novel family of proteins.
Streptomyces coelicolor, a gram-positive, sporulating bacterium, is phylogenetically a close relative of Mycobacterium tuberculosis, with a similarly high GC content (65 to 70%). This organism follows a differentiation cycle in which young colonies send hyphal extensions into the agar and later-generation cells form white aerial hyphae which become pigmented and produce spores (4). Mutants which show an arrest in the development of mature spores and pigment formation remain white, and mutations leading to this phenotype have been described to map to eight independent loci (5, 12). The first-characterized nonsporulating mutant was shown to be defective in whiB, a gene encoding a basic polypeptide 87 amino acids long with a putative helix-turn-helix (HTH) motif (8). whiB mutants are viable but incapable of sporulating. Secondary-structure prediction and mutational analysis suggest that WhiB is a DNA binding protein (8). In addition, whiB mutants show reduced expression of the whiE cluster encoding spore pigment, indicating that this gene is likely to be a transcription factor.
The large database of DNA sequences of members of the order Actinomycetales has revealed a family of genes encoding proteins related to WhiB. The list is currently comprised of 121 bacterial proteins (http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF02467) exclusive to the high-GC-content actinomycete genera, including Streptomyces, Rhodococcus, and Mycobacterium. Each of these proteins contains four invariant cysteine residues arranged as Cys-X14-22-Cys-X2-Cys-X5-Cys and a conserved HTH-like motif characterized by the 7-residue signature (FYG)-G-(VI)-W-G-G-(LVIM) in the putative β-turn and is relatively short in length (76 to 139 residues). Although this motif does not comprise a typical HTH motif (1), it is believed to be involved in DNA binding (21). These proteins all possess a high overall hydrophilicity suggestive of a cytoplasmic location. Despite an overall negative charge, these proteins have positively charged regions near their carboxy termini (17) and are predicted to contain extensive α-helical structures with a central β-sheet region between the first and second α-helices (21). Analysis of the genome sequence (7) indicates that M. tuberculosis contains seven whiB homologues (whiB1 to whiB7) (7, 21) which show all the hallmark features of the members of the WhiB family. The presence of four cysteine residues suggests that these proteins may be sensitive to redox changes, perhaps through a bound metal atom or through direct sensitivity to oxidation via disulfide bond formation. A recent report demonstrated that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster (13) and that all four cysteine residues are essential for WhiD activity. A second recent article showed that the whcE gene of Corynebacterium glutamicum, also a member of the whiB family, is important for survival following heat and oxidative stress (14). Both these observations lend support to the hypothesis that the members of this family of proteins are likely to be associated with intracellular redox-sensing pathways.
Although the most C-terminal α-helix of the putative HTH-like domain contains a segment rich in basic residues likely to be involved in DNA binding (24), there are no published reports demonstrating the interaction of any of the WhiB family of proteins with DNA. In addition, the significance of the 10 conserved amino acid residues and the conserved predicted secondary structural elements in all the WhiB-like proteins remains unknown.
WhmD (WhiB2 in M. tuberculosis), which by amino acid similarity is the closest Mycobacterium smegmatis orthologue of Streptomyces coelicolor WhiB, is encoded by whmD, an essential mycobacterial gene required for proper septation and cell division. In M. smegmatis, this gene could be disrupted only in the presence of a plasmid supplying whmD in trans. In a conditionally complemented system, on withdrawal of the inducer, the mutant exhibited irreversible filamentous branched growth with diminished septum formation and aberrant septal placement (10). Computer algorithm-based secondary-structure analysis predicted that, like all WhiB-like proteins, WhmD is composed largely of helical and coiled regions with two probable sheet regions, the second of the two being a part of the putative HTH-like motif. Using WhmD as a prototype, we have examined the role of the conserved residues as well as the predicted structural modules in WhiB function. Using a combination of deletion and site-specific mutation analysis followed by genetic complementation, we have mapped regions and residues essential to the function of WhmD in M. smegmatis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli strain DH5α {F′ endA1 hsdR17(rK− mK+) glnV44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15]} procured from Stratagene, La Jolla, CA, was used for cloning purposes. E. coli BL21(DE3) [F− ompT hsdSB(rB− mB−) gal dcm (DE3)] used for protein expression was purchased from Novagen, Madison, WI. M. smegmatis mc26 1-2c was kindly provided by Bill Jacobs, Albert Einstein College of Medicine, New York, NY, and M. tuberculosis CDC1551 genomic DNA was obtained from Colorado State University. The vector pET-22b(+) was purchased from Novagen, Madison, WI. Luria-Bertani (LB) broth and LB agar were used to culture E. coli. 7H9 broth and 7H10 agar from Difco Laboratories (Becton Dickinson) were supplemented with albumin dextrose complex (5 g/liter bovine serum albumin, 2 g/liter dextrose, 0.85 g/liter NaCl), 0.2% glycerol and were used for culturing mycobacteria. Both E. coli and mycobacteria were grown at 37°C with shaking at 200 rpm. The following antibiotics were added when necessary: ampicillin (200 μg/ml), kanamycin (50 μg/ml for E. coli and 15 μg/ml for mycobacteria), hygromycin (200 μg/ml for E. coli and 50 μg/ml for mycobacteria), apramycin (30 μg/ml), and Zeocin (25 μg/ml for E. coli and 50 μg/ml for mycobacteria).
DNA techniques.
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (NEB), Beverly, MA, and Taq polymerase was purchased from Invitrogen Corporation, CA. Pfu DNA polymerase was purchased from Stratagene, CA. The Klenow fragment of DNA polymerase was from NEB. Protocols for DNA manipulations, including plasmid DNA preparation, restriction endonuclease digestion, agarose gel electrophoresis, isolation and ligation of DNA fragments, E. coli transformation, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were performed as described by Sambrook et al. (20). Mycobacterial strains were transformed by electroporation. PCR amplifications were carried out according to the manufacturer's specifications (Bio-Rad Laboratories, CA). Each of the 30 cycles was carried out at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a final extension cycle at 72°C for 10 min. DNA fragments used for cloning and for labeling reaction products were purified by using the QIAGEN gel extraction kit (QIAGEN) per the manufacturer's specifications.
Construction of plasmid pBP10 zeo and test alleles for complementation.
Plasmid pBP10 zeo was constructed by excising a 1.1-kb fragment from the vector pER10 (E. Rubin, unpublished data) using the enzymes EcoRI and XbaI, end-filling using the Klenow fragment of E. coli DNA polymerase, and inserting the resulting fragment into the EcoRV site of pBP10. whmD containing 187 bp of its 5′ untranslated region (UTR) was amplified from M. smegmatis genomic DNA using the PCR primers pBP10whmD-F (5′ AAAACTGCAGGAATTCGCGCCCTGGAGC 3′) and pBP10whmD-R (5′ GGACTAGTCTAGATGATGCCGCGCTT 3′) and cloned at the PstI-SpeI sites of pBP10 zeo to generate pBP10 zeo whmD. The S. coelicolor whiB test plasmid was generated by PCR amplifying the whiB gene, including its promoter sequence, from S. coelicolor A(3)2 genomic DNA using the primers pBP10ScwhiB-F (5′ AAAACTGCAGGACTTCATTGGTATGTCG 3′) and pBP10ScwhiB-R (5′ GGACTAGTCTGCACGGACATCGAGGT 3′), followed by cloning into the PstI and SpeI sites of pBP10 zeo. whiB2 was amplified from M. tuberculosis CDC1551 genomic DNA by using the PCR primers pBP10MtwhiB2-F (5′AAAACTGCAGTTACGAGATGATATGGAA 3′) and pBP10MtwhiB2-R (5′ GGACTAGTTCAGATGATCCCGCGTTT 3′) and cloned into the PstI and SpeI sites of pBP10 zeo to generate pBP10 zeo whiB2. All clones generated as described above were confirmed by sequencing.
Construction of mutant alleles of WhmD.
All deletion mutants of WhmD were constructed using a PCR-based strategy. To generate the N-terminal deletion mutants, the whmD promoter fragment, amplified using the PCR primers pBP10 whmD-F (5′ AAAACTGCAGGAATTCGCGMLCTGGAGC 3′) and PwhmDMet-R (5′ CGCGGATCCCATATCMLCCGCCTCCTC 3′), was fused to a fragment of whmD carrying the appropriate deletion and cloned at the PstI-SpeI sites of pBP10 zeo. The fusion, mediated by a BamHI site, introduces a Gly-Ser dipeptide at the junction of the Met encoded by the start codon and the WhmD fragment carrying the N-terminal deletion. The addition of this dipeptide does not interfere with WhmD protein function in the complementation assay. The following forward primers, in conjunction with the whmD reverse primer pBP10whmD-R (5′ GGACTAGTCTAGATGATGCCGCGCTT 3′), were used to generate the following N-terminal deletions: whmDΔN42-F (5′ CGCGGATCCCTGAGTCTGGTGCCCGAT 3′), whmDΔN57-F (5′ CGCGGATCCGAAGACCAATGGCAGGAG 3′), whmDΔN62-F (5′ CGCGGATCCGAGCGTGCCCTGTGCGCG 3′), and whmDΔN67-F (5′ CGCGGATCCGCGCAAACTGACCCGGAG 3′). C-terminal deletion mutants were generated by PCR amplifying whmD using the forward primer pBP10whmD-F (5′ AAAACTGCAGGAATTCGCGCCCTGGAGC 3′) and a combination of reverse primers depending on the required length of the deletion, following which the products were cloned at the PstI-SpeI sites of pBP10 zeo. The reverse primers used to generate the C-terminal deletions were as follows: whmDΔC12-R (5′GGACTAGTCTACGACAGACCGCCCCA 3′), whmDΔC23-R (5′ GGACTAGTCTAATGCGCGAGCGCGTA 3′), and whmDΔC30-R (5′ GGACTAGTCTAGCACGCGTCACGAAC 3′). All site-directed mutants were generated using the Quik-Change mutagenesis strategy (Stratagene) in the plasmid pBP10 zeo whmD with Pfu DNA polymerase and primer pairs carrying the desired mutation. The following primer pairs were used: C67A-1 (5′ GCAGGAGCGTGCCCTGGCCGCGCAAACTGACCCG 3′) and C67A-2 (3′ CGTCCTCGCACGGGACCGGCGCGTTTGACTGGGC 5′); C90A-1 (5′ GAGGCCAAGCGCATCGCCCAGGGGTGCGAAGTTCG 3′) and C90A-2 (3′ CTCCGGTTCGCGTAGCGGGTCCCCACGCTTCAAGC 5′); C93A-1 (5′ CGCATCTGCCAGGGGGCCGAAGTTCGTGACGCG 3′) and C93A-2 (3′ GCGTAGACGGTCCCCCGGCTTCAAGCACTGCGC 5′); C99A-1 (5′ CGAAGTTCGTGACGCGGCCCTGGAATACGCGCTCG 3′) and C99A-2 (3′ GCTCGCGCATAAGGTCCGGGCGCAGTGCTTGAAGC 5′); Y102G-1 (5′ GACGCGTGCCTGGAAGGCGCGCTCGCGCATGATG 3′) and Y102G-2 (3′ CTGCGCACGGACCTTCCGCGCGAGCGCGTACTAC 5′); Y102P-1 (5′ GACGCGTGCCTGGAACCGGCGCTCGCGCATGATG 3′) and Y102P-2 (3′ CTGCGCACGGACCTTGGCCGCGAGCGCGTACTAC 5′); G111P-1 (5′ GCATGATGAGCGCTTCCCGATCTGGGGCGGTCTG 3′) and G111P-2 (3′ CGTACTACTCGCGAAGGGCTAGACCCCGCCAGAC 5′); R122G-1 (5′ CTGTCGGAGCGTGAGCGCGGCCGCCTCAAGCGCGGC 3′) and R122G-2 (3′ GACAGCCTCGCACTCGCGGGCGCGGAGTTCGCGCCG 5′); and R122P-1 (5′ CTGTCGGAGCGTGAGCGCCCGCGCCTCAAGCGCGGC 3′) and R122P-2 (3′ GACAGCCTCGCACTCGCGGGCGCGGAGTTCGCGCCG 5′). All deletion and site-directed whmD mutants were confirmed by sequencing using the primer whmDseq (5′ MLTCAACTGAGTCTGGTGCC 3′).
M. smegmatis 628-53 complementation assays.
For complementation analysis, the test allele and the control plasmid pBP10 zeo were transformed into M. smegmatis 628-53 and selected on 0.2% acetamide-supplemented 7H10 agar plates containing apramycin, hygromycin, and Zeocin. Acetamide withdrawal was performed as follows: transformants were cultured in 7H9 broth supplemented with 0.2% acetamide, grown to an optical density of 1.0, washed twice with 7H9 broth, resuspended in 7H9 broth, and grown overnight (∼16 h) in the absence of acetamide. Following inducer withdrawal, cultures were washed and resuspended in phosphate-buffered saline, heat fixed on slides, and stained with carbolfuchsin for 5 min. After the excess dye was washed off with distilled water, the slides were dried and examined at a ×600 or ×1,000 magnification on a Nikon Eclipse E800 microscope under oil. Images were captured using an onboard digital still camera (model DXM1200) and edited using the software package ACT-1 version 2. Cell length measurements were made to determine the extent of complementation of each allele. Transformants with a mean cell length of ≤7.5 μm were scored as complemented.
Western blotting.
To quantitate protein levels of the point mutants and C-terminal deletion mutants of WhmD, transformants of M. smegmatis 628-53 containing each complementing allele and the controls pBP10 zeo and pBP10 zeo whmD were grown to an optical density of 1.0 in the presence of 0.2% acetamide, washed twice with 7H9, resuspended in 7H9 broth, and grown overnight (∼16 h) in the absence of acetamide. Following inducer withdrawal, cultures were washed and resuspended in phosphate-buffered saline and lysed by bead beating on a mini bead beater (Biospec Products, Bartlesville, OK) and the amount of protein was quantitated by the Bradford assay (3). Thirty micrograms of total cell lysate proteins and 3 μg of purified WhmD (rWhmD) were electrophoresed on a 12% SDS-PAGE system and transferred to nitrocellulose. For Western blotting, WhmD antiserum was used at a 1:200 dilution. Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G at a 1:3,500 dilution and a chemiluminescent substrate (Amersham) were used to detect the presence of WhmD and its mutant alleles.
Purification of M. smegmatis WhmD.
The gene encoding WhmD was PCR amplified from M. smegmatis chromosomal DNA using the gene-specific primers pET22bwhmD-F (5′ GGGAATTCCATATGTCTTATGAGAGCGGC 3′) and pET22bwhmD-R (5′ CCGCTCGAGGATGATGCCGCGCTTGAG 3′). The amplified product was cloned into the expression vector pET-22b(+) at the NdeI and XhoI sites, and the recombinant plasmid was used to transform E. coli BL21(DE3). The transformant culture was grown to exponential phase, induced with 1 mM isopropyl-thio-β-d-galactoside (IPTG) for 2 h at room temperature, and lysed by sonication. C-terminal hexahistidine-tagged WhmD was purified from the soluble fraction by Ni-nitrilotriacetic acid chromatography according to the manufacturer's protocol (QIAGEN, Valencia, CA). The purity of the protein was analyzed by SDS-PAGE. To overexpress the cysteine-to-alanine point mutants of WhmD, the mutant open reading frames (ORFs) were amplified from their cognate mutant constructs in pBP10 zeo whmD using the primers pET22bwhmD-F and pET22bwhmD-R and cloned into pET-22b(+). The expression and purification regimens were as described above. Native PAGE and SDS-PAGE was performed as described previously (20). The 4× SDS-PAGE sample buffer used either contained or lacked 5 mM β-mercaptoethanol.
Iron staining.
Staining was carried out as described by Kuo and Fridovich (15). All procedures were carried out at room temperature. Briefly, 9 μg of purified WhmD was electrophoresed on a 10% native PAGE system and immersed in a solution of 50 mM sodium acetate, pH 5.0. H2O2 from a 30% (8 M) stock solution was added to 40 mM, and diaminobenzoic acid dihydrochloride from a freshly prepared 0.8 M solution was added to a final concentration of 80 mM. The gel was gently agitated for 30 min, following which the staining solution was decanted, twice rinsed with water, and then placed in 7% acetic acid. The gel was photographed following the appearance of visible staining.
Sequence analysis.
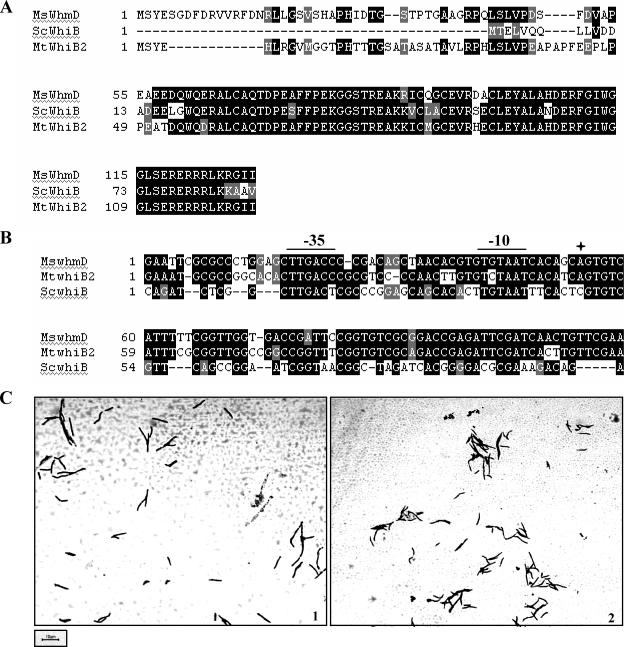
All sequence alignments were performed with the BCM search launcher in the Multiple Sequence Alignment package (Baylor College of Medicine) using the ClustalW 1.8 algorithm. The output files were imported into Boxshade 3.21 (www.ch.embnet.org) to generate the formatted alignments shown in Fig. 2. The secondary-structure prediction for WhmD was carried out with the NPS@ package on the Pole BioInformatique Lyonnais server (http://npsa-pbil.ibcp.fr) and the PSIPRED protein structure prediction server (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html). The boundary coordinates of the 5′ UTR sequences shown in the alignment in Fig. 2B, with reference to their locations upstream of the start codon, are as follows: for M. smegmatis whmD, −187 to −70; for M. tuberculosis whiB2, −185 to −68; and for S. coelicolor whiB, −114 to −13.
FIG. 2.
Orthologues of M. smegmatis whmD rescue M. smegmatis 628-53. (A) Protein sequence alignments of M. smegmatis WhmD (MsWhmD), S. coelicolor WhiB (ScWhiB), and M. tuberculosis WhiB2 (MtWhiB2). (B) Alignments of the 5′ UTRs of M. smegmatis whmD, S. coelicolor whiB, and M. tuberculosis whiB2, showing the positions of the promoter elements. The four-pointed star marks the transcription start site as determined by 5′ rapid amplification of cDNA ends. (C) Morphology upon inducer withdrawal of M. smegmatis 628-53 transformed with plasmids containing S. coelicolor whiB (1) and M. tuberculosis whiB2 (2).
RESULTS
A wild-type copy of M. smegmatis whmD rescues the conditionally complemented whmD mutant M. smegmatis 628-53, providing a genetic assay for screening nonfunctional mutants of WhmD.
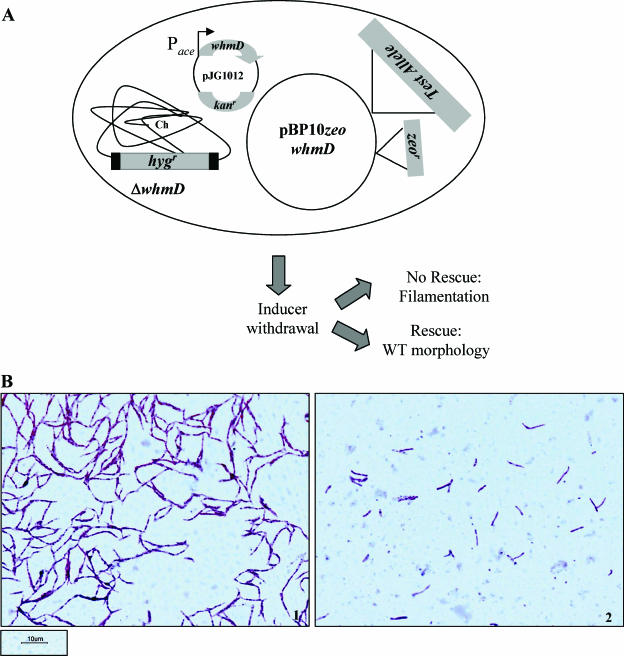
M. smegmatis 628-53, the conditionally complemented whmD deletion mutant, undergoes filamentation under nonpermissive conditions (10). In order to establish a genetic assay to identify mutations that disrupt WhmD function, we developed a complementation system based on M. smegmatis 628-53. To determine if a wild-type (WT) copy of whmD rescues the filamentation defect of the mutant, the gene was cloned under the control of its own promoter into the vector pBP10. This vector exists as a single-copy episome in mycobacterial cells and contains an origin of replication compatible with the pAL5000 replicon (2). This was critical since M. smegmatis 628-53 contains pJG1012, a pAL5000-based plasmid expressing whmD under the control of the acetamidase promoter (Pace). To facilitate selection, a gene encoding Zeocin (zeo) resistance was inserted into pBP10. As depicted schematically in Fig. 1A, transformants of M. smegmatis 628-53 containing either pBP10 zeo whmD or pBP10 zeo alone were cultured in acetamide-containing medium and then subjected to inducer withdrawal as described in the experimental procedures. Following acetamide withdrawal, cells were stained with carbolfuchsin and visualized by light microscopy. Transformants containing the control plasmid were highly filamentous (Fig. 1B, left), whereas no filamentation was seen in those containing the extra copy of whmD (Fig. 1B, right). Cell length measurements were made to quantitate the extent of complementation. The appreciable differences in cell length in control versus WT whmD-complemented cells (Table 1) provided us with a robust system for examining complementation phenotypes of various alleles of whmD.
FIG. 1.
A wild-type copy of M. smegmatis whmD rescues the conditionally complemented whmD mutant M. smegmatis 628-53. (A) Schematic of assay system to screen nonfunctional mutants of WhmD. Ch, chromosome. (B) Morphology upon inducer withdrawal of vector-transformed (1) and whmD-transformed (2) M. smegmatis 628-53.
TABLE 1.
Cell length measurements of transformants of M. smegmatis 628-53 containing the complementing alleles generated in this study
| Vector, complementing allele, or WhmD mutation | Cell length (μm)a | % of cells below cutoffb |
|---|---|---|
| Vector (pBP10 zeo) | 23.28 ± 2.2 | 0 |
| M. smegmatis WhmD | 5.16 ± 0.31 | 100 |
| S. coelicolor WhiB | 7.33 ± 0.48 | 65 |
| M. tuberculosis WhiB2 | 6.95 ± 0.5 | 80 |
| M. smegmatis WhmD mutations | ||
| ΔN42 | 4.55 ± 0.39 | 90 |
| ΔN57 | 5.35 ± 0.46 | 90 |
| ΔN62 | 4.39 ± 0.53 | 90 |
| ΔN67 | 7.12 ± 0.44 | 65 |
| ΔC12 | 6.14 ± 0.33 | 76.6 |
| ΔC23 | 10.51 ± 0.6 | 20 |
| ΔC30 | 22.6 ± 1.59 | 0 |
| C67A | 6.43 ± 0.4 | 63 |
| C90A | ND | |
| C93A | 20.1 ± 1.73 | 0 |
| C99A | 19.44 ± 2.49 | 0 |
| D71A | 24.29 ± 2.29 | 3.33 |
| Y102G | 6.95 ± 0.63 | 66.6 |
| Y102P | 5.66 ± 0.38 | 83.3 |
| G111P | 25.7 ± 1.65 | 0 |
| L116P | 13.2 ± 1.18 | 16.6 |
| R122G | 6.12 ± 0.54 | 83.3 |
| R122P | 5.23 ± 0.22 | 93.3 |
Values represent means ± standard errors of the means (n = 30). ND, not determined.
Percentage of cells below the cell length cutoff of ≤7.5 μm, which defines a filament.
Orthologues of M. smegmatis whmD rescue M. smegmatis 628-53.
The homologues of M. smegmatis whmD in M. tuberculosis and S. coelicolor show extensive identity at the protein level. WhmD shares 70% identity with M. tuberculosis WhiB2 and 69% identity with S. coelicolor WhiB (Fig. 2A). In addition, the three genes also share a high degree of homology in their 5′ and 3′ UTRs (data not shown). To examine if the two genes are truly orthologous to M. smegmatis whmD, the S. coelicolor whiB and M. tuberculosis whiB2 ORFs, including 200 bp of their 5′ UTRs, were cloned into pBP10 zeo and put through the complementation assay described above. As shown in Fig. 2C and Table 1, both S. coelicolor WhiB and M. tuberculosis WhiB2 functionally complement M. smegmatis 628-53, as assessed by rescue of filamentation, implying that these genes are true orthologues of M. smegmatis whmD. The fact that complementation was observed also indicates that the promoter sequences driving the expression of S. coelicolor whiB and M. tuberculosis whiB2 are functional in M. smegmatis. A sequence alignment of the 5′ UTRs of the three genes shows that the putative promoter elements are highly conserved (Fig. 2B).
The N-terminal extension of WhmD is not essential for its activity.
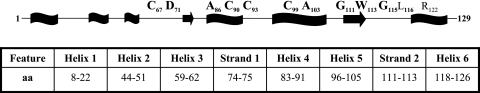
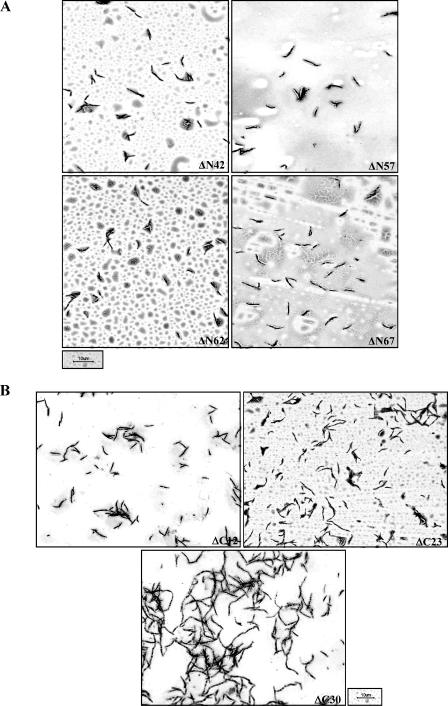
Sequence alignment of WhmD and its orthologues (Fig. 2A) shows that WhmD and WhiB2 carry N-terminal extensions missing in S. coelicolor WhiB and Mycobacterium leprae WhiB2 (data not shown). To clarify if the N-terminal extension contributes significantly to WhmD function, we generated sequential N-terminal deletions in WhmD in pBP10 using a PCR-based strategy. Three of the deletion mutants created (WhmD ΔN42, WhmD ΔN57, and WhmD ΔN62) carried nested deletions of the first three predicted helices. The largest deletion (ΔN67) lacks the first 67 amino acids, including the first of the four conserved cysteine residues, C67 (Fig. 3). All these mutants were tested for their ability to rescue the filamentation phenotype of M. smegmatis 628-53. None of the four mutants were compromised in their ability to complement the mutant (Fig. 4A; Table 1), suggesting that the N-terminal half of WhmD is dispensable for activity, at least under the conditions tested. It was surprising that deletion of the first conserved cysteine had no effect on WhmD function, an observation which was later confirmed by site-directed mutagenesis.
FIG. 3.
Schematic representation of WhmD showing the coordinates of the predicted secondary-structure elements (wavy boxes represent helices, arrows represent β strands, and the line represents the coil) and the amino acid residues mutagenized in this study. Residues in bold are those conserved in all WhiB-like proteins. aa, amino acids.
FIG. 4.
Complementation phenotypes of N-terminal (A) and C-terminal (B) deletion mutants of WhmD.
Truncations in the putative HTH motif of WhmD are deleterious to WhmD function.
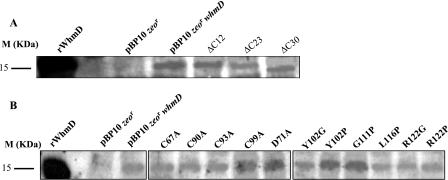
The C terminus of WhmD is predicted to contain a putative HTH-like domain, believed to be involved in DNA binding (21). As shown in Fig. 3, this domain is likely to be comprised of predicted helices 5 and 6, interspersed with a β-turn. To investigate the significance of this domain, we constructed three C-terminal WhmD truncations which progressively deleted portions of this domain and tested them in the complementation assay described above. We observed a progressive debilitation in protein function as deletions in the C terminus (and as a consequence, into the HTH) got larger. The mutant lacking 12 amino acids from the C terminus, which deletes predicted helix 6, retained activity (Fig. 4B, top left, and Table 1), whereas the ΔC23 mutant, which carries a deletion in the predicted turn as well, showed a partial complementation phenotype (Fig. 4B, top right, and Table 1). ΔC30, the largest deletion, which removed part of helix 5 as well as the turn and helix 6, failed to complement the mutant (Fig. 4B, bottom, and Table 1). These observations together suggest that an intact HTH motif is required for the optimal functioning of WhmD. To ensure that the lack of complementation of the deletion mutants tested was not due to the instability of the mutant proteins, we probed cell lysates of M. smegmatis 628-53 transformants containing each complementing allele with polyclonal antisera to WhmD. The autoradiograph in Fig. 5A shows that, under conditions of inducer withdrawal, the levels of each of the C-terminal deletion alleles of whmD were comparable to the levels of these alleles in WT whmD in the positive control, indicating that all these alleles were stable in vivo.
FIG. 5.
Estimation of the stability of complementing mutant alleles of WhmD. Western blot analysis of complementing C-terminal deletion mutant alleles (A) and point mutant alleles (B) of WhmD following inducer withdrawal. M, molecular mass marker; rWhmD, purified recombinant WhmD.
Three of the four conserved cysteine residues and a conserved aspartate are critical to WhmD function.
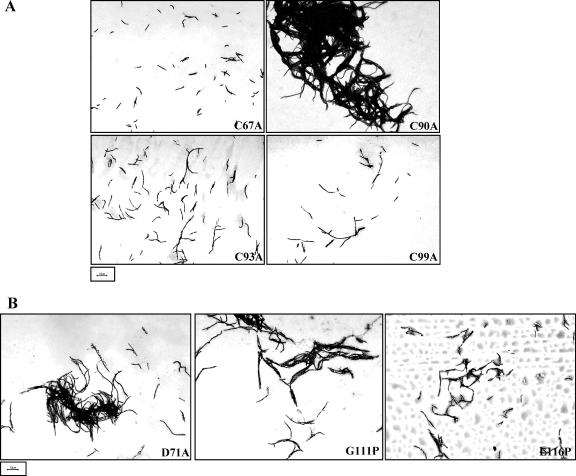
Having broadly mapped the essential boundaries of WhmD, we adopted a site-specific mutagenesis approach to systematically identify residues critical to WhmD activity. The targeted residues included 6 of the 10 residues conserved in all the WhiB-like proteins sequenced to date, as well as a few residues within predicted helices 5 and 6 of the putative DNA binding motif (Fig. 3). To determine the essentiality of the four conserved cysteine residues, we mutagenized each to an alanine residue using the Quik-Change mutagenesis technique and evaluated the ability of each mutant to rescue the filamentation phenotype of the conditionally complemented whmD mutant. Converting C90, C93, and C99 to alanine was clearly deleterious to WhmD activity, whereas the same did not hold true for C67, the N-terminal cysteine (Fig. 6A, top left; Table 1). The C90A mutant, though filamentous, showed an increased propensity to clump, and the few filaments of which measurements could be made had an average cell length of ∼30 μm. The retention of functionality of WhmD C67A was consistent with the observation that the ΔN67 mutant lacking C67 was able to functionally complement M. smegmatis 628-53. Interestingly, D71, a conserved aspartate residue in close proximity to C67, was found to be essential, since a D71A mutation led to inactivation of WhmD (Fig. 6B, left; Table 1).
FIG. 6.
Phenotypic consequences of mutagenizing conserved residues in WhmD. (A) Complementation phenotypes of the four cysteine-to-alanine mutants of WhmD. (B) The inactivation of a conserved aspartate (left) and a glycine (middle) inactivate WhmD. The L116P mutation mimics the inactive S. coelicolor whiB70 allele (right).
Phenotypic effects of helix-destabilizing mutations in putative helices 5 and 6.
Complementation analysis of the C-terminal deletion mutants of WhmD suggested that predicted helix 6 was expendable for WhmD function in the filamentation assay. To confirm this hypothesis, we introduced the helix-disrupting mutations R122G and R122P into WhmD and tested the mutants in the filamentation rescue assay. Both mutants remained functional (Table 1), lending credence to the hypothesis of the nonessentiality of this helix in WhmD activity. Moreover, helix 5 was also able to tolerate the helix-disrupting mutations Y102G and Y102P (Table 1).
The L116P mutation mimics the inactive S. coelicolor whiB70 allele.
whiB70 is an allele of S. coelicor whiB which is inactive due to the conversion of a leucine residue to a proline residue at position 74 in WhiB (8). The corresponding leucine residue in WhmD occurs at position 116 and lies just upstream from a predicted α-helical region. In light of the extraordinarily high degree of identity between WhmD and S. coelicolor WhiB in their C termini, it is realistic to speculate that the two proteins share conserved structural features. To examine if making a cognate mutation to the whiB70 allele leads to inactivation of WhmD, L116 was mutagenized to a proline residue, and the activity of the mutant was assessed. WhmD L116P was only partially functional (Fig. 6B, right, and Table 1), signifying that the perturbations in structure in both M. smegmatis WhmD and S. coelicolor WhiB arising as a consequence of this mutation are likely to be similar. We also observed that a G111P mutation which presumably disrupts the β-turn between the two terminal helices also led to WhmD inactivation (Fig. 6B, middle, and Table 1), implying that this structural element is of functional significance.
Western blot analysis indicated that all the tested point mutant alleles were stable in vivo (Fig. 4B), ruling out the possibility that the protein instability of certain alleles was responsible for their inability to complement M. smegmatis 628-53.
WhmD structure is sensitive to treatment by a reducing agent.
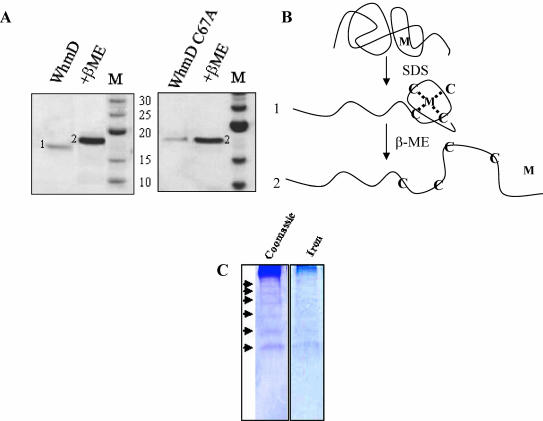
The presence of four conserved cysteine residues suggests that the WhiB-like proteins may be sensitive to redox changes, perhaps through a bound metal atom or through direct sensitivity to oxidation via disulfide bond formation. To determine if either of these possibilities is likely, the whmD ORF was cloned and expressed in the vector pET22b(+) with a C-terminal hexahistidine tag. Following purification from the soluble fraction using conventional metal affinity chromatography, WhmD was electrophoresed on a denaturing SDS-PAGE system. Curiously, a difference in protein mobility was observed between samples lacking or containing the reducing agent β-mercaptoethanol (Fig. 7A, left, lane 1 versus lane 2). The same difference was observed when WhmD was expressed with an N-terminal glutathione S-transferase fusion, indicating that the phenomenon was not an artifact of the expression system (data not shown). We hypothesized that perhaps the reducing agent disrupted the coordination between the cysteine residues and a metal ion, leading to a loss of compaction in protein conformation and a reduction in mobility (Fig. 7B). On treatment of the protein with 1 mM EDTA followed by SDS-PAGE, both forms of the protein were observed (data not shown), consistent with the presence of a bound metal. No difference in mobility was observed when WhmD C67A was electrophoresed under identical conditions (Fig. 7A, right), a result implicit in the fact that the cysteine residues coordinate a metal ion. The same was observed for the three other cysteine mutations, C90A, C93A, and C99A (data not shown). Iron-specific staining (15) of WT WhmD electrophoresed on a 10% native PAGE system (Fig. 7C) strongly suggested that the metal ion coordinated by the cysteine residues was iron. Interestingly, the native gel electrophoretic profile indicated that WhmD forms oligomers and iron staining was observed to be uniform over the entire profile.
FIG. 7.
WhmD structure is sensitive to treatment by a reducing agent. (A) Coomassie staining of purified WhmD (left) or WhmD C67A (right) electrophoresed on a 12% denaturing PAGE gel in the absence (lane 1) and presence (lane 2) of β-mercaptoethanol (β-ME). M, molecular mass markers (kDa). (B) Schematic representation of the effect of the reducing agent on WhmD conformation. Cysteine residues are denoted C, and M is the metal ion. Mobilities of predicted conformations 1 and 2 are indicated in panel A. (C) WhmD contains bound iron. Coomassie blue (left) and Fe-specific (right) staining of WhmD following native gel electrophoresis is shown. Oligomeric forms of WhmD are indicated by the arrowheads.
DISCUSSION
The WhiB-like proteins have been associated with a myriad of functions, including sporulation in S. coelicolor (5, 8), septum formation in M. smegmatis (10), pathogenesis in Mycobacterium marinum (19), transcription in M. tuberculosis (22), antibiotic resistance in the mycobacteria and streptomyces (16), and survival following oxidative stress in C. glutamicum (14). Despite advances in our understanding of the physiological roles of the WhiB-like proteins in the actinomycetes, their structure-function relationships as well as the significance of the conserved set of residues in all these proteins are poorly understood. To address the above issues, we set out to map the essential regions and residues of WhmD and the WhiB-like protein, essential for septation in M. smegmatis. A single-copy complementation assay was developed for M. smegmatis 628-53 to allow screening for nonfunctional mutants of WhmD.
M. tuberculosis whiB2 and S. coelicolor whiB were found to complement the M. smegmatis whmD mutant. The three proteins shared extensive identity towards their C termini, so this observation was no surprise. In addition, since the two homologous genes were cloned into the complementation vector under the control of their own promoters, this indicated that the two heterologous promoter sequences were active in M. smegmatis. Alignment of the three 5′ UTR sequences suggested that this was likely to be due to the near identity of the predicted −10 and −35 elements. Transcription start site mapping confirmed that, for M. smegmatis whmD and M. tuberculosis whiB2, the actual promoter elements are the predicted hexamers shown here (our unpublished data). The complementation analysis allowed us to conclude that S. coelicolor whiB and M. tuberculosis whiB2 are truly orthologous to M. smegmatis whmD. The corollary of this observation is that the results obtained from the functional analysis of WhmD can in principle be extended to the two orthologous proteins as well.
Large deletions in the N-terminal region of WhmD seemed not to compromise protein function, implying that the extended N terminus is apparently superfluous. It is conceivable that this observation is specific to the conditions under which the complementation was performed, and the extension might be required in an independent assay system. It is also possible that WhmD plays other roles beyond regulating septation and that the N terminus is important for one of these nonseptation functions. The result from the largest deletion, which included C67, a conserved residue, was partly unexpected, since we anticipated that all four cysteines would be essential for activity. This observation was initially thought to be an artifact but was later confirmed using a site-directed mutagenesis approach, where three of the four cysteines were found to be indispensable, and WhmD C67A retained its ability to complement M. smegmatis 628-53. In S. coelicolor WhiD, the four cysteine residues are believed to be involved in binding a [4Fe-4S] cluster (13). The functional importance of this cluster was emphasized by the observation that none of the four whiD alleles carrying mutations at these cysteine residues was able to complement the whiD mutant phenotype in S. coelicolor. In this study, we observed that the mobility of purified WhmD changed in response to a reducing agent, an effect not seen in all four mutants of WhmD with cysteine-to-alanine mutations. In addition, WhmD also displayed iron-specific staining, strongly suggesting that in all probability the protein coordinates an iron-sulfur cluster. In light of the nonessentiality of C67, it was interesting that WhmD D71A was inactive, implying that this aspartate was functionally essential. It is conceivable that in the absence of C67, D71 could play a role in binding the cluster along with C90, C93, and C99, as seen in ferredoxin III from Desulfovibrio africanus (9). It is also intriguing that, of the 121 bacterial WhiB-like proteins in the Pfam database, the only protein which shows a deviation from the four-cysteine conservation (Tropheryma whipplei TW 636, Pfam entry Q83NC4) is missing the first cysteine residue. We initially hypothesized, based on the conservation of the aspartate residue, that WhmD could be part of a two-component sensor-response regulator system (6) and that D71 might serve as a phosphor acceptor. This possibility is unlikely since such systems are usually modular (23), and WhmD possesses no sequence resembling the consensus CheY-like phosphorylation site seen in most two-component response regulators (25). Indeed, the present study suggests that WhmD Asp71, while essential, might be required in order to coordinate a metal ion rather than serve as a phosphate acceptor.
Structural evidence strongly suggests that the WhiB-like proteins are DNA binding proteins. The four-conserved-cysteine signature is a common motif in metal-coordinating DNA binding proteins such as Zn-binding GAL4, Fe-binding SoxR, or Hg-binding MerR (18). Secondary-structure analysis of S. coelicolor WhiB showed a potential DNA binding HTH-like motif from residues 64 to 84, and the importance of this motif was emphasized by the loss of WhiB activity in the whiB70 allele in which Leu74 is replaced by a proline residue. In M. smegmatis WhmD, the predicted HTH lies between residues 93 and 126, and the cognate L116P mutation in WhmD led to partial inactivation of protein function, underscoring the importance of this motif. Additional support for the significance of this motif was provided by the observations that a deletion into helix 5 and a mutation in the turn (G111P) were not tolerated. We were puzzled by the retention of activities of the mutants carrying helix-disrupting substitutions in helix 5 (Y102G, Y102P) and helix 6 (R122G, R122P), since these helices are integral parts of the HTH motif. A simplistic explanation is that the protein retains its functional conformation despite the kinks generated by inserting proline residues in the putative helices, as seen in the Saccharomyces cerevisiae heat shock transcription factor (11). The nondeleterious effect of ΔC12 on WhmD was surprising as well, since the deletion removes the terminal helix. If WhmD functions by directly regulating the expression of genes involved in septum formation, then in principle, this deletion mutant should have been unable to rescue filamentation in M. smegmatis 628-53. Since WhmD has the propensity to oligomerize (Fig. 7C), it is plausible that this C-terminal deletion mutant could form hetero-oligomers with other WhiB-like proteins and compensate for the DNA binding defect. All the above issues can be unequivocally resolved by examining the biochemical properties of the mutants constructed in this study.
From the above analyses, we have delineated the essential region of WhmD as lying between residues 68 and 106. This region includes 6 of the 10 residues conserved in all seven M. tuberculosis WhiBs as well as in most members of the WhiB family identified to date. To the best of our knowledge, this is the first comprehensive report documenting the structure-function relationships in the WhiB-like protein family.
Acknowledgments
We gratefully acknowledge the support of NIH grants AI37856, AI36973, AI43846, and AI51668. T.R.R. was the recipient of a postdoctoral fellowship from the Heiser Program.
We thank Keith F. Chater, Helen Kieser, and their colleagues at the John Innes Center, Norwich, United Kingdom, for their invaluable assistance. We thank Rashna Bhandari for helpful discussions and Nisheeth Agarwal for assistance with purification of the cysteine mutants of WhmD.
REFERENCES
- 1.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231-262. [DOI] [PubMed] [Google Scholar]
- 2.Bachrach, G., M. J. Colston, H. Bercovier, D. Bar-Nir, C. Anderson, and K. G. Papavinasasundaram. 2000. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology 146:297-303. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Champness, W. C., and K. F. Chater. 1994. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp., p. 61-94. In P. Piggot, C. P. Moran, Jr., and P. Youngman (ed.), Regulation of bacterial differentiation. ASM Press, Washington, D.C.
- 5.Chater, K. F. 1972. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 72:9-28. [DOI] [PubMed] [Google Scholar]
- 6.Cho, H. S., J. G. Pelton, D. Yan, S. Kustu, and D. E. Wemmer. 2001. Phosphoaspartates in bacterial signal transduction. Curr. Opin. Struct. Biol. 11:679-684. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Davis, N. K., and K. F. Chater. 1992. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol. Gen. Genet. 232:351-358. [DOI] [PubMed] [Google Scholar]
- 9.George, S. J., F. A. Armstrong, E. C. Hatchikian, and A. J. Thomson. 1989. Electrochemical and spectroscopic characterization of the conversion of the 7Fe into the 8Fe form of ferredoxin III from Desulfovibrio africanus. Identification of a [4Fe-4S] cluster with one non-cysteine ligand. Biochem. J. 264:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez, J. E., and W. R. Bishai. 2000. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc. Natl. Acad. Sci. USA 97:8554-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy, J. A., and H. C. Nelson. 2000. Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor. Protein Sci. 9:2128-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopwood, D. A., H. Wildermuth, and H. M. Palmer. 1970. Mutants of Streptomyces coelicolor defective in sporulation. J. Gen. Microbiol. 61:397-408. [DOI] [PubMed] [Google Scholar]
- 13.Jakimowicz, P., M. R. Cheesman, W. R. Bishai, K. F. Chater, A. J. Thomson, and M. J. Buttner. 2005. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe4S] cluster. J. Biol. Chem. 280:8309-8315. [DOI] [PubMed] [Google Scholar]
- 14.Kim, T. H., J. S. Park, H. J. Kim, Y. Kim, P. Kim, and H. S. Lee. 2005. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem. Biophys. Res. Commun. 337:757-764. [DOI] [PubMed] [Google Scholar]
- 15.Kuo, C. F., and I. Fridovich. 1988. A stain for iron-containing proteins sensitive to nanogram levels of iron. Anal. Biochem. 170:183-185. [DOI] [PubMed] [Google Scholar]
- 16.Morris, R. P., L. Nguyen, J. Gatfield, K. Visconti, K. Nguyen, D. Schnappinger, S. Ehrt, Y. Liu, L. Heifets, J. Pieters, G. Schoolnik, and C. J. Thompson. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 102:12200-12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulder, N. J., H. Zappe, and L. M. Steyn. 1999. Characterization of a Mycobacterium tuberculosis homologue of the Streptomyces coelicolor whiB gene. Tuber. Lung Dis. 79:299-308. [DOI] [PubMed] [Google Scholar]
- 18.O'Halloran, T. V. 1993. Transition metals in control of gene expression. Science 261:715-725. [DOI] [PubMed] [Google Scholar]
- 19.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Soliveri, J. A., J. Gomez, W. R. Bishai, and K. F. Chater. 2000. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146:333-343. [DOI] [PubMed] [Google Scholar]
- 22.Steyn, A. J., D. M. Collins, M. K. Hondalus, W. R. Jacobs, Jr., R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 99:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki, M. 1993. Common features in DNA recognition helices of eukaryotic transcription factors. EMBO J. 12:3221-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]