Abstract
Deinococcus geothermalis E50051 forms tenuous biofilms on paper machine surfaces. Field emission electron microscopy analysis revealed peritrichous appendages which mediated cell-to-surface and cell-to-cell interactions but were absent in planktonically grown cells. The major protein component of the extracellular extract of D. geothermalis had an N-terminal sequence similar to the fimbrial protein pilin annotated in the D. geothermalis DSM 11300 draft sequence. It also showed similarity to the type IV pilin sequence of D. radiodurans and several gram-negative pathogenic bacteria. Other proteins in the extract had N-terminal sequences identical to D. geothermalis proteins with conservative motifs for serine proteases, metallophosphoesterases, and proteins whose function is unknown. Periodic acid-Schiff staining for carbohydrates indicated that these extracellular proteins may be glycosylated. A further confirmation for the presence of glycoconjugates on the cell surface was obtained by confocal laser scanning imaging of living D. geothermalis cells stained with Amaranthus caudatus lectin, which specifically binds to galactose residues. The results indicate that the thread-like appendages of D. geothermalis E50051 are glycosylated type IV pili, bacterial attachment organelles which have thus far not been described for the genus Deinococcus.
Deinococcus geothermalis is an important primary biofilm former found in paper machine water (17, 35). It forms tenuous biofilms on abiotic surfaces and is difficult or impossible to remove from the surfaces using industrial washing procedures (18). The attachment of D. geothermalis involves thread-like structures that connect the cells to the abiotic surface while allowing sliding movement, to escape mechanical stress (18).
Capsules and slimes are involved in the adhesion of bacteria onto living and nonliving substrates (6, 15). However, adhesion of D. geothermalis seems to occur in the absence of either of these (18, 29). Another attachment mode by means of pili has been described for several gram-negative pathogenic bacteria, including, for example, Neisseria meningitidis, N. gonorrhoeae (22, 36), and Pseudomonas aeruginosa (38), and for some gram-positive bacteria, such as Enterococcus faecalis (11), Actinomyces spp. (19), and Ruminococcus albus (28). So far there is no experimental evidence that members of the genus Deinococcus produce flagella or pili, even though several genes encoding pilus-associated functions have been found in the genome of D. radiodurans (20). Within the phylum Deinococcus-Thermus, only the Thermus thermophilus strain HB27 has been reported to express pili during natural transformation (8).
This study describes the ultrastructure of the thread-like appendages expressed by the industrially relevant strain D. geothermalis E50051. It is proposed that they represent pili and glycoconjugates necessary for adhesion and biofilm formation of D. geothermalis.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Deinococcus geothermalis E50051 (HAMBI 2411) was originally isolated from paper machine biofilms (35). Sessile cells in this study were harvested from modified R2 (5) agar plates containing the following (per liter): casein digest (0.75 g), soluble starch (0.5 g), yeast extract (0.5 g), K2HPO4 (0.3 g), sodium pyruvate (0.3 g), meat extract (0.25 g), and MgSO4 (0.024 g) at pH 7.0 with the addition of 1.5% agar and grown at 45°C for 48 h. Alternatively, the cells were grown on glass coupons as described below. Planktonic cells were harvested from tryptic soy broth (TSB) grown under agitation (160 rpm) at 45°C for 24 h. D. geothermalis did not form a biofilm in undiluted TSB and only did so in modified R2. For a control, bacteria were also grown on R2 glucose agar plates where starch was replaced by glucose (0.5 g liter−1).
Isolation of cell surface materials.
Plate-grown cells (0.2 g [wet weight]) were collected from the agar surface and suspended in 5 ml of cold TE-PMSF (1 mM EDTA, 0.1 M Tris-HCl, 14 μM phenylmethylsulfonyl fluoride, pH 7.0). The suspension was vigorously vortexed for 4 min and centrifuged (4°C, 20 min, 8,300 × g), and the supernatant was collected. Proteins were then precipitated from the supernatant with 10% trichloroacetic acid at 4°C overnight and pelleted by centrifugation (8,300 × g, 60 min, 4°C). TSB-grown planktonic cells (1.0 g wet weight) were harvested by centrifugation and treated as described above.
Protein identification.
The precipitated proteins were redissolved in sample loading buffer (0.313 M Tris-HCl, pH 6.8, 10% sodium dodecyl sulfate [SDS], 0.05% bromophenol blue, 50% glycerol, 5% mercaptoethanol) and separated on PAGEr Duramide precast 4 to 20% Tris glycine gels (Cambrex, Rockland, Maine), or 7% polyacrylamide gels, at 120 V. Molecular weights of the detected protein bands were calculated with the Quantity One software (Bio-Rad) based on the low-range SDS-polyacrylamide gel electrophoresis (SDS-PAGE) molecular weight marker (Bio-Rad). Gels were electroblotted onto ProBlott membranes (Applied Biosystems) at 10 V for 3 h (4 to 20% gels) or 6 h (7% gels). The membranes were stained with Coomassie brilliant blue R-250 (Bio-Rad) for 1 min and destained in 50% methanol (vol/vol). The N-terminal sequences were analyzed with the 492 Procise protein sequencer (Applied Biosystems). The first 10 N-terminal amino acids were used for matching with the Basic Local Alignment Search Tool provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/).
Glycosylation analysis.
Parallel SDS gels were periodic acid-Schiff (PAS) stained for carbohydrates with a commercial kit (Glyco-Pro; Sigma Aldrich, St. Louis, Mo.) according to the manufacturer's instructions.
FESEM analysis.
Glass coupons (∼1 cm2) were cleaned with detergent (Nelli soap; Farmos, Turku, Finland), rinsed with acetone and distilled water, and then autoclaved. The sterile coupons were mounted in the wells of a 12-well polystyrene plate. The wells were filled with 1 ml of 10× diluted TSB and seeded with the D. geothermalis strain E50051 grown on modified R2A. The polystyrene plate was covered with a lid and shaken (160 rpm) for 1 day at 45°C in the dark. The coupons were removed from the wells, rinsed under running water, and prepared for field emission scanning electron microscopy (FESEM) as described by Raulio et al. (29). Planktonic D. geothermalis E50051 cells were grown in oligotrophic R2 medium (5) for 6 h (45°C, 160 rpm). Cells were harvested by centrifugation (5 min, 1,600 × g), washed with sterile tap water, and prepared for FESEM imaging.
Confocal laser scanning microscopy (CLSM) and fluorescent lectin-binding analysis.
Glass coupons with D. geothermalis biofilms were grown as described for FESEM. Planktonic cells were also grown as described for FESEM (see above) and harvested on membrane filters (pore size, 0.2 μm; Isophore GTBP; Millipore, Ireland). Lectins from Amaranthus caudatus, Erythrina cristagalli, Phaseolus lunatus, and Arachis hypogaea (EY, San Mateo, Calif.) conjugated with fluorescein isothiocyanate were used for detecting surface-expressed glycoconjugates according to the procedure of Neu et al. (23). In brief, the coupons or membranes were covered with 100 μl of lectin solution (100 μg ml−1 in phosphate-buffered saline), incubated for 20 min in the dark at room temperature, rinsed three times with tap water to remove unbound lectins, and then stained with the nucleic acid stain Syto 60 (Molecular Probes, Eugene, Oreg.). An upright TCS SP1 confocal microscope (Leica, Heidelberg, Germany) was used to record fluorescence in the green channel (excitation using an Ar 488-nm laser) and in the far-red channel (excitation using a He/Ne 633-nm laser) and a 63× 0.9 numerical aperture water-immersible lens. An inverted TCS SP2 confocal microscope (Leica) with Ar 488-nm and He/Ne 633-nm lasers, and the 63× 1.4 numerical aperture oil objective was used for recording images of planktonic cells.
RESULTS
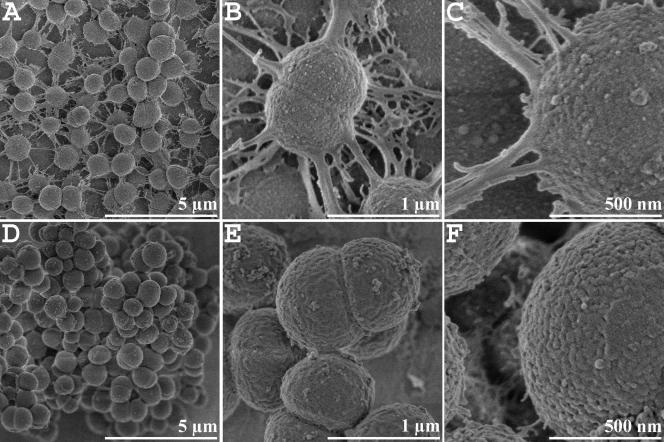
Cells from biofilms of D. geothermalis E50051 and their planktonic counterparts show different ultrastructures in FESEM (Fig. 1). Cells growing as a biofilm on a glass surface displayed numerous appendages of variable lengths and thicknesses (Fig. 1A to C). A network of thread-like structures seems to mediate attachment of the Deinococcus cells to the abiotic surface and to the neighboring cells as well. The appendages were present on dividing and nondividing sessile cells (Fig. 1B), but no appendages were detected on dividing or nondividing planktonic cells (Fig. 1D to F).
FIG. 1.
FESEM images of sessile (A, B, and C) and planktonic (D, E, and F) cells of D. geothermalis E50051. Planktonic cells lack adhesion threads, which mediate surface and cell-to-cell attachment of D. geothermalis. The sessile cells were grown on glass slides as described in Materials and Methods.
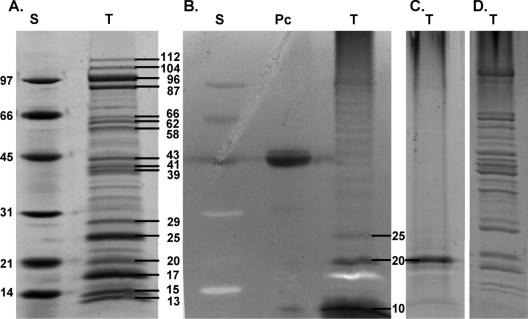
Adhesion threads were also detected by FESEM on cells grown on agar plates (not shown). As such, cells could easily been grown in larger amounts; therefore, agar cultures were used for further characterization of these structures. The adhesion threads could be extracted from agar cultures of D. geothermalis E50051 by suspending the cells in TE-PMSF and vigorous vortexing, followed by sedimentation of the cells. The supernatants were further analyzed by one-dimensional SDS-PAGE, followed by protein and carbohydrate staining. Surprisingly, the extracts contained a number of proteins with molecular masses ranging from 13 to 112 kDa (Fig. 2A and D). In contrast, these proteins were not detected in extracts from planktonic cells which were prepared in the same way (data not shown). Therefore, we can conclude that these proteins represent material based on the characteristic surface structures of the sessile cells of D. geothermalis. Carbohydrate-specific PAS labeling stained most intensively the protein bands with molecular masses of 20 kDa and 25 kDa (Fig. 2B). Additionally, 14 bands, ranging in size from 35 to 112 kDa, were also positively stained, though less intensely. The staining procedure employed has a detection limit of about 25 to 100 ng of carbohydrates, depending on the nature and the degree of glycosylation of proteins (according to the supplier's information). Based on the data shown in Fig. 1 and 2, we propose that the thread-like materials of sessile cells consist of several proteins, some of which may be glycosylated.
FIG. 2.
Protein and carbohydrate analyses of extracts obtained by mechanical agitation of sessile cells of D. geothermalis 50051 isolated from agar plates. The extracts (T) were prepared from 200 mg (wet weight) of bacterial cells and separated with SDS-PAGE. (A) Coomassie brilliant blue-stained gel; (B and C) parallel gel stained with PAS dye. The gel in panel D was stained with both Coomassie blue and PAS. D. geothermalis was grown in modified R2A with starch (A and B) or in R2A where starch was replaced by glucose (C and D). Molecular masses (in kDa) were calculated based on a molecular mass standard (S, low range; Bio-Rad). Horseradish peroxidase (Sigma Aldrich) was used as positive control for glycosylated protein (Pc).
Interestingly, a major band of about 10 kDa was heavily stained with PAS but was not stained with Coomassie blue. This band was absent in a control experiment, where D. geothermalis was cultivated on R2A plates with glucose as carbon source instead of starch (Fig. 2C). Also, this band was not detected from extracts prepared from noninoculated R2A plates; thus, it was not a component of the starch-containing culture medium. Since the other protein bands were detected from cells which were grown in R2 medium with starch and glucose as well, we conclude that the 10-kDa band corresponds to a starch-derived deinococcal metabolite.
N-terminal sequencing was performed for all protein bands, which are labeled with their molecular weights in Fig. 2A, in order to obtain more information about the nature of these proteins. Six different N-terminal sequences were obtained from the 16 bands analyzed (Table 1). The most abundant protein bands, ranging from 17 kDa to 29 kDa, shared the same N-terminal sequence, MTLIELLIVI. For the bands at 25 and 29 kDa, a longer N-terminal sequence, MTLIELLIVIAIIGILAAVL, was obtained. Both sequences are highly similar to the fimbrial protein pilin sequence (ZP_00395377) found in the Deinococcus geothermalis DSM 11300 draft sequence (GenBank access code NZ_AAHE01000000) (30), differing only by the first amino acid, which is methionine in the D. geothermalis E50051 protein but phenylalanine in the draft sequence of strain DSM 11300. The obtained sequences are also highly similar to two putative type IV pilins annotated in the genome of D. radiodurans R1 and differ only by the first amino acid (NP_294956) or by the eighth amino acid, which is valine instead of isoleucine in D. radiodurans (NP_294271). Based on these data, we suggest that at least some of the threads observed in sessile D. geothermalis E50051 represent type IV pili.
TABLE 1.
Identification of N-terminal amino acid sequences of the protein bands shown in Fig. 2A
| Protein band | Sequence identification according to annotated D. geothermalis DSM 11300 draft sequencea
|
|||
|---|---|---|---|---|
| Calculated size (kDa) | N-terminal sequence | Identity | Sequence similarity (%) | GenBank accession no. |
| 87, 96, 104, 112 | APTPNTTSTL | Protein of unknown function DUF11b | 100 | ZP_00397476 |
| 87 | MFYAKGMTGA | Peptidases S8 and S53, subtilisin, kexin, sedolisin; protease-associated; protein of unknown function DUF1034 | 100 | ZP_00396844 |
| 58, 62, 66 | APLTVTILHT | Metallophosphoesterase, 5′-nucleotidase | 100 | ZP_00397507 |
| 25, 29 | MTLIELLIVIAIIGILAAVL | Fimbrial protein pilin | 95 | ZP_00395377 |
| PilA (Thermus thermophilus HB27) | 90 | YP_004829 | ||
| Pilin, type IV, putative (D. radiodurans R1) | 95 | NP_294271 | ||
| 17, 20 | MTLIELLIVI | Fimbrial protein pilin | 90 | ZP_00395377 |
| PilA (Thermus thermophilus HB27) | 90 | YP_004829 | ||
| Pilin, type IV, putative (D. radiodurans R1) | 90 | NP_294956 | ||
| 15 | ASVGAYFGTD | Hypothetical protein DgeoDRAFT_2095 | 100 | ZP_00395407 |
| 13 | INLSLTSGVG | Hypothetical protein DgeoDRAFT_2025 | 100 | ZP_00395337 |
GenBank accession no. NZ_AAHE01000000.
High homology to other surface lipoproteins from gram-positive bacteria.
The extracellular extract of the sessile cells also contained other proteins. According to their N-terminal sequences, they are family members of serine proteases and metallophosphoesterases, based on a 100% homology to proposed proteins in the D. geothermalis DSM 11300 draft sequence. Also, the 13- and 15-kDa proteins as well as the four bands (87 kDa, 96 kDa, 104 kDa, and 112 kDa) with the same N-terminal sequence, APTPNTTSTL, had counterparts in the draft sequence, but their function is thus far unknown. The 87-kDa band consisted of two comigrating proteins which were separated as individual proteins on a 7% SDS gel (not shown). Analysis of 43-kDa, 41-kDa, and 39-kDa bands failed due to a blocked N terminus, possibly caused by glycosylation.
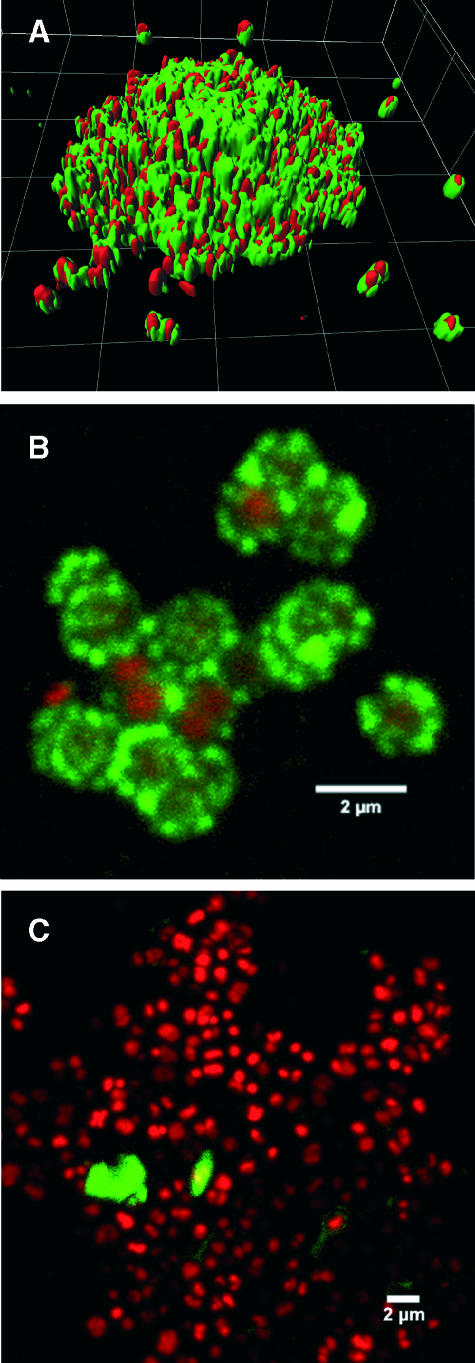
Living, fully hydrated biofilms of D. geothermalis strain E50051 were analyzed by CLSM using fluorescent lectin-binding analysis. After lectin staining of cell surface glycoconjugates, the bacterial cells were counterstained with a general nucleic acid-specific fluorochrome to locate the cell body. Figure 3A shows that Amaranthus caudatus lectin interacts strongly with D. geothermalis microcolonies. The lectin indicated glycoconjugates with a petal-like appearance around the coccal bacterial cells (Fig. 3B). The appendages on the deinococcal cell surface appear bulkier in CLSM than in FESEM, reflecting the fully hydrated state of the CLSM sample. As a control, Fig. 3C shows lectin- and nucleic acid-stained planktonic cells. This image reveals the complete absence of lectin-reactive components on the cell surface. The lectins of Erythrina cristagalli, Phaseolus lunatus, and Arachis hypogaea did not bind to the deinococcal biofilms.
FIG. 3.
CLSM images of D. geothermalis E50051 cells grown on glass surfaces (A and B) and in liquid medium (C). The specimens were double stained with lectin (Amaranthus caudatus-fluorescein isothiocyanate [green]) and Syto 60 (red). Lectin binding indicates the locations of glycoconjugates, and Syto 60 is specific for nucleic acids. Grid size in panel A, 5 μm.
DISCUSSION
FESEM and CLSM after lectin staining showed that the surface structures of sessile cells of D. geothermalis E50051 differ from their planktonic counterparts. The images show that the existence of cell surface appendages is connected to the bacterial growth mode, namely to adhesion and biofilm formation, but they were not observed either in growing or stationary-phase planktonic cells. Recently, Raulio et al. (29) proved the importance of these appendages for attachment by FESEM analysis. A destruction of these appendages by photolytic activity resulted in massive detachment of the cells growing as a biofilm on coated steel surfaces.
A detailed analysis of the protein composition showed that the material released by mechanical agitation from the surface of sessile D. geothermalis cells contained four pilin proteins. This is concluded from the high homology of their N-terminal sequences to the putative pilins predicted in the genome sequences of D. geothermalis DSM 11300 (95% homology [reference 30]) and D. radiodurans (90% homology) and the homology to the pilA gene of Thermus thermophilus (90% homology). Pilins are structural components of type IV pili (14, 25). The N-terminal domain of pilin is conserved interspecifically (12, 14, 33), which explains why the homology searches also showed significant similarity to the type IV pilins of gram-positive bacteria as well as to several gram-negative pathogens. Typically, the primary translation product (prepilin) includes a short leader sequence which is cleaved prior to assembly into the fimbrial strand, an unusually modified amino acid as the first residue of mature pilin, which functions as a cleavage site, and a conserved glutamate residue at position 5 in the mature protein (14). Similarly to known pilins, the N-terminal sequences of the described proteins lack seven amino acids included in the prepilin open reading frame of D. geothermalis DSM 11300 and have glutamate as the fifth amino acid. Interestingly, the N-terminal sequence of the analyzed proteins contained as the first amino acid methionine, instead of modified phenylalanine, which has been reported for most type IV pilins (1, 14, 33). However, a variety of hydrophobic amino acids can be tolerated at this position (33), e.g., methionine is also the first amino acid after the cleavage site of one of the type IV pilins of D. radiodurans (NP_294271) and of the Vibrio cholerae (AAL09686) pilin.
We could not detect the proteins with the typical N-terminal sequence for type IV pili in planktonic cells. Therefore, we conclude that D. geothermalis produces these proteins only when growing as sessile cells. This is well in agreement with the data from electron microscopy, which show the typical appendages only on sessile cells (Fig. 1). Therefore, it is likely that these appendages that mediate surface attachment of D. geothermalis are type IV pili. This concurs with the current view that IV pili possess versatile functions in connection with colonization and adhesion to both biotic and abiotic surfaces (9, 24). Type IV pili are also involved in bacterial motility (21, 37), DNA transfer (7, 8, 14), and bacteriophage infection (2). Additionally, type IV pili are closely related to the type II protein secretion system (26, 31). Interestingly, the extracellular extracts of D. geothermalis contained also a serine-like protease and three forms of a 5′-nucleotidase. Such proteins are known to be secreted by type II secretion systems and are expressed during nutrient limitation (31). The very high homology with many other bacterial 5′-nucleotidases suggests that this enzyme may be engaged in peptidoglycan and exopolysaccharide synthesis. The detected metallophosphoesterase with 5′-nucleotidase activity also might be engaged in the pili retraction system, allowing the motility of cells with type IV pili. Recently, it was confirmed that a PilT homolog of Aquifex aeolicus has nucleotide phosphorylase activity (13). PilT is proposed to be included in a rotary system which is hypothesized to work similarly as the F1-ATPase rotary motor (a model proposed by Oster was reported in reference 16).
The 20- and 25-kDa pilin-like proteins stained positively in response to PAS, indicating that they are glycosylated. In contrast, the pilin-like protein band at 17 kDa gave a clear negative signal (white band). Attached carbohydrates increase the molecular mass of the protein and may have thus led to a slower migration on SDS gels and may explain the deviation from the predicted molecular mass (13.4 kDa). Aside from different glycosylation levels, the 25- and 29-kDa pilin-like proteins might represent dimers, which were not resolved under the experimental conditions used for their monomers.
Confocal laser scanning microscopy and fluorescent lectin-binding analysis of live cells confirmed the glycoconjugate nature of the deinococcal cell surface. CLSM images of D. geothermalis showed that Amaranthus caudatus lectin strongly associated with adhesion threads of deinococcal cells (Fig. 3) and, therefore, the images indicate that the surface appendages of live cells are the location of glycoconjugates both in individual surface-adhered cells and inside biofilm colonies. Previous studies (18, 29) showed that an extensive slime matrix is absent in D. geothermalis biofilms, which discards the possibility that lectins may bind matrix exopolysaccharides. According to the manufacturer, Amaranthus caudatus lectin has specificity for oligosaccharides containing galactosyl Galβ(1,3)GalNAc α-units. Previously, galactose-containing glycoconjugates have been reported from pili of Neisseria meningitidis (10). A number of reports have described glycosylation of type IV pilins (4, 25, 27, 36), which is considered to be variable and a highly regulated posttranslational modification (34) that enhances adherence and also protects the cell from environmental stress (25, 32). Apart from the pili described, cell surface glycoconjugates may be also important for Deinococcus attachment and biofilm development.
The detection of type IV pili in Deinococcus is interesting in light of the hypothesis recently discussed by Nudleman and Kaiser (24) that pili which are widely distributed in the β-, γ-, and δ-proteobacteria may have originated from a common ancestor (also suggested by Alm and Mattick [1]). Deinococcus is one of the most ancient bacterial groups. Together with recent investigations which show that a PilT homolog that possesses nucleotide phosphorylase activity (typical for PilT in type IV pili complexes [13]) also exists in an even older thermophilic eubacterium, Aquifex aeolicus, these findings seem to support the ancestor hypothesis.
Although Deinococcaceae are considered nonmotile, a previous study described the ability of D. geothermalis E50051 to slide away from mechanical stress (18). Similarly, adhesion pili from uropathogenic Escherichia coli are capable of a structural transition which allows the pilus to unravel and extend plastically (3). Glycosylated pili may additionally protect the cells sterically from hostile environments (29). In conclusion, it is suggested that pilus and glycoconjugate production of sessile D. geothermalis cells can explain the firm attachment to surfaces, the persistence of this bacterium in biofilms, and the gliding ability and finally may offer a target for the design of more efficient antifouling treatments.
Acknowledgments
This work was kindly supported by the PINTA program of the Finnish Funding Agency for Technology and Innovation (TEKES; Biofouling and Shine-Pro projects).
We thank Hongmin Tu from the Biocenter Oulu core facility for the N-terminal sequencing.
REFERENCES
- 1.Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. [DOI] [PubMed] [Google Scholar]
- 2.Bradley, D. E. 1974. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology 58:149-163. [DOI] [PubMed] [Google Scholar]
- 3.Bullitt, E., and L. Makowski. 1995. Structural polymorphism of bacterial adhesion pili. Nature 373:164-167. [DOI] [PubMed] [Google Scholar]
- 4.Castric, P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141:1247-1254. [DOI] [PubMed] [Google Scholar]
- 5.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton. 2005. Standard methods for the examination of water and wastewater, p. 9-36. American Public Health Association, Washington, D.C.
- 6.Costerton, J. W., R. T. Irvin, and K. J. Cheng. 1981. The bacterial glycocalyx in nature and disease. Annu. Rev. Microbiol. 35:299-324. [DOI] [PubMed] [Google Scholar]
- 7.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich, A., J. Rumszauer, A. Henne, and B. Averhoff. 2003. Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl. Environ. Microbiol. 69:3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giltner, C. L., E. J. van Schaik, G. F. Audette, D. Kao, R. S. Hodges, D. J. Hassett, and R. T. Irvin. 2006. The Pseudomonas aeruginosa type IV pilin receptor binding domain functions as an adhesin for both biotic and abiotic surfaces. Mol. Microbiol. 59:1083-1096. [DOI] [PubMed] [Google Scholar]
- 10.Hamadeh, R. M., M. M. Estabrook, P. Zhou, G. A. Jarvis, and J. M. Griffiss. 1995. Anti-Gal binds to pili of Neisseria meningitidis: the immunoglobulin A isotype blocks complement-mediated killing. Infect. Immun. 63:4900-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handley, P. S., and A. E. Jakobs. 1981. Some structural and physiological properties of fimbria of Streptococcus faecalis. J. Gen. Microbiol. 127:289-293. [DOI] [PubMed] [Google Scholar]
- 12.Hazes, B., P. A. Sastry, K. Hayakawa, R. J. Read, and R. T. Irvin. 2000. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J. Mol. Biol. 299:1005-1017. [DOI] [PubMed] [Google Scholar]
- 13.Herdendorf, T. J., D. R. McCaslin, and K. T. Forest. 2002. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J. Bacteriol. 184:6465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 15.Jones, H. C., I. L. Roth, and W. M. Sanders III. 1969. Electron microscopic study of a slime layer. J. Bacteriol. 99:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser, D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:R777-R780. [DOI] [PubMed] [Google Scholar]
- 17.Kolari, M., J. Nuutinen, and M. S. Salkinoja-Salonen. 2001. Mechanisms of biofilm formation in paper machine by Bacillus species: the role of Deinococcus geothermalis. J. Ind. Microbiol. Biotechnol. 27:343-351. [DOI] [PubMed] [Google Scholar]
- 18.Kolari, M., U. Schmidt, E. Kuismanen, and M. S. Salkinoja-Salonen. 2002. Firm but slippery attachment of Deinococcus geothermalis. J. Bacteriol. 184:2473-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, T., M. K. Khah, S. Slavnic, I. Johansson, and N. Stromberg. 2001. Different type 1 fimbrial genes and tropisms of commensal and potentially pathogenic Actinomyces spp. with different salivary acidic proline-rich protein and statherin ligand specificities. Infect. Immun. 69:7224-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 22.Merz, A. J., and M. So. 2000. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16:423-457. [DOI] [PubMed] [Google Scholar]
- 23.Neu, T., G. D. Swerhone, and J. R. Lawrence. 2001. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 147:299-313. [DOI] [PubMed] [Google Scholar]
- 24.Nudleman, E., and D. Kaiser. 2004. Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 7:52-62. [DOI] [PubMed] [Google Scholar]
- 25.Parge, H. E., K. T. Forest, M. J. Hickey, D. A. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378:32-38. [DOI] [PubMed] [Google Scholar]
- 26.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 27.Power, P. M., L. F. Roddam, M. Dieckelmann, Y. N. Srikhanta, Y. C. Tan, A. W. Berrington, and M. P. Jennings. 2000. Genetic characterization of pilin glycosylation in Neisseria meningitidis. Microbiology 146:967-979. [DOI] [PubMed] [Google Scholar]
- 28.Rakotoarivonina, H., G. Jubelin, M. Hebraud, B. Gaillard-Martinie, E. Forano, and P. Mosoni. 2002. Adhesion to cellulose of the gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiology 148:1871-1880. [DOI] [PubMed] [Google Scholar]
- 29.Raulio, M., V. Pore, S. Areva, M. Ritala, M. Leskela, M. Linden, J. B. Rosenholm, K. Lounatmaa, and M. Salkinoja-Salonen. 2006. Destruction of Deinococcus geothermalis biofilm by photocatalytic ALD and sol-gel TiO2 surfaces. J. Ind. Microbiol. Biotechnol. 33:261-268. [DOI] [PubMed] [Google Scholar]
- 30.Richardson, P. 2005. Annotation of the draft genome assembly of Deinococcus geothermalis DSM 11300. [Online.] htpp://genome.ornl.gov/microbial/dgeo/15apr04/dgeo_N_cogs.html.
- 31.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smedley, J. G., III, E. Jewell, J. Roquskie, J. Horzempa, A. Syboldt, D. B. Stoltz, and P. Castric. 2005. Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect. Immun. 73:7922-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom, M. S., and S. Lory. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa. J. Biol. Chem. 266:1656-1664. [PubMed] [Google Scholar]
- 34.Tuomanen, E. I. 1996. Surprise? Bacteria glycosylate proteins too. J. Clin. Investig. 98:2659-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Väisänen, O. M., A. Weber, A. Bennasar, F. A. Rainey, H. J. Busse, and M. S. Salkinoja-Salonen. 1998. Microbial communities of printing paper machines. J. Appl. Microbiol. 84:1069-1084. [DOI] [PubMed] [Google Scholar]
- 36.Virji, M., H. Kayhty, D. J. Ferguson, C. Alexandrescu, J. E. Heckels, and E. R. Moxon. 1991. The role of pili in the interactions of pathogenic Neisseria with cultured human endothelial cells. Mol. Microbiol. 5:1831-1841. [DOI] [PubMed] [Google Scholar]
- 37.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 38.Woods, D. E., D. C. Straus, W. G. Johanson, Jr., V. K. Berry, and J. A. Bass. 1980. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect. Immun. 29:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]