Abstract
The Smu0630 protein (AtlA) was recently shown to be involved in cell separation, biofilm formation, and autolysis. Here, transcriptional studies revealed that atlA is part of a multigene operon under the control of at least three promoters. The morphology and biofilm-forming capacity of a nonpolar altA mutant could be restored to that of the wild-type strain by adding purified AtlA protein to the medium. A series of truncated derivatives of AtlA revealed that full activity required the C terminus and repeat regions. AtlA was cell associated and readily extractable from with sodium dodecyl sulfate. Of particular interest, the surface protein profile of AtlA-deficient strains was dramatically altered compared to the wild-type strain, as was the nature of the association of the multifunctional adhesin P1 with the cell wall. In addition, AtlA-deficient strains failed to develop competence as effectively as the parental strain. Mutation of thmA, which can be cotranscribed with atlA and encodes a putative pore-forming protein, resulted in a phenotype very similar to that of the AtlA-deficient strain. ThmA was also shown to be required for efficient processing of AtlA to its mature form, and treatment of the thmA mutant strain with full-length AtlA protein did not restore normal cell separation and biofilm formation. The effects of mutating other genes in the operon on cell division, biofilm formation, or AtlA biogenesis were not as profound. This study reveals that AtlA is a surface-associated protein that plays a critical role in the network connecting cell surface biogenesis, biofilm formation, genetic competence, and autolysis.
Bacteria produce a variety of enzymes involved in the modification and degradation of peptidoglycan, including N-acetyglucosaminidases, N-acetylmuramidases, N-acetylmuramyl-l-alanine amidases, endopeptidases, and transglycosylases (31, 54, 71, 80). Some of these enzymes are known as autolysins because they digest the cell wall when cells are exposed to unfavorable conditions (70, 71, 80). The presence of these enzymes may not be sufficient for lysis, and additional factors may be required to activate or regulate the activity of the autolysins (44), presumably to protect the cells from the lytic activity of the enzymes. Peptidoglycan hydrolases have also been demonstrated to play critical roles in cell wall turnover, cell growth, antibiotic resistance, cell-to-surface adhesion, genetic competence, and protein secretion (9, 23, 28, 51, 74), as well as contributing to virulence (8, 86). In many bacteria, a correlation of a lack of peptidoglycan hydrolase(s) activity and a failure in cell separation has been reported (35, 60, 77, 87).
An increasing number of peptidoglycan hydrolases have been identified in gram-positive bacteria, including Bacillus subtilis (37, 39, 48, 59, 74), Lactococcus lactis (11, 12, 22, 34, 75), Listeria monocytogenes (57, 79, 86), Staphylococcus aureus (19, 36, 53, 78), Enterococcus faecalis (15, 16, 55, 56, 72, 73), and Streptococcus pneumoniae (8, 18, 20, 83). Recently, during a search for surface proteins of Streptococcus mutans involved in biofilm formation, we identified a structurally interesting protein (Smu0630) that was shown to be essential for efficient formation of stable biofilms by this organism (10). Smu0630 contained a typical signal sequence and two sets of large repeated domains in the N-terminal two-thirds of the protein. The C-terminal domain contained a glycohydrolase-like domain, and apparent orthologues of the protein were found only in a few streptococci (10). In addition to the profound effect on biofilm formation, loss of Smu0630 was revealed to cause excessive chaining of the bacteria. Subsequently, Shibata et al. identified the same protein in a screen for peptidoglycan-degrading enzymes, confirmed our observations on biofilm formation and chaining, and provided evidence that the protein was an autolysin that they designated atlA (69). Here, we dissect the transcriptional organization of a four-gene operon containing atlA and examine structure-function relationships in the AtlA protein. We also disclose the involvement of gene products in the atlA operon in AtlA biogenesis and maturation and reveal a critical role for AtlA in generation of a normal cell surface. This study provides novel insights into the expression and regulation of an apparent autolytic enzyme that is essential for cellular processes required for virulence expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Escherichia coli DH10B was grown in Luria broth, and S. mutans strain UA159 and its derivatives were grown in brain heart infusion (BHI) broth (Difco). For selection of antibiotic-resistant colonies after genetic transformation, ampicillin (100 μg ml−1 for E. coli), erythromycin (300 μg ml−1 for E. coli or 10 μg ml−1 for S. mutans), kanamycin (50 μg ml−1 for E. coli or 1 mg ml−1 for S. mutans), or spectinomycin (1 mg ml−1 for S. mutans) were added to the media. For biofilm formation assays, S. mutans strains were grown in the semidefined biofilm medium [BM; 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4)2SO4, 35 mM NaCl, 0.8% (wt/vol) glucose, 0.2% (wt/vol) casamino acids, and 100 mM MnCl2 · 4H2O (pH 7.4)] (47) supplemented with glucose at a final concentration of 20 mM. Plasmid pDL278 (40), an E. coli-Streptococcus shuttle vector carrying a spectinomycin resistance (Spr) gene that confers resistance to spectinomycin in both organisms, was used to measure transformation efficiency.
Construction of mutant strains.
Primers used for deletion mutagenesis are listed in Table 1. To make deletions in the smu0631, pepT, and thmA, genes, 5′ and 3′ flanking regions of each gene were amplified from chromosomal DNA of S. mutans UA159, ligated together using BamHI sites designed into each primer set, and cloned into a pGEM-T Easy vector (Promega, Madison, WI). Plasmids were digested with BamHI and ligated to a nonpolar (NPKm) or polar (ΩKm) kanamycin cassette from pALH124 or pVT924, respectively, that was digested with the same enzyme (2). Also, a plasmid was digested with BamHI, blunt-ended with T4 DNA polymerase, and replaced by a polar erythromycin cassette (Emr) gene (1). The mutagenic plasmids were used to transform S. mutans UA159 or 630NP, which carries a nonpolar deletion/insertion in the atlA gene (10). Transformants were selected on BHI agar containing kanamycin or erythromycin or both, and double-crossover recombination into each gene was confirmed by PCR and sequencing. The mutant strains of S. mutans constructed in this study are listed in Table 2.
TABLE 1.
Primers used for construction of deletion mutants, amplification of the putative promoters for cat fusion, real-time PCR, and construction of histidine-tagged AtlA derivatives in this study
| Primer for 5′ end amplicon | Target | Primer for 3′ end amplicon
|
||
|---|---|---|---|---|
| Name and function | Nucleotide sequence (5′-3′) | Name | Nucleotide sequence (5′-3′) | |
| For deletion mutants | ||||
| ciaR-flanking-RV | GCCAATGAGTCTCTTCCATGA | Deletion of smu0631 | ciaH-BamHI-C | CAAGGTATTGGATCCGAGGATAAGA |
| ciaR-BamHI-B | TATTGACATGGATCCTGACATTCTC | ciaH-D2 | TTGCCAGAGACATTTGGAAAG | |
| ciaH-A | TCGAAATATCCCAAGTCATGC | Deletion of pepT | ciaH-BamHI-C | CAAGGTATTGGATCCGAGGATAAGA |
| ciaH-BamHI-B | TATTGACATGGATCCTGACATTCTC | ciaH-D1 | TGAGAAAGACTTGCCAAATATGTTA | |
| ciaR-flanking-RV | GCCAATGAGTCTCTTCCATGA | Deletion of thmA | ciaR-BamHI-C | TAGATAGACGGATCCCGTCTTCTAC |
| ciaR-BamHI-B | AGTGTAGGAGGATCCTTGAAGGAT | ciaR-flanking-FW | CCACCCTTTTGTCGTGTTCT | |
| For amplification of the putative promoters | ||||
| PatlA-EcoRI-FW | TTGAGTAATGAATTCGAAAGGAAAC | PatlA (promoter) | PatlA-BamHI-RV | GCTTTTCATGGATCCTCC TAATTT |
| PpepT-EcoRI-FW | GCCAGCCAAGAATTCCTTATCTAAT | PpepT (promoter) | PpepT-BamHI-RV | ATAAGTCATGGATCCCCTCTTACC |
| PthmA-EcoRI-FW | GGAACAGATGAATTCAAAATTTCCT | PthmA (promoter) | PthmA-BamHI-RV | CTTAATCACGGATCCTCCTCCATG |
| For RT-PCR or real-time RT-PCRa | ||||
| 630-sense | TCAACATCGTCTGCTCCTTTGG | atlA (amplicon) | 630-antisense | GTGCCTGTACTTGCTTGTCTTG |
| pepT-sense | CGTTGATGGCGGTCCTTTAGG | pepT (amplicon) | pepT-antisense | GCTGTACCCGGATGAACATTGC |
| thmA-sense | AGAGGTAACAAAGTCAACTGTG | thmA (amplicon) | thmA-antisense | TCTGGATGAAGTGCCAAAGC |
| For construction of His-tagged AtlA derivativesb | ||||
| pET630-S-BamHI-FW | GACCCTATTGGATCCTGCTAATAAA | pET630-RV | CAAACGCCTCCTAGGTTTACTGTT | |
| pET630-2R-BamHI-FW | TGGACAAAGGGATCCTGACAATAAA | pET630 + 1R-AvrII-RV | CGACAATTTCCTAGGAATACCACTT | |
| pET630-1R-BamHI-FW | ACCTTAAGTGGGATCCACCAAAAATT | pET630 + 2R-AvrII-RV | AAGCAGCTACCTAGGCTTCTTTGA | |
Additional primers used for RT-PCR were as follows: smu0630-rt-FW, GGATATACGCGGTGGGCAAG; smu0631-rt-FW, GGTCTGACAACCCCTGGAATG; smu0631-rt-RV, GGGAGCAAGGGTTTCCTCGT.
Primer sets were employed as follows: pET630-S-BamHI-FW/pET630-RV for 630D1; pET630-S-BamHI-FW/pET630 plus 1R-AvrII-RV for 630D2; pET630-S-BamHI-FW/pET630 + 2R-AvrII-RV for 630D3; pET630-2R-BamHI-FW Forward/pET630-RV for 630D4, and pET630-1R-BamHI-FW/pET630RV for 630D5.
TABLE 2.
The S. mutans strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| UA159 | Wild type | |
| 630NP | atlA::NPKmr | |
| 630P | atlA::ΩKmr | |
| SAB61 | Δsmu0631::ΩKmr | This study |
| SAB66 | ΔpepT::ΩKmr | This study |
| SAB67 | ΔpepT::NPKmr | This study |
| SAB71 | ΔthmA::ΩKmr | This study |
| SAB78 | ΔthmA::EmratlA::NP Kmr | This study |
| SJ256 | UA159:PatlA-cata | This study |
| SJ257 | UA159:PpepT-cata | This study |
| SJ258 | UA159:PthmA-cata | This study |
Promoter fused to cat gene.
To construct a reporter gene fusion for measuring the transcription from the 5′ regions of atlA, pepT, and thmA, fragments containing the putative promoter regions of these genes were cloned into the EcoRI and BamHI sites in front of a promoterless chloramphenicol acetyltransferase (CAT) gene (cat) in pGEM3cat (1). The gene fusions were released using EcoRI and HindIII digestion, treated with T4 DNA polymerase, and then cloned into the integration vector pBGK2 (81, 82) at the SmaI site. The resulting CAT fusion vectors were integrated into the gtfA gene in single copy to create strains SJ256, SJ257, and SJ258, carrying atlA, pepT, and thmA promoters, respectively. Double-crossover recombination of the reporter gene fusion into the S. mutans chromosome was confirmed by PCR amplification using primers internal to gtfA. GtfA is a sucrose phosphorylase that is not required for virulence, normal growth, adherence, or biofilm formation.
Growth phenotypes.
Growth of strains in BHI medium was monitored using a Bioscreen C Labsystem (Helsinki, Finland) (2). For complementation tests using purified, recombinant AtlA derivatives, the mutant strains 630NP and SAB71 were grown in BHI broth supplemented with the proteins at various concentrations, and the extent of clumping of cultures was observed. The cultures were also observed by phase-contrast microscopy to record chain length. Cultures (50 ml) to which 630D1 protein (one of a series of recombinant AtlA derivative proteins [see Fig. 3A]) was added at a final concentration of 2.5 ng/ml were also used for protein analysis. To assess the ability of an antibody raised against AtlA to affect the growth phenotypes of S. mutans, anti-630D1 or preimmune sera (100 μl) or purified immunoglobulin G (IgG; final concentration of 0.25 mg ml−1) was added to cultures of S. mutans UA159 growing in BHI broth.
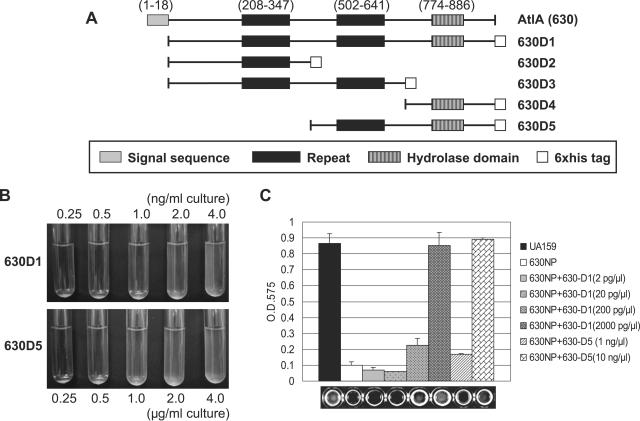
FIG. 3.
Restoration activity of histidine-tagged derivatives of AtlA (630). Schematic illustration of the different AtlA constructs (A). Amino acid coordinates are shown in parentheses. Restoration of 630NP to wild-type behavior is shown for culture confluence (B) and biofilm formation (C) after treatment of a strain lacking AtlA (630NP) with various concentrations of the 630D1 and 630D5 proteins. The 630NP strain was grown in BHI medium for the observation of planktonic growth and in BM supplemented with glucose at a final concentration of 20 mM for 24 h to monitor biofilm formation. Biofilms were assayed in polystyrene microtiter plates by staining with crystal violet and were quantified by adding an ethanol-acetone mixture and reading the optical density at 575 nm. Data are representative of at least two separate experiments performed in triplicate.
Biofilm, transformation, and CAT assays.
The ability to form stable biofilms in microtiter plates and to be transformed genetically were measured as previously described (2). CAT activity expressed from the promoter fusion constructs was measured by a spectrophotometric method (68) using the colorimetric substrate 5,5′-dinitro-bis-nitrobenzoic acid (Boehringer Mannheim, Indianapolis, IN). One unit of CAT activity was defined as the amount of enzyme needed to acetylate 1 nmol of chloramphenicol min−1. Protein concentrations were determined by a bicinchoninic acid assay (Sigma).
Cloning, expression, and purification of His-tagged proteins.
Recombinant plasmids expressing AtlA and its derivatives, but excluding the predicted signal sequence, were constructed by PCR cloning of the relevant DNA fragments into the vector pET-45b(+) (Novagen, Darmstadt, Germany), such that the coding sequence of the AtlA derivatives was in frame with an N-terminal His6 tag. The primers used are shown in Table 1. Forward and reverse primers incorporated BamHI and AvrII sites, respectively, for compatibility with sites in the vector. For expression, E. coli BL21(DE3) cells (Novagen) were transformed with the resulting plasmids. Each transformant was grown at 37°C to an optical density at 600 nm of 0.6. Expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside. The cells were harvested 3 h after induction, and the proteins were purified from E. coli cultures via their His6 tag using Ni-nitrilotriacetic acid affinity chromatography under denaturing conditions. It was not possible to purify these proteins under native conditions. Denaturants were slowly removed by dialysis to allow protein refolding. The purity of the proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Production and purification of antisera.
The purified His-tagged 630D1 protein (400 μg) was electrophoresed on two SDS-PAGE gels, and the protein was located by staining a strip from each gel with Coomassie blue. The areas containing the desired protein from the unstained gels were excised and used for raising antibodies in rabbits by Lampire Biological Laboratories (Pipersville, PA). Antibodies were purified from immune and preimmune sera by affinity chromatography on a protein A column (Amersham Bioscience, Piscataway, N.J.).
Preparation of protein fractions from S. mutans.
Various fractions of proteins from S. mutans were prepared from cell pellets that were harvested from BHI cultures at an optical density at 600 nm of 0.5, centrifuged, and washed twice with Tris-buffered saline (10 mM Tris, 0.9% NaCl, pH 7.4). Whole-cell lysates for protein analysis were obtained by homogenization with a bead beater (Biospec, Bartlesville, Okla.) in SDS boiling buffer (60 mM Tris, pH 6.8, 10% glycerol, and 5% SDS) in the presence of glass beads, as previously described (13). The extracts prepared by homogenizing cells in 20 mM Tris buffer (pH 7.4) was designated as the soluble fraction. In other cases, bacterial cells were suspended in 4% SDS and incubated for 30 min at room temperature. After centrifugation, the supernatant was used as the SDS extract (69, 89). To extract surface-associated proteins (84), the cells were suspended in phosphate-buffered saline (PBS) containing 0.2% (wt/vol) N-dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (Zwittergent; Sigma), incubated at 28°C with shaking at 80 rpm for 1 h, and centrifuged. Culture supernatant proteins were obtained by passing the supernatant fluid through a 0.45-μm-pore-size filter and concentrating the proteins 60-fold by precipitation with ammonium sulfate at 80% saturation.
Protein electrophoresis and Western blotting.
Proteins (10 μg) were separated by SDS-PAGE with a 10% polyacrylamide gel with a 4.5% stacking gel, as described by Laemmli (38). The proteins were then stained with Coomassie blue or blotted onto Immobilon-P membranes (Millipore, Bedford, Mass.) and subjected to Western blot analysis by standard techniques (65). Membranes were incubated with the anti-630D1 polyclonal antiserum or an anti-P1 monoclonal antibody (4), which was kindly provided by Jeannine Brady, University of Florida. The protein concentration of samples was determined by a bicinchoninic acid assay (Sigma).
Transcriptional analysis.
The potential for cotranscription of two genes was examined by reverse transcriptase PCR (RT-PCR). Levels of pepT and thmA mRNA were quantified by real-time RT-PCR. Extraction of RNA, RT-PCR, and real-time RT-PCR were performed as previously described (1). The primers used for reverse transcription reactions and real-time PCR are shown in Table 1.
RESULTS
Transcriptional studies.
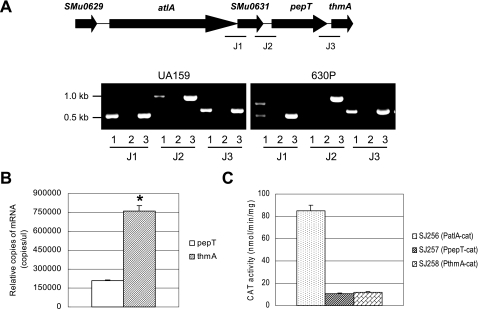
Many autolysins have been shown to function in conjunction with the products of linked genes, so the possibility of a functional connection of atlA with the genes immediately downstream was evaluated. The atlA region includes at least four genes, atlA, smu0631, pepT, and thmA, and all are transcribed in the same direction. The putative autolysin is encoded by atlA, smu0631 encodes a predicted secreted lipoprotein, pepT encodes a peptidase, and thmA encodes a predicted pore-forming protein. A transcriptional analysis by RT-PCR showed that the genes atlA and smu0631 and the genes pepT and thmA could be cotranscribed (Fig. 1A). The two loci, altA-smu0631 and pepT-thmA, could also be transcribed as a four-gene operon (Fig. 1A). Transcription through the region between smu0631 and pepT appears to be due to inefficient termination of the atlA-smu0631 transcript, as an RT-PCR product was not detected in the mutant carrying a strongly polar insertion in the atlA gene (630P). However, the amount of read-through transcript between smu0631 and pepT was only about eightfold less abundant than the atlA-smu0631 mRNA as measured by real-time PCR (data not shown), so there is a meaningful contribution of the atlA promoter to transcription of the downstream genes. Interestingly, real-time PCR experiments revealed that the thmA transcript was 3.6-fold more abundant than that of pepT (P = 0.0016) (Fig. 1B). Putative promoter sequences could be identified 20 bp upstream from the start codon for thmA (TAAAAAT-N16-AATAAT) and 27 bp upstream of pepT (CAGATAA-N16-TATAAT). Promoter activity of sequences 5′ to pepT and thmA was confirmed by creating a transcriptional fusion of these regions to cat (Fig. 1C). Both regions could drive transcription of cat at similar levels, although the CAT activity expressed from these promoters was about eightfold lower than that driven from the atlA promoter. Collectively, these results confirm that pepT and thmA can be expressed from their own promoters and can also be cotranscribed as part of a four-gene operon with atlA and smu0631.
FIG. 1.
Transcriptional analysis of the atlA region of wild-type and the atlA polar mutant (630P) of S. mutans UA159. (A) Schematic diagram of the atlA locus and RT-PCR in the junctions J1 (between atlA and smu0631), J2 (between smu0631 and pepT), and J3 (between pepT and thmA). Following reverse transcription with reverse primer smu0631-rt-RV (for J1), pepT-antisense (for J2), and thmA-antisense (for J3), PCR amplification was performed with primer sets of smu0630-rt-FW/smu0631-rt-RV, smu0631-rt-FW/pepT-antisense, and pepT-sense/thmA-antisense. The PCR products were run on Tris-acetate-EDTA gels. Lane 1, RT-PCR product; lane 2, negative control lacking RT; lane 3, positive control of PCR from chromosomal DNA of UA159. The higher Mr band in lane 1 of the 630P strain appears to arise from amplification of a product generated by low levels of read-through from the polar kanamycin cassette. We have measured read-through from the polar cassette by real-time PCR at about 1% of the atlA-smu0631 cotranscript (data not shown). (B) Real-time PCR. For measuring pepT and thmA mRNAs, RNA from UA159 was used for RT with a thmA-antisense primer. (C) Promoter activity of atlA, pepT, and thmA genes as measured by CAT assays. The promoter fusion of each gene with the cat gene was inserted in a single copy into the chromosome of the wild-type strain. Data presented are means ± standard deviations (error bars) of three independent experiments. *, P < 0.005 (Student's t test).
Construction of mutant strains.
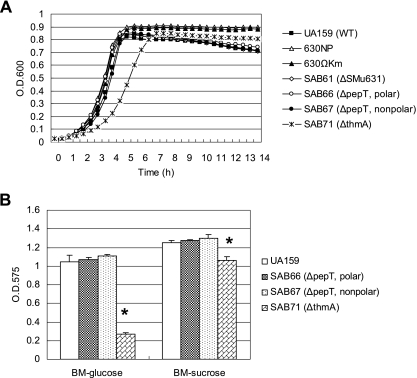
To examine the possible involvement of smu0631, pepT, and thmA genes in the biogenesis or function of AtlA, the genes were disrupted by deletion and insertion of a polar kanamycin cassette to create strains SAB61, SAB66, and SAB71, respectively (Table 2). Additionally, the pepT gene was disrupted by deletion and insertion of a nonpolar marker (SAB67), because pepT and thmA can be cotranscribed. No differences were observed in the growth rate of SAB61, SAB66, or SAB67 using BHI medium, but the thmA deletion mutant (SAB71) showed a significantly slower growth rate (Fig. 2A). The ThmA-deficient strain had characteristics very similar to the 630P or 630NP strains, which lack AtlA. SAB71 showed a moderate resistance to autolysis in late stationary phase (Fig. 2A) and formed clumps (data not shown). Light-microscopic observations revealed that SAB71 formed significantly longer chains compared to those formed by the parent, albeit not as long those of 630P or 630NP (data not shown). Disruption of thmA caused a significant reduction (75%) in biofilm formation in BM supplemented with glucose and in BM-sucrose medium (15.4%), as assessed by crystal violet staining of biofilms (Fig. 2B). Also of note, the colonies formed by SAB71 displayed a mucoid appearance on BHI agar plates (data not shown), which is not a characteristic of AtlA-deficient strains. The SAB66 and SAB67 strains, as well as SAB61 (data not shown), did not show an obvious difference in biofilm formation in the same medium. The polar pepT mutant (SAB66) did not show reduced biofilm formation, consistent with the observation that thmA can be expressed from its own promoter (Fig. 1A).
FIG. 2.
Growth phenotypes of S. mutans strains. (A) Growth in BHI broth was monitored in a Bioscreen C system. Data points are averages of triplicate samples. (B) Biofilm formation of S. mutans UA159 (wild type) and its derivatives (SAB66, SAB67, and SAB71) in BM supplemented with glucose or sucrose. See text for more details. Data are representative of at least two separate experiments. The error bars represent standard deviations. *, P < 0.005 (Student's t test).
Characterization of AtlA activity.
A histidine-tagged AtlA protein was constructed that contained the full-length protein without the predicted signal sequence (Fig. 3A, 630D1). The most obvious phenotypes of the atlA mutant reported in previous studies (10, 69) were the formation of longer chains, clumping in broth culture, and a dramatic reduction in the ability of cells to form biofilms. Somewhat surprisingly, the defects of the mutant were completely corrected to wild-type levels simply by adding purified 630D1 protein to the culture of the atlA mutant (630NP) (Fig. 3B and C). To identify the domains of AtlA required for restoration of the phenotypes, we constructed a series of recombinant AtlA derivatives as shown in Fig. 3A. Restoration of normal chain length and biofilm formation was observed only with 630D5, which included the second repeat region and the hydrolase domain (Fig. 3B and C). However, the amount of 630D5 protein required for restoration of the defects was between 5- and 500-fold greater than for full-length AtlA, depending on the phenotype. Extensive clumping by the mutant culture was alleviated by the addition of 2.0 ng ml−1 of 630D1, but 1.0 μg ml−1 of 630D5 was needed to elicit the same effect. Under these conditions, the cultures treated with pure AtlA showed the same chain length as the wild type (data not shown). Also, the ability of the 630NP mutant to form biofilms was restored to that of the wild-type strain by inclusion of 2.0 μg ml−1 of 630D1 in the biofilm medium, but about 10.0 μg ml−1 of 630D5 was required for the same effect. These results are consistent with the idea that the catalytic center of the AtlA protein is the hydrolase domain in the C terminus (10) and also demonstrate that the two repeats in the middle of the protein significantly impact the function of AtlA. The 630D4 protein showed no activity even when mixed with 630D2 or 630D3, and 630D2 or 630D3 showed no synergistic effect on the activity of 630D5 (data not shown). Also of interest, the 630D1 protein restored the autolytic activity of the mutant strain but had no impact on growth and did not cause lysis of the wild-type strain, even at concentrations as high as 800 ng ml−1 in planktonic culture (data not shown), roughly 400-fold higher than what was needed to restore normal chain length. As noted above, cells grew well and formed biofilms equivalent to the parental strain with as much as 2 μg ml−1 of 630D1.
Surface localization of AtlA.
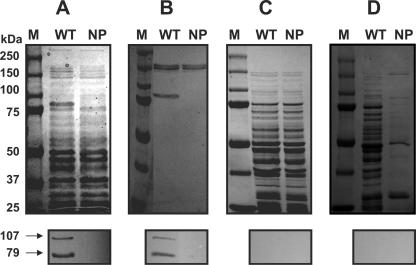
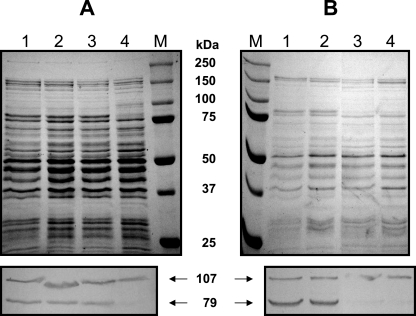
Proteins of both the wild-type and mutant (630NP) strains were extracted by different methods and subjected to SDS-PAGE and Western blot analysis (Fig. 4). In a Western blot profile of whole-cell lysates, which was prepared by homogenizing cells in the presence of SDS, the wild type showed two major AtlA bands, corresponding to 107 and 79 kDa (Fig. 4A). The 79-kDa band was previously reported to be cleaved by processing of the 107-kDa form (69) and, interestingly, appears to be one of most abundant proteins in S. mutans, as is evident in the Coomassie-stained gel (Fig. 4A). The two major bands were readily detected in the 4% SDS extract (Fig. 4B) but were not detectable in the soluble fraction prepared by bead beating in Tris buffer without SDS (Fig. 4C), suggesting that AtlA is rapidly degraded in the absence of denaturants in the cell lysates. When both cultures were extracted with PBS containing the nonionic detergent Zwittergent (0.2%), which has been reported to enrich for surface-associated proteins lacking covalent anchors (84), no AltA protein was extracted (Fig. 4D). When the cell pellets that had been treated with Zwittergent were subjected to SDS treatment, the intact forms of the AtlA protein were still present (data not shown), suggesting a fairly tight association of AtlA with the wall or other surface components. When the wild-type strain was grown with the anti-630D1 serum or purified immune IgG, the cells displayed characteristics of the atlA mutant, forming clumps and longer chains (data not shown), albeit not to the extent noted in the 630NP mutant. Similarly, biofilm formation was inhibited by antibodies to AtlA (data not shown). Preimmune IgG did not affect the phenotypes.
FIG. 4.
SDS-PAGE analysis of different cell extracts from wild-type (WT) and 630NP (NP) strains of S. mutans: bead-beaten SDS-boiling extracts (A), 4% SDS extracts (B), bead-beaten Tris extracts (C), and surface-associated fractions extracted with 0.2% (wt/vol) Zwittergent in PBS (D). Following SDS-PAGE, proteins were either stained with Coomassie blue (top) or transferred onto a nitrocellulose membrane and subjected to Western blotting using an anti-630D1 polyclonal antiserum at the dilution of 1:350 (bottom). Lane M, size marker.
Processing of AtlA requires ThmA.
SDS-PAGE and Western blot analysis using whole-cell lysates revealed that disruption of thmA, but not pepT, dramatically impacted the proteolytic processing of AtlA. Specifically, the amount of the processed form (79 kDa) of AtlA was significantly reduced in the thmA mutant (SAB71) compared to UA159, whereas the pepT mutants showed no differences in AtlA processing (Fig. 5A). No impact on AtlA expression or processing was noted in the Smu0631 mutant (data not shown). Importantly, the reduced processing in the ThmA-deficient strain does not appear to result from a reduction in transcription of atlA, as there were no obvious differences in the amount of altA mRNA in SAB71 and UA159 as measured by real-time PCR (data not shown). To further assess the requirement for ThmA in processing of AtlA, purified 630D1 was added into the cultures of 630NP (deficient in atlA) and SAB78 (deficient in atlA and thmA); the cells were harvested immediately after the addition of the protein (0 h) or after 1 h of incubation, whole-cell lysates were prepared, and the proteins were subjected to SDS-PAGE and Western blotting (Fig. 5B). In all cases, the recombinant protein rapidly became associated with the cells, but the strain lacking ThmA did not efficiently convert the 107-kDa form to the mature form of AtlA. An interesting observation is that the processing occurred very quickly, within minutes after the protein made contact with the cell surface (data not shown). Also of interest, the biofilm and chain length phenotypes of the thmA mutation could not be restored by treatment of cells with exogenous 630D1 (data not shown), unlike the atlA mutant. Thus, the conversion to the 79-kDa form is a critical event affecting biofilm formation, chain length, and clumping.
FIG. 5.
Effect of ThmA on the conversion of AtlA from a 107-kDa to a 79-kDa peptide. (A) Protein profiles in Coomassie-stained SDS-PAGE gel of UA159 (wild type; lane 1) and its derivatives, SAB66 (ΔpepT, polar mutant; lane 2), SAB67 (ΔpepT, nonpolar mutant; lane 3), and SAB71 (ΔthmA mutant; lane 4). (B) Effect of ThmA on the processing of exogenous 630D1. Two strains, 630NP (lane 1 and 2) and SAB78 (lanes 3 and 4), were grown in BHI broth; samples either were left untreated (lanes 1 and 3), or purified 630D1 was added to a final concentration of 20 ng/ml culture for 1 h (lanes 2 and 4). Following SDS-PAGE, proteins were stained with Coomassie blue (top) or transferred onto a nitrocellulose membrane and subjected to Western blotting using an anti-630D1 polyclonal antiserum (dilution 1:350) (bottom). Lane M, size marker. WT, wild type.
AtlA is required for biogenesis of a normal cell surface.
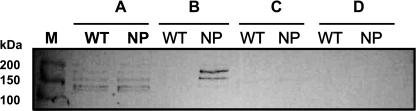
As can be observed in the Coomassie-stained gel in Fig. 4D, the surface protein profiles of the wild-type strain were dramatically different from those of the mutant. Specifically, a large number of proteins could be extracted from the parent strain with nonionic detergent, whereas very few proteins were released from cells of the mutant by the same treatment. The same effect was not observed in cells treated with 4% SDS. Thus, AtlA appears to have a profound impact on expression or localization of surface proteins or on the nature of the association of those proteins with the cell envelope. To investigate this further, the protein extracts were subjected to Western blot analysis using monoclonal antibody to detect the major surface protein adhesin P1, which contains an LPXTG anchor and is coupled to peptidoglycan under normal conditions. Western blotting revealed multiple immunoreactive bands in whole-cell lysates of both the wild-type and 630NP strains (Fig. 6A), typical of P1 (42), and no P1 was detected in the 4% SDS extracts or supernatant fractions (Fig. 6C and D). Notably, the P1 protein could be extracted from the mutant with nonionic detergent, but none was extractable from the wild-type strain (Fig. 6B). This suggests that the absence of AtlA may affect sortase-mediated anchoring of surface proteins or enzymatic release of wall-anchored proteins or that factors involved in P1 turnover are not properly localized to the cell surface.
FIG. 6.
Western blot analysis of different cell extracts from wild-type (WT) and 630NP (NP) strains of S. mutans: bead-beaten SDS-boiling extracts (A), surface-associated fractions extracted with 0.2% (wt/vol) Zwittergent in PBS (B), 4% SDS extracts (C), and supernatant fraction, concentrated approximately 60-fold by ammonium sulfate (D). Following SDS-PAGE, proteins were transferred onto a nitrocellulose membrane and subjected to Western blotting using an anti-P1 monoclonal antiserum at the dilution of 1:700. Lane M, size marker.
AtlA and genetic competence.
The possibility that autolysins are involved in genetic competence has been suggested in previous studies with gram-positive bacteria, such as Bacillus and Streptococcus (5, 17, 32). To assess the contribution of the S. mutans AtlA protein to competence, the efficiency of transformation of the atlA mutant (630NP) was evaluated by transforming with plasmid pDL278 (Table 3). The mutant strain showed a significant reduction in the number of cells that could develop competence in the absence and presence of exogenous competence-signaling peptide (CSP). To determine if the reduction in transformation by the mutant was due to aberrant expression of genes involved in the development of competence, mRNA levels of the competence-associated genes comD, ciaR, comX, comYA, and htrA were measured using real-time PCR. The data revealed significantly lower expression of these genes in the mutant, suggesting that reduction of transformation in the mutant may, at least in part, be due to effects exerted at the transcriptional level (data not shown).
TABLE 3.
Transformability of Smu0630 mutant compared to UA159
| Culture conditions | % Transformabilitya
|
|
|---|---|---|
| UA159 | 630NP | |
| Without CSP | 7.37E-04 | 1.31E-04b |
| With CSP | 0.38 | 0.09b |
The percent transformability was determined by the ratio of the number of transformants of the mutant (630NP) versus that of the wild type (UA159), multiplied by 100. The transformation was performed in 200 μl culture with or without 5 μl of synthetic CSP solution (1 μmol ml−1). See the text for more details. The data shown are representative of two independent experiments performed in triplicate.
*, P < 0.01 (Student's t test).
Distribution in oral streptococci of proteins cross-reactive with anti-AtlA antibodies.
Previously, we reported that apparent orthologues of atlA could only be tentatively identified by BLAST searches in Streptococcus gordonii and Streptococcus agalactiae. SDS-PAGE and Western blot analysis with the anti-630D1 serum revealed that the two major AtlA bands were detected in stains of S. mutans, including NG8, LT11, and GS5, but not in S. gordonii, Streptococcus oralis, Streptococcus rattus or Streptococcus salivarius (data not shown). Also, the purified recombinant AtlA proteins had no obvious effect on biofilm formation of those oral streptococci (data not shown).
DISCUSSION
We originally identified AtlA as a predicted surface protein that was required for maturation of biofilms (10). Disruption of atlA also resulted in resistance to autolysis and excessive chaining of cells (10, 69), and Shibata et al. (66) determined that the protein had peptidoglycan hydrolase activity in zymograms. Our present study reveals that AtlA is also required for biogenesis of a normal cell surface and full expression of genetic competence by S. mutans. Thus, AtlA appears to play a central role connecting cell wall remodeling, biofilm formation, genetic competence, and autolysis. These networks have often been shown to overlap with the stress regulon, but the AtlA protein does not appear to be required for acid tolerance, since no obvious differences were observed in the growth rate of the atlA mutant in acidified BHI broth (pH 6.4 or 5.4) (data not shown). However, it is possible that AtlA may be involved in other stress responses, because autolysin-mediated cell wall turnover is known to be influenced by diverse external environments (31, 44, 74). For example, nutrient limitation and induction of the stringent response have been reported to indirectly control murein turnover and the rate of phospholipid synthesis (62). Notably, the expression of atlA in S. mutans does not appear to be regulated by several two-component systems, such as CiaRH (21, 58, 90), ComED (3, 24, 26, 27, 45), and LytST (61, 88) (data not shown), which are known to regulate biofilm formation, genetic competence, and autolysis. The atlA gene is also not under control of AI-2-mediated quorum sensing, because no obvious differences were observed in the expression of atlA in the wild-type and a luxS mutant (data not shown). This finding contrasts with a recent study in S. pneumoniae demonstrating that LuxS is involved in the control of LytA-dependent autolysis, as well as in cell-cell communication culminating in repression of competence (63).
The fact that the addition of a large excess of purified AltA had no impact on the growth or viability of S. mutans indicates that the activity or expression of AltA must be strictly regulated, which is logical if the cells are to remain viable. This is especially notable, given that addition of purified AtlA in concentrations as low as 20 pM is sufficient to ameliorate the defects in chaining and clumping. The potential for tight genetic regulation of atlA expression seems high, given that the four genes in the atlA region are transcribed by at least three promoters, as revealed in this study. Surprisingly, our preliminary information suggests that atlA expression is constitutive throughout the growth cycle. Thus, the complex genetic organization of the atlA operon may have evolved to finely tune the stoichiometry of the gene products. Along these lines, it is interesting to note that when the atlA defect was complemented with an intact atlA gene on a plasmid (SAB40) (10), only a limited quantity of AtlA was shown to be converted to the mature form (our unpublished data), implying that the coordinated synthesis of AtlA and other factors produced from the atlA operon, and possibly elsewhere in the genome, is critical to proper production of this enzyme. It is also noteworthy that AtlA appears to constitute such a tremendous proportion of the total cell-associated protein, as is evident in Fig. 4A. In light of the abundance and constitutive production of AtlA, it seems, therefore, that the cells must tightly regulate the biochemical activity or access of the protein to its cognate substrate to avoid rapid degradation of cells walls.
Shibata el al. (69) have shown that both the 79-kDa and 107-kDa forms of AtlA can hydrolyze cell walls in zymograms, implying that it is not proteolytic processing to the mature form that controls AtlA activation. Also, the localization of AtlA does not appear to be influenced by growth stage, as the protein is found almost exclusively in the SDS-extractable surface fractions regardless of growth phase (data not shown). However, the idea that conversion to the 79-kDa form is not required for activation of the enzyme conflicts with our observations that ThmA-deficient strains, which fail to properly convert either endogenously produced or exogenously added AtlA to the 79-kDa form, form long chains, clump, and are poor biofilm formers. Thus, there must be major differences in the activity of the two forms of the protein. It may also be significant that AtlA is highly toxic when expressed in E. coli (data not shown). From these observations and the expression data, the most logical conclusion is that AtlA activity is controlled through an association with factors that sequester it from a substrate under particular conditions. One can speculate that these factors may differ for the two forms of the protein.
There are a number of other interesting aspects of AtlA biogenesis and function besides the processing and control of its activity. Perhaps the most intriguing of these is that disruption of thmA elicited phenotypes similar to the atlA mutants. Disruption of the other two genes, smu0631 (10) and pepT, in the atlA region had no effect on biofilm formation, chaining, or clumping, so the role and relationship of Smu0631 and PepT to AtlA remains enigmatic. Apparent homologues of PepT have been inactivated in gram-positive bacteria, such as L. lactis and B. subtilis (52, 67, 69), with no obvious changes in growth characteristics. The role of pepT, encoding a putative peptidase, has not been defined in this study, though it is noteworthy that pepT can be cotranscribed with atlA and thmA. Also, AtlA has a conserved glycohydrolase domain, implying that it probably cleaves the polysaccharide backbone in peptidoglycan. In light of these observations and the predicted activity of PepT, it seems reasonable to suggest that PepT participates in degradation of products liberated by AtlA, perhaps cleaving amino acids from the peptide cross-links in the wall. The smu0631 gene product is predicted to encode a putative lipoprotein and to be located outside of the cell, with the N terminus covalently coupled to the membrane. Thus, Smu0631 could interact with AtlA on the cell surface, with the potential to play a role in localization of AtlA or regulation of its activity. However, no changes in the amounts or ratios of the forms of AtlA were evident in Smu0631-deficient mutants or in the localization of the proteins. In addition, inactivation of the genes immediately flanking the atlA operon is not lethal and does not affect growth or chaining to any appreciable extent (data not shown).
There are a number of observations to support the importance of ThmA in AtlA biogenesis and activity. The thmA mutant has phenotypes similar to the strain lacking AtlA, and ThmA is required for efficient processing of AtlA to its lower-molecular-weight form. Exogenously added 630D1 protein was still able to be become cell associated and processed by strains lacking ThmA, albeit the rate of processing was much slower than in strains producing ThmA; so ThmA probably does not catalyze the cleavage of the 107-kDa form of AtlA. Therefore, a more likely role for ThmA may be in either translocation or targeting of AtlA to specific sites on the cell surface. According to the program HMMTOP (http://www.enzym.hu/hmmtop/), ThmA contains two transmembrane helices (positions 18 to 37 and 46 to 62) near its N terminus, and most of ThmA, except for the region between the two helices, is predicted to be located at the inner membrane. Although the best BLAST hits for ThmA are with putative pore-forming proteins, ThmA has some structural similarities to the LktD transport protein in the lkdCABD leukotoxin operon of gram-negative bacteria, including Actinobacillus actinomycetemcomitans and Pasteurella hemolytica (25, 29, 30, 46, 76). The lktA gene encodes the inactive protoxin, lktC is required for posttranslational activation of the toxin prior to secretion, and the lktB and lktD genes are required for secretion of the toxin from the organism, with LktD being postulated to help in toxin localization. When either lktD or both lktB and lktD were inactivated, the level of leukotoxin protein in the cell was reported to significantly decrease with no effect on the levels of leukotoxin RNA (25). Also of relevance, Bensing and Dunny (7) have described a protein (EbsA) that is structurally similar to LktD but which we found to share some weak amino acid similarities with ThmA. EbsA was postulated to assist with the presentation of factors required for intercellular aggregation during mating in E. faecalis. Interestingly, Bensing and Dunny also noted that EbsA shared some structural similarity with endolysins of phages of gram-positive bacteria (7). Thus, ThmA may be part of a complex required for proper localization and activation of the wall-degrading activities of AtlA.
The fact that the treatment of the atlA mutant (630NP) with exogenous His-tagged AtlA protein resulted in complete restoration of normal chain length was somewhat remarkable in itself and also provided useful insights into localization of and structure-function relationships in AtlA. Coupled with the observation that antibody against AtlA induces a phenotype similar to inactivation of atlA and that AtlA can be readily extracted with 4% SDS, it appears that AtlA is localized to, and functions at, the surface of the cell and that covalent association with the cell is neither apparent nor required for activity. Our assay using a series of recombinant AtlA derivatives demonstrated that the activity of AtlA is dependent on the repeat domains, with one domain being adequate for activity but possession of two domains being required for full activity. Although the repeat domains are necessary, they are not sufficient, and the C-terminal glycohydrolase domain is absolutely required for AtlA function (10, 69). We suggest that the repeats play a role in targeting of AtlA onto a particular surface structure (6, 66, 85) and that these associations are critical to the activity of AtlA. It is also noteworthy that the restoration of biofilm formation was also achieved by treatment with exogenous AtlA. There are a number of possible explanations for the effects of AtlA on biofilm formation by S. mutans. The first is that AtlA can directly mediate intercellular adhesion. Notably, the glucan binding protein B (GbpB) of S. mutans was reported to share homology with a putative peptidoglycan hydrolase from group B streptococcus (14, 49, 50). The possession of the large repeat regions, such as those found in AtlA, is characteristic of surface proteins involved in ligand binding, and the peptidoglycan hydrolase domain may also mediate cell wall binding, possibly in a more complex manner. Addition of recombinant AtlA to S. mutans lacking the protein, even in quantities that far exceed those that complement the aberrant phenotypes of the atlA mutant (Fig. 3C) and even though the added AtlA rapidly becomes cell associated (Fig. 6B), does not cause coaggregation. A more likely explanation is that the presence of AtlA results in modifications to the cell surface that enhance the capacity to form biofilms. In this regard, the finding that the number of proteins and amount of protein extractable from the surface of the AtlA-deficient strain are dramatically reduced may, in fact, entirely explain the biofilm formation defect. However, we cannot exclude that changes in gene expression brought on by surface modifications or other environmental signals in the atlA or thmA strains also adversely affect biofilm formation. For example, our findings that com gene expression is altered in the mutants emphasizes the need to consider alternative explanations, since competence gene expression has been linked to biofilm formation (28, 51).
Indeed, one of the most remarkable findings in this study is that the Zwittergent extracts of cells lacking AtlA have a dramatically reduced number of proteins compared to extracts prepared from the wild-type strain. There is no evidence that the proteins are shed into the culture fluid, since the protein profiles of an 80% ammonium sulfate cut of the culture fluid from the mutant and wild-type strain are indistinguishable (data not shown). These findings suggest a critical role for AtlA in the biogenesis of a variety of other surface-associated proteins. In light of recent findings that there exists a specialized structure for protein export in Streptococcus pyogenes called the ExPortal (64), it is tempting to speculate that components of the atlA operon may constitute a portion of an apparatus that creates a portal for movement of multiple surface proteins across the membrane. Such a structure might require significant remodeling of the cell surface, which could be catalyzed in part by AtlA. This idea would also be consistent with the tight regulation of AtlA activity and the failure of relatively large quantities of recombinant AtlA to lyse cells. Notably, lack of AtlA did not result in decreases in the total cellular concentrations of the major adhesin P1. However, as is evident from the data presented in Fig. 6, the nature of the association of P1 with the cell wall is altered in the AtlA mutant, since P1, which is normally covalently coupled to the cell wall (33, 41, 43), was readily extracted with a nonionic detergent. This finding could imply that other proteins required for the anchoring of P1, such as sortase (41), require a functional AtlA for proper localization and function or that there are changes in the cell wall structure that preclude efficient sortase-mediated anchoring. Still, it seems more likely that the defect is at the level of secretion, given a lack of change in the secreted protein profiles in the supernatant fluid. Studies are under way to understand why loss of AtlA causes radical changes in the cell-surface protein profile.
In a previous study showing bacteriolytic enzyme profiles of oral streptococci (89), S. mutans showed a strong bacteriolytic activity against Streptococcus sobrinus, a weak activity against Streptococcus mitis and S. salivarius, and no activity against Streptococcus sanguis, suggesting that the difference in susceptibility to autolysins (immunity) may be species or strain specific. BLAST searches and our antibody screening results suggest that AtlA may be peculiar to S. mutans, and homologues are missing in other oral streptococci. We have also found that recombinant AtlA has no effect on the lysis of, or biofilm formation by, other oral Streptococcus species we have tested, although it is not clear whether this is attributable to differences in cell wall structure or the possession of factors that modulate AtlA activity (data not shown). Efforts to identify factors that interact with AtlA are under way.
The results presented herein provide insights into novel roles and regulation of AtlA, a suspected autolysin of S. mutans that profoundly impacts the biofilm-forming capacity of the cells. Apparently, the biogenesis and activity of AtlA is tightly controlled by the cells, as would be expected for an enzyme that contributes to the autolytic potential of the cells and that appears to be critical for biogenesis of a normal cell surface. Experiments are under way to define the subcellular localization of these proteins and how they interact to modulate the composition of the bacterial cell surface.
Acknowledgments
We thank Paula Crowley for input on experiments involving the P1 adhesion.
This study was supported by NIH-NIDCR DE13239.
REFERENCES
- 1.Ahn, S. J., J. A. Lemos, and R. A. Burne. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187:3028-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74:1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 4.Ayakawa, G. Y., L. W. Boushell, P. J. Crowley, G. W. Erdos, W. P. McArthur, and A. S. Bleiweis. 1987. Isolation and characterization of monoclonal antibodies specific for antigen P1, a major surface protein of mutans streptococci. Infect. Immun. 55:2759-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azoulay-Dupuis, E., V. Rieux, C. Rivier, and M. C. Trombe. 1998. Pleiotropic mutations alter the kinetics of calcium transport, competence regulation, autolysis and experimental virulence in Streptococcus pneumoniae. Res. Microbiol. 149:5-13. [DOI] [PubMed] [Google Scholar]
- 6.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 17:4639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensing, B. A., and G. M. Dunny. 1993. Cloning and molecular analysis of genes affecting expression of binding substance, the recipient-encoded receptor(s) mediating mating aggregate formation in Enterococcus faecalis. J. Bacteriol. 175:7421-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry, A. M., R. A. Lock, D. Hansman, and J. C. Paton. 1989. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackman, S. A., T. J. Smith, and S. J. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144:73-82. [DOI] [PubMed] [Google Scholar]
- 10.Brown, T. A., Jr., S. J. Ahn, R. N. Frank, Y. Y. Chen, J. A. Lemos, and R. A. Burne. 2005. A hypothetical protein of Streptococcus mutans is critical for biofilm formation. Infect. Immun. 73:3147-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buist, G., H. Karsens, A. Nauta, D. van Sinderen, G. Venema, and J. Kok. 1997. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl. Environ. Microbiol. 63:2722-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buist, G., G. Venema, and J. Kok. 1998. Autolysis of Lactococcus lactis is influenced by proteolysis. J. Bacteriol. 180:5947-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Y. Y., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chia, J. S., L. Y. Chang, C. T. Shun, Y. Y. Chang, Y. G. Tsay, and J. Y. Chen. 2001. A 60-kilodalton immunodominant glycoprotein is essential for cell wall integrity and the maintenance of cell shape in Streptococcus mutans. Infect. Immun. 69:6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornett, J. B., B. E. Redman, and G. D. Shockman. 1978. Autolytic defective mutant of Streptococcus faecalis. J. Bacteriol. 133:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornett, J. B., and G. D. Shockman. 1978. Cellular lysis of Streptococcus faecalis induced with triton X-100. J. Bacteriol. 135:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 18.Diaz, E., R. Lopez, and J. L. Garcia. 1992. Role of the major pneumococcal autolysin in the atypical response of a clinical isolate of Streptococcus pneumoniae. J. Bacteriol. 174:5508-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, S. J. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia, J. L., J. M. Sanchez-Puelles, P. Garcia, R. Lopez, C. Ronda, and E. Garcia. 1986. Molecular characterization of an autolysin-defective mutant of Streptococcus pneumoniae. Biochem. Biophys. Res. Commun. 137:614-619. [DOI] [PubMed] [Google Scholar]
- 21.Giammarinaro, P., M. Sicard, and A. M. Gasc. 1999. Genetic and physiological studies of the CiaH-CiaR two-component signal-transducing system involved in cefotaxime resistance and competence of Streptococcus pneumoniae. Microbiology 145:1859-1869. [DOI] [PubMed] [Google Scholar]
- 22.Govindasamy-Lucey, S., P. K. Gopal, P. A. Sullivan, and C. J. Pillidge. 2000. Varying influence of the autolysin, N-acetyl muramidase, and the cell envelope proteinase on the rate of autolysis of six commercial Lactococcus lactis cheese starter bacteria grown in milk. J. Dairy Res. 67:585-596. [DOI] [PubMed] [Google Scholar]
- 23.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guiral, S., V. Henard, C. Granadel, B. Martin, and J. P. Claverys. 2006. Inhibition of competence development in Streptococcus pneumoniae by increased basal-level expression of the ComDE two-component regulatory system. Microbiology 152:323-331. [DOI] [PubMed] [Google Scholar]
- 25.Guthmiller, J. M., D. Kolodrubetz, and E. Kraig. 1995. Mutational analysis of the putative leukotoxin transport genes in Actinobacillus actinomycetemcomitans. Microb. Pathog. 18:307-321. [DOI] [PubMed] [Google Scholar]
- 26.Havarstein, L. S. 1998. Identification of a competence regulon in Streptococcus pneumoniae by genomic analysis. Trends Microbiol. 6:297-300. [DOI] [PubMed] [Google Scholar]
- 27.Havarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change phenotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 29.Highlander, S. K., M. Chidambaram, M. J. Engler, and G. M. Weinstock. 1989. DNA sequence of the Pasteurella haemolytica leukotoxin gene cluster. DNA. 8:15-28. [DOI] [PubMed] [Google Scholar]
- 30.Highlander, S. K., M. J. Engler, and G. M. Weinstock. 1990. Secretion and expression of the Pasteurella haemolytica Leukotoxin. J. Bacteriol. 172:2343-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holtje, J. V. 1995. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch. Microbiol. 164:243-254. [DOI] [PubMed] [Google Scholar]
- 32.Horne, D., and A. Tomasz. 1985. Competence-specific autolysis in Streptococcus sanguis. J. Gen. Microbiol. 131:533-541. [DOI] [PubMed] [Google Scholar]
- 33.Huang, D., Z. Luo, and X. Zhou. 2000. Effect of calcium on adherence of Streptococcus mutans MT6R(serotype c) surface protein P1. Hua Xi Kou Qiang Yi Xue Za Zhi 18:163-164,180. [In Chinese.] [PubMed] [Google Scholar]
- 34.Huard, C., G. Miranda, F. Wessner, A. Bolotin, J. Hansen, S. J. Foster, and M. P. Chapot-Chartier. 2003. Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149:695-705. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa, S., Y. Hara, R. Ohnishi, and J. Sekiguchi. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol. 180:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kajimura, J., T. Fujiwara, S. Yamada, Y. Suzawa, T. Nishida, Y. Oyamada, I. Hayashi, J. Yamagishi, H. Komatsuzawa, and M. Sugai. 2005. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58:1087-1101. [DOI] [PubMed] [Google Scholar]
- 37.Kuroda, A., and J. Sekiguchi. 1991. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J. Bacteriol. 173:7304-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 39.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 40.LeBlanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 41.Lee, S. F., and T. L. Boran. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, S. F., A. Progulske-Fox, and A. S. Bleiweis. 1988. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect. Immun. 56:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, S. F., A. Progulske-Fox, G. W. Erdos, D. A. Piacentini, G. Y. Ayakawa, P. J. Crowley, and A. S. Bleiweis. 1989. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II). Infect. Immun. 57:3306-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo, R. Y., C. A. Strathdee, and P. E. Shewen. 1987. Nucleotide sequence of the leukotoxin genes of Pasteurella haemolytica A1. Infect. Immun. 55:1987-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margot, P., C. Mauel, and D. Karamata. 1994. The gene of the N-acetylglucosaminidase, a Bacillus subtilis 168 cell wall hydrolase not involved in vegetative cell autolysis. Mol. Microbiol. 12:535-545. [DOI] [PubMed] [Google Scholar]
- 49.Mattos-Graner, R. O., S. Jin, W. F. King, T. Chen, D. J. Smith, and M. J. Duncan. 2001. Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect. Immun. 69:6931-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattos-Graner, R. O., K. A. Porter, D. J. Smith, Y. Hosogi, and M. J. Duncan. 2006. Functional analysis of glucan binding protein B from Streptococcus mutans. J. Bacteriol. 188:3813-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercier, C., C. Durrieu, R. Briandet, E. Domakova, J. Tremblay, G. Buist, and S. Kulakauskas. 2002. Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol. Microbiol. 46:235-243. [DOI] [PubMed] [Google Scholar]
- 52.Mierau, I., A. J. Haandrikman, O. Velterop, P. S. Tan, K. L. Leenhouts, W. N. Konings, G. Venema, and J. Kok. 1994. Tripeptidase gene (pepT) of Lactococcus lactis: molecular cloning and nucleotide sequencing of pepT and construction of a chromosomal deletion mutant. J. Bacteriol. 176:2854-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perkins, H. R. 1980. The bacterial autolysins. Chapman and Hall, London, United Kingdom.
- 55.Pooley, H. M., and G. D. Shockman. 1970. Relationship between the location of autolysin, cell wall synthesis, and the development of resistance to cellular autolysis in Streptococcus faecalis after inhibition of protein synthesis. J. Bacteriol. 103:457-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pooley, H. M., G. D. Shockman, M. L. Higgins, and J. Porres-Juan. 1972. Some properties of two autolytic-defective mutants of Streptococcus faecalis ATCC 9790. J. Bacteriol. 109:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popowska, M., M. Kloszewska, S. Gorecka, and Z. Markiewicz. 1999. Autolysis of Listeria monocytogenes. Acta Microbiol. Pol. 48:141-152. [PubMed] [Google Scholar]
- 58.Qi, F., J. Merritt, R. Lux, and W. Shi. 2004. Inactivation of the ciaH Gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 72:4895-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rashid, M. H., M. Mori, and J. Sekiguchi. 1995. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology 141:2391-2404. [DOI] [PubMed] [Google Scholar]
- 60.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rice, K. C., T. Patton, S. J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 186:3029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodionov, D. G., A. G. Pisabarro, M. A. de Pedro, W. Kusser, and E. E. Ishiguro. 1995. Beta-lactam-induced bacteriolysis of amino acid-deprived Escherichia coli is dependent on phospholipid synthesis. J. Bacteriol. 177:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romao, S., G. Memmi, M. R. Oggioni, and M. C. Trombe. 2006. LuxS impacts on LytA-dependent autolysis and on competence in Streptococcus pneumoniae. Microbiology 152:333-341. [DOI] [PubMed] [Google Scholar]
- 64.Rosch, J. W., and M. G. Caparon. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol. Microbiol. 58:959-968. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 66.Sanchez-Puelles, J. M., J. M. Sanz, J. L. Garcia, and E. Garcia. 1990. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene 89:69-75. [DOI] [PubMed] [Google Scholar]
- 67.Schrogel, O., O. Krispin, and R. Allmansberger. 1996. Expression of a pepT homologue from Bacillus subtilis. FEMS Microbiol. Lett. 145:341-348. [DOI] [PubMed] [Google Scholar]
- 68.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 69.Shibata, Y., M. Kawada, Y. Nakano, K. Toyoshima, and Y. Yamashita. 2005. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect. Immun. 73:3512-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shockman, G. D., and J. F. Barrett. 1983. Structure, function, and assembly of cell walls of gram-positive bacteria. Annu. Rev. Microbiol. 37:501-527. [DOI] [PubMed] [Google Scholar]
- 71.Shockman, G. D., and J.-V. Holtje. 1994. Microbial peptidoglycan (murein) hydrolases. Elsevier Science Publishers B. V., Amsterdam, The Netherlands.
- 72.Shockman, G. D., J. S. Thompson, and M. J. Conover. 1967. The autolytic enzyme system of Streptococcus faecalis. II. Partial characterization of the autolysin and its substrate. Biochemistry 6:1054-1065. [DOI] [PubMed] [Google Scholar]
- 73.Shungu, D. L., J. B. Cornett, and G. D. Shockman. 1979. Morphological and physiological study of autolytic-defective Streptococcus faecium strains. J. Bacteriol. 138:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 75.Steen, A., E. Palumbo, M. Deghorain, P. S. Cocconcelli, J. Delcour, O. P. Kuipers, J. Kok, G. Buist, and P. Hols. 2005. Autolysis of Lactococcus lactis is increased upon d-alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol. 187:114-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strathdee, C. A., and R. Y. Lo. 1989. Cloning, nucleotide sequence, and characterization of genes encoding the secretion function of the Pasteurella haemolytica leukotoxin determinant. J. Bacteriol. 171:916-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sugai, M., H. Komatsuzawa, T. Akiyama, Y. M. Hong, T. Oshida, Y. Miyake, T. Yamaguchi, and H. Suginaka. 1995. Identification of endo-beta-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J. Bacteriol. 177:1491-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takahashi, J., H. Komatsuzawa, S. Yamada, T. Nishida, H. Labischinski, T. Fujiwara, M. Ohara, J. Yamagishi, and M. Sugai. 2002. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46:601-612. [DOI] [PubMed] [Google Scholar]
- 79.Tyrrell, E. A. 1973. Autolysis of Listeria monocytogenes. J. Bacteriol. 113:1046-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ward, J. B., and R. Williamson. 1984. Bacterial autolysins: specificity and function. Elsevier Science Publishers B. V., Amsterdam, The Netherlands.
- 81.Wen, Z. T., and R. A. Burne. 2001. Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45:31-36. [DOI] [PubMed] [Google Scholar]
- 82.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Whatmore, A. M., and C. G. Dowson. 1999. The autolysin-encoding gene (lytA) of Streptococcus pneumoniae displays restricted allelic variation despite localized recombination events with genes of pneumococcal bacteriophage encoding cell wall lytic enzymes. Infect. Immun. 67:4551-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilkins, J. C., D. Beighton, and K. A. Homer. 2003. Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl. Environ. Microbiol. 69:5290-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wren, B. W. 1991. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol. Microbiol. 5:797-803. [DOI] [PubMed] [Google Scholar]
- 86.Wuenscher, M. D., S. Kohler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamada, S., M. Sugai, H. Komatsuzawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang, S. J., K. C. Rice, R. J. Brown, T. G. Patton, L. E. Liou, Y. H. Park, and K. W. Bayles. 2005. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J. Bacteriol. 187:5893-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshimura, G., H. Komatsuzawa, J. Kajimura, T. Fujiwara, M. Ohara, K. Kozai, and M. Sugai. 2004. Zymographic characterization of bacteriolytic enzymes produced by oral streptococci. Microbiol. Immunol. 48:465-469. [DOI] [PubMed] [Google Scholar]
- 90.Zahner, D., K. Kaminski, M. van der Linden, T. Mascher, M. Meral, and R. Hakenbeck. 2002. The ciaR/ciaH regulatory network of Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 4:211-216. [PubMed] [Google Scholar]