Abstract
The physiological changes in an F1-ATPase-defective mutant of Escherichia coli W1485 growing in a glucose-limited chemostat included a decreased growth yield (60%) and increased specific rates of both glucose consumption (168%) and respiration (171%). Flux analysis revealed that the mutant showed approximately twice as much flow in glycolysis but only an 18% increase in the tricarboxylic acid (TCA) cycle, owing to the excretion of acetate, where most of the increased glycolytic flux was directed. Genetic and biochemical analyses of the mutant revealed the downregulation of many TCA cycle enzymes, including citrate synthase, and the upregulation of the pyruvate dehydrogenase complex in both transcription and enzyme activities. These changes seemed to contribute to acetate excretion in the mutant. No transcriptional changes were observed in the glycolytic enzymes, despite the enhanced glycolysis. The most significant alterations were found in the respiratory-chain components. The total activity of NADH dehydrogenases (NDHs) and terminal oxidases increased about twofold in the mutant, which accounted for its higher respiration rate. These changes arose primarily from the increased (3.7-fold) enzyme activity of NDH-2 and an increased amount of cytochrome bd in the mutant. Transcriptional upregulation appeared to be involved in these phenomena. As NDH-2 cannot generate an electrochemical gradient of protons and as cytochrome bd is inferior to cytochrome bo3 in this ability, the mutant was able to recycle NADH at a higher rate than the parent and avoid generating an excess proton-motive force. We discuss the physiological benefits of the alterations in the mutant.
The elucidation of the regulatory mechanism of glycolytic flux is critical for the development of effective fermentation processes for the production of useful metabolites by microorganisms. Glycolytic flux in Escherichia coli is controlled primarily by the ATP demand of the cells rather than by glycolytic enzymes (22). For example, defects in the activity of F1Fo-ATP synthase that impair oxidative phosphorylation (21, 38, 40) or increased ATPase activity in hydrolyzing ATP (22), which both lead to a reduced ATP/ADP ratio (21, 22), result in enhanced rates of glucose consumption. The enhancement of glucose consumption by defective F1Fo-ATP synthase activity has also been reported for the gram-positive bacteria Bacillus subtilis (32) and Corynebacterium glutamicum (34), which are industrially important. Several attempts have been made to apply these findings to the production of useful metabolites from glucose by fermentation. Our group reported the first successful application of pyruvate production, using an E. coli mutant with a defective F1-ATPase (40). In this case, enhanced pyruvate production was achieved with an increased rate of glucose consumption. The effectiveness of F1Fo-ATP synthase defects for the production of acetate (9), as well as pyruvate (8), has also been reported for different E. coli mutants. Recently, we demonstrated that the mutation also works for the improvement of glutamate production in C. glutamicum (1).
Although the ATP/ADP ratio is well accepted as a controlling factor of glycolysis, the underlying mechanisms by which enhanced glucose metabolism is established in response to an energy shortage are still not well understood. The allosteric activation of the key enzymes in the glycolytic pathway, i.e., phosphofructokinase I (2) and pyruvate kinase II (23), under a reduced ATP/ADP ratio is thought to contribute to this phenomenon. However, previous works (21, 38) have suggested the possibility that qualitative changes in certain cell components, such as an increase in b-type cytochrome contents, as well as allosteric control, are involved in the mechanism of enhanced glucose metabolism. To address this important question, we investigated the alterations in cellular physiology that occur in E. coli in response to impaired oxidative phosphorylation due to a defective F1-ATPase. To avoid any metabolic distortion from unnecessary genetic background, we constructed a simple F1-ATPase-defective mutant from wild-type E. coli W1485. Glucose-limited chemostat culture was employed to ensure that cell samples grew at the same rate in the exponential phase. We conducted detailed analyses of metabolic flux, gene expression profiles, and central carbon metabolic and respiratory chain enzyme activities to elucidate the mechanism(s) of enhanced glucose metabolism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The wild-type strain E. coli W1485 (ATCC 12435) was used. An F1-ATPase-defective mutant, HBA-1 (atpA401 bgl+), was constructed by the P1kc transduction of atpA401 (7), a defective gene for the α subunit of F1-ATPase, into strain W1485. This mutant allele was first isolated in 1971 by Butlin et al. (7) as a gene (uncA401) that causes uncoupling of phosphorylation associated with electron transport. The E. coli K-12 strain carrying uncA401 showed negligible activity of Ca2+/Mg2+-activated ATPase (7). A series of intensive investigations of this mutant allele has located this mutation in the α subunit of F1-ATPase (13), and the sequence analysis has revealed a single base change that resulted in the replacement of Ser 373 with Phe (29). This mutant F1-ATPase has been shown to have virtually no ATPase activity yet retain the same subunit (α, β, γ, δ, and ɛ) organization in terms of molecular weight, stoichiometry (α3β3γδɛ), and arrangement (5). This mutant F1-ATPase has been suggested to bind to ATPase-depleted membranes and maintain the proton impermeability of the membrane (5). This was further confirmed in our preliminary experiment, in which similar levels of valinomycin-induced artificial membrane potential were monitored using inside-out membrane vesicles prepared from strains W1485 and HBA-1 as monitored by the fluorescence quenching method {probe, bis-(1,3-dibutylbarbituric acid)pentamethine oxonol [DiBAC4(5)]} (data not shown). Therefore, the membrane of strain HBA-1 has also been confirmed to be sealed and maintain a normal level of proton impermeability. To obtain transductants effectively, atpA401 was cotransduced with bgl+, as described previously, using AN718bgl-7 as the donor strain (40). Almost no ATPase activity was detected in strain HBA-1 when enzyme activity was measured as described previously (40). Both strains were cultured in a glucose-limited chemostat in modified M9 minimal medium containing trace elements to stabilize the continuous culture. The medium contained 14.7 g/liter Na2HPO4 12H2O, 3.0 g/liter KH2PO4, 0.5 g/liter NaCl, 1.0 g/liter NH4Cl, 1.0 mM MgSO4, 0.1 mM CaCl2, 1.0 μM FeCl3, 0.03 μM (NH4)6Mo7O24, 4 μM H3BO3, 0.3 μM CoCl2, 0.1 μM CuSO4, 0.8 μM MnCl2, 0.1 μM ZnSO4, and 2 g/liter glucose as a carbon source. The continuous chemostat culture was conducted at a dilution rate of 0.2 h−1, with a working volume of 750 ml, in a 2-liter jar fermentor. The cultures were aerated at 1.5 liter/min, with stirring at 700 rpm. Dissolved oxygen in the culture broth of both parent and mutant was monitored by a dissolved oxygen electrode and was maintained at about 90%. The culture temperature was controlled at 37°C, and the pH was adjusted to 7.0 with NaOH.
Fermentation analysis.
Growth was measured by the spectrophotometric absorbance of the culture broth at 660 nm. The concentration of glucose remaining in the culture broth was determined by the glucose oxidase method, using Glucose C2 (Wako Pure Chemical Industries, Ltd., Osaka, Japan). Organic acids in the culture broth were determined by high-pressure liquid chromatography (column, AMINEX HPX-87H; mobile phase, 0.01 N H2SO4; flow rate, 0.6 ml/min; detection, absorbance at 210 nm; Bio-Rad Laboratories, Hercules, CA). The respiration rate of the bacterial cells during chemostatic culture was measured using a dissolved oxygen analyzer (model MD-1000; Iijima Electronics Corporation, Gamagori, Aichi, Japan) equipped with a Clark-type oxygen electrode. Measurements were conducted at 37°C in the airtight chamber within the range yielding a linear relationship between the cell concentration and the oxygen consumption rate. Our calculation assumed the oxygen solubility in the 37°C medium to be 0.214 mM. The results were expressed in mmol O2 h−1 g−1 (dry cell weight). The dry cell weights of strains W1485 and HBA-1 were determined from the correspondence of one optical density unit at 660 nm to 0.414 mg and 0.411 mg (dry cell weight) per ml, respectively.
Flux analysis.
The metabolic fluxes of the wild-type strain and the mutant were estimated using the stoichiometric approach described by Holms (19). This method provides the way to calculate metabolic fluxes within the central metabolic pathways in E. coli growing on various single carbon sources at a constant growth rate. The idea is to balance the metabolic events in the conversion of feedstock (glucose) to biomass and by-products by using the defined metabolic pathways and the experimental data for growth rate, glucose consumption, by-product formation, and biomass production. The kinetic parameters (specific rates of glucose consumption and metabolite production) in chemostat culture and the amounts of precursor metabolites required for the biosynthesis of building blocks (27) were used to calculate the fluxes in the central metabolic pathways.
Extraction of total RNA.
Cells in the chemostat culture were withdrawn and immediately mixed with crushed ice prepared at −80°C. The mixtures were centrifuged at 8,000 × g at 4°C for 10 min, and the supernatants were discarded. The RNA was isolated from the cell pellet with ISOGEN (Nippon Gene Co., Ltd., Toyama, Tomaya, Japan), according to the manufacturer's instructions. The RNA was treated with RQ1 RNase-free DNase (Promega Corporation, Madison, WI) and extracted again with ISOGEN. The concentration and quality of the total RNA yield were determined spectrophotometrically and by agarose gel electrophoresis. The extracted RNA was kept at −80°C until used.
DNA array analysis.
For E. coli-specific primed cDNA synthesis, 2 μg total RNA and 4 μl E. coli cDNA-labeling primers (Sigma-Aldrich Corporation, St. Louis, MO) were added to the transcription mixture (6 μl 5× first-strand buffer and 1 μl each of 10 mM dATP, 10 mM dGTP, and 10 mM dTTP), and the total volume was adjusted to 26.5 μl with RNase-free water. The samples were incubated at 90°C for 2 min and were kept at 42°C for 20 min. Then, 0.5 μl RNase inhibitor (20 U RNase OUT; Invitrogen Corporation, Carlsbad, CA), 1 μl reverse transcriptase (200 U, SuperScript II; Invitrogen), and 2 μl [α-33P]dCTP (20 μCi; GE Healthcare Bio-Sciences Corp., Piscataway, NJ) were added to the reaction mixture. After incubation at 42°C for 2.5 h, the labeled cDNA was purified on a Sephadex G-25 spin column (GE Healthcare Bio-Sciences). The purified cDNA was denatured at 94°C for 10 min and immediately chilled on ice. The cDNA probe thus prepared was used to perform the hybridization experiment with Panorama E. coli gene arrays (Sigma-Aldrich) as described in the manufacturer's instructions. After hybridization, the arrays were exposed to imaging plates (Fuji Photo Film Co., Ltd., Minami-Ashigara, Kanagawa, Japan) for 48 h. The exposed imaging plates were scanned with BAS-5000 (Fuji Photo Film). Data analysis was performed with Array Gauge software (v 1.2; Fuji Photo Film). The data were calculated as the averages and standard deviations for eight independent experiments and expressed as a fraction of the total hybridization signal on each DNA array filter. A two-tailed Student's t test P value of <0.05 was considered statistically significant.
Northern blot analysis.
The extracted total RNA was separated by formaldehyde-agarose gel electrophoresis (9% formaldehyde, 1× MOPS [morpholinepropanesulfonic acid] buffer, pH 5.0, 5 mM sodium acetate, 1 mM EDTA, 1% agarose). The separated RNA was transferred onto a Hybond-N+ membrane (GE Healthcare Bio-Sciences) by the capillary method. To detect hns gene expression with the hybridization probe, a 0.41-kb DNA fragment was amplified by PCR with the following primer set: 5′-CGAAGCACTTAAAATTCTGA-3′ and 5′-TTATTGCTTGATCAGGAAAT-3′. Northern hybridization was carried out using AlkPhos Direct and ECF substrate (GE Healthcare Bio-Sciences). The signals were quantified by Typhoon 8600 (GE Healthcare Bio-Sciences) and ImageQuant software (v 5.2; Molecular Dynamics, Sunnyvale, CA).
Real-time PCR analysis.
The reaction mixture containing 5 μg total RNA, 1 μl random primers (300 ng; Invitrogen), and 1 μl 10 mM deoxynucleoside triphosphate mixture in a total volume of 12 μl was incubated at 65°C for 5 min and immediately chilled on ice. Then, 4 μl of 5× first-strand buffer, 2 μl 0.1 M dithiothreitol (DTT), and 1 μl RNase inhibitor (40 U RNase OUT; Invitrogen) were added, and the mixture was incubated at 25°C for 10 min and then at 42°C for 2 min. After that, 1 μl reverse transcriptase (200 U, SuperScript II; Invitrogen) was added, and the mixture was incubated at 42°C for 90 min and then 70°C for 15 min. The real-time PCR was carried out in a 50-μl (total volume) mixture containing 25 μl 2× TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 900 nM each of forward and reverse primers, 200 nM TaqMan probe specific for the target gene, and 5 μl of the cDNA sample. The amplification and detection of specific products were performed with the ABI PRISM 7000 sequence detection system (Applied Biosystems) using the following profile: incubation at 50°C for 2 min and 95°C for 10 min, 40 cycles at 95°C for 15 s, and incubation at 60°C for 1 min. Data analysis was performed using the ABI PRISM 7000 sequence detection system software (v 1.0; Applied Biosystems). Each sample was analyzed in duplicate. The sequences for the TaqMan probes and primers for the target genes were as follows: ndh (probe, 5′-FAM [6-carboxyfluorescein]-CTGCTGCGGCCCACAACGAG-TAMRA [6-carboxytetramethylrhodamine]-3′; forward primer, 5′-TGCGTTACCGCCACGTATC-3′; reverse primer, 5′-ACGCGAACGCCAAGTTTC-3′), cyoA (probe, 5′-FAM-TTCCCGCAATCTTGATGGCT-TAMRA-3′; forward primer, 5′-GGCCTGATGTTGATTGTCGTT-3′; reverse primer, 5′-GGTACTTCCAGGCGAAACCA-3′), cydA (probe, 5′-FAM-TTGCCTTGACCGCGATGTACCACTTC-TAMRA-3′; forward primer, 5′-TCGAACTGTCGCGCTTACAG-3′; reverse primer, 5′-CGAGCGTCAGTGGCACAA-3′). For the endogenous control, the 16S rRNA gene rrsA was used (probe, 5′-FAM-CCGGGCCTTGTACACACCGCC-TAMRA-3′; forward primer, 5′-GAATGCCACGGTGAATACGTT-3′; reverse primer, 5′-ACCCACTCCCATGGTGTGA-3′). A relative standard curve method was used to calculate the relative expression level of the target gene. The expression ratio was obtained by dividing the relative expression level of the mutant by that of the parent.
Measurement of enzymes in central carbon metabolism.
Cells were harvested by centrifugation, washed with an appropriate buffer, and kept at −20°C until use. The cells were disrupted by sonication in the same buffer, and the cell debris was removed by centrifugation at 39,000 × g at 4°C for 40 min. The supernatant was gel filtered using a PD-10 column (GE Healthcare Bio-Sciences) with the same buffer to remove low-molecular-weight materials. The eluate was used as the crude enzyme for the assay. The composition of the buffer system is included in the description of the assay conditions for the respective enzymes. Enzyme activity was monitored spectrophotometrically using a Beckman DU 7400 spectrophotometer (Beckman Coulter, Inc., Fullerton, CA) at 25°C. The protein concentration of the crude enzyme was determined using the Bio-Rad protein assay (Bio-Rad Laboratories), with bovine serum albumin as the standard. The specific activity of each enzyme under the assay conditions was expressed in nmol min−1 mg protein−1. For pyruvate dehydrogenase (PDH), 50 mM potassium phosphate buffer (pH 8.1) was used as the washing and extraction buffer. The reaction mixture consisted of 50 mM potassium phosphate buffer (pH 8.1), 0.05 mM coenzyme A (CoA), 3 mM l-cysteine, 2.33 mM NAD+, 0.2 mM thiamine pyrophosphate, 1 mM MgSO4, 2 mM sodium pyruvate, and the crude enzyme. The reaction was initiated by the addition of sodium pyruvate, and the NADH concentration increase was monitored at 340 nm (39). For acetate kinase, 50 mM of imidazole-HCl buffer (pH 7.3) containing 10 mM MgCl2 was used as the washing and extraction buffer. The reaction mixture consisted of 50 mM imidazole-HCl buffer (pH 7.3), 10 mM MgCl2, 12 mM acetyl phosphate, 5 mM ADP, 10 mM glucose, 1.6 mM NADP, hexokinase (56 U/ml), and glucose 6-phosphate-dehydrogenase (1.5 U/ml). The reaction was initiated by the addition of ADP, and ATP formation was measured by monitoring the increase of the NADPH concentration at 340 nm (31). For citrate synthase, 20 mM Tris-HCl (pH 8.0) containing 10 mM MgCl2 and 1 mM EDTA was used as the washing and extraction buffer. Activity was measured in the reaction mixture, which contained 100 mM Tris-HCl buffer (pH 8.0), 0.16 mM acetyl-CoA, 0.2 mM oxaloacetic acid, and 0.1 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). The reaction was initiated by the addition of oxaloacetic acid. The CoA yield was measured by monitoring the absorbance increase at 412 nm. The molecular extinction coefficient of 13,600 M−1 cm−1 for 5-mercapto-2-nitrobenzoic acid was used to calculate the enzyme activity (36). For succinyl-CoA synthetase (6), 20 mM potassium phosphate buffer (pH 7.2) containing 20 mM MgCl2 was used as the washing and extraction buffer. The reaction mixture contained 50 mM Tris-HCl buffer (pH 7.2), 10 mM MgCl2, 100 mM KCl, 10 mM sodium succinate, 0.1 mM CoA, and 0.4 mM ATP. The reaction was initiated by the addition of ATP, and the formation of succinyl-CoA was measured by monitoring the absorbance increase at 230 nm. For succinyl-CoA, we used the molar extinction coefficient of 4,900 M−1 cm−1 at 230 nm to calculate enzyme activity. For malate dehydrogenase, 7 mM potassium phosphate buffer (pH 7.0) containing 30% glycerol and 3.5 mM DTT was used as the washing and extraction buffer. The reaction mixture consisted of 100 mM potassium phosphate buffer (pH 7.2), 0.13 mM NADH, and 0.33 mM oxaloacetic acid. The reaction was initiated by the addition of oxaloacetic acid. The decrease of NADH, coupled with the formation of malate, was monitored at 340 nm (26).
Measurement of enzymes in the respiratory chain.
Cells were washed with 50 mM potassium phosphate buffer (pH 7.5) containing 5 mM MgSO4, 1 mM DTT, and 10% glycerol, resuspended in the same buffer, and then disrupted twice using a French pressure cell (Ohtake Works, Tokyo, Japan) at 16,000 lb/in2. The mixtures were centrifuged at 8,000 × g at 4°C for 10 min, and the supernatant was ultracentrifuged at 120,000 × g, at 0°C for 2 h. The membrane fraction was suspended by homogenization with a Teflon-coated homogenizer in the same buffer and used as the crude enzyme for the NADH dehydrogenase (NDH) assay. The activities of the NDHs were measured by monitoring the decrease of the NADH or deamino-NADH concentration at 340 nm. The reaction mixture consisted of 50 mM potassium phosphate buffer (pH 7.5), 5 mM MgSO4, and 0.125 mM of either NADH or deamino-NADH substrate. The reaction was initiated by the addition of the crude enzyme. As deamino-NADH is the substrate for NDH-1 but not for NDH-2 (25), the NDH-2 activity was calculated by subtracting the deamino-NADH oxidase activity (NDH-1 activity) from the NADH oxidase activity (total NDH activity). The protein concentration of the crude enzyme was determined using the Bio-Rad protein assay (Bio-Rad Laboratories), with bovine serum albumin as the standard. The molar extinction coefficient of 6,220 M−1 cm−1 at 340 nm for both substrates was used to calculate the specific activity, which was expressed in nmol min−1 mg protein−1. The aerobic respiratory chain of E. coli contains two types of terminal oxidases, cytochrome bo3 oxidase and cytochrome bd oxidase. The bo3-type oxidase is more efficient (2H+/e−) than the bd-type oxidase (1H+/e−) in creating the electrochemical gradient of protons. The activities of these oxidases cannot be measured separately, so the total activity was measured as ubiquinol-2 (Q2H2) oxidase activity. The crude enzyme for the assay of Q2H2 oxidase activity was prepared in the same manner as that described for the NDHs, except that DTT and glycerol were omitted from the buffer used for cell washing and disruption. The measurement was conducted with 30 μM Q2H2 in 50 mM potassium phosphate buffer (pH 7.5) containing 0.1% Tween 20. The absorbance increase at 275 nm was monitored after the enzyme was added to start the reaction. The protein concentration of the crude enzyme was measured by the modified Lowry method, with bovine serum albumin as the standard (12). The molar extinction coefficient of 12,250 M−1 cm−1 at 275 nm was used to calculate the specific activity, which was expressed in nmol min−1 mg protein−1.
Immunoblot analysis of the terminal oxidases.
Immunoblot analysis was conducted to investigate the abundance of each of the terminal oxidases. The membrane preparations used for the Q2H2 oxidase assay were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (23 μg protein per lane). After electrophoresis, the protein bands in the gel were transferred electrophoretically onto a polyvinylidene fluoride membrane (Millipore Corporation, Billerica, MA) at 100 mA for 4 h. After being blocked with 3% gelatin and washed, the membrane was incubated for 2 h with anti-cytochrome bo3 or anti-cytochrome bd antibody. After being incubated for 2 h with protein A-peroxidase, the protein bands were visualized by the addition of color reagents and H2O2. Prestained marker proteins (Bio-Rad Laboratories) were used to estimate the relative molecular weights. Anti-cytochrome bo3 serum against the cytochrome bo3 purified from E. coli (K. Matsushita, unpublished) was obtained and was used at a 50-fold dilution. Tatsushi Mogi (ATP System Project, ERATO, JST) kindly supplied the anti-cytochrome bd serum. The following pretreatment was carried out before use: the anti-cytochrome bd serum (0.1 ml) was mixed with 1 ml (∼10 mg/ml) of the membrane suspension prepared from E. coli GO103 (ΔcydAB′) (30) and then incubated at 30°C for 2 h. The mixture was centrifuged at 10,000 × g for 10 min to obtain the supernatant, which was used as the antibody after a 20-fold dilution.
RESULTS
Enhanced glucose metabolism in an F1-ATPase-defective mutant, HBA-1, as revealed by chemostat cultures.
To evaluate the effects of an F1-ATPase defect on glucose metabolism, the HBA-1 mutant and its parent, W1485, were cultured in M9 minimal medium in a glucose-limited chemostat at the same growth rate (D = 0.2 h−1), and the fermentation parameters for both strains were calculated. As shown in Table 1, the mutant exhibited reduced biomass production (60%), with increased specific rates of both glucose consumption (168%) and respiration (171%), compared with the parent. Analysis of the fermentation products revealed a substantial excretion of acetate by the mutant, whereas no acetate excretion was observed for the parent. These results clearly demonstrated that glucose metabolism by the F1-ATPase-defective mutant was enhanced, in agreement with previous observations for different atp mutants (21, 40).
TABLE 1.
Parameters for the chemostat culturesa
| Strain | YCELLb | Fluxc
|
Respirationd | ||
|---|---|---|---|---|---|
| Glucose | Acetate | 2-OGA | |||
| W1485 | 0.056 (1) | 3.57 (1) | 0.00 | 0.20 | 14.4 (1) |
| HBA-1 | 0.033 (0.6) | 6.00 (1.68) | 4.30 | 0.10 | 24.6 (1.71) |
An F1-ATPase-defective mutant, HBA-1, and its parent strain, W1485, were cultured under glucose-limited chemostat conditions in M9 minimal medium at a dilution rate of 0.2 h−1, as described in Materials and Methods. Values in parentheses indicate differences (n-fold) between strains W1485 and HBA-1.
Cell yield in g (dry cell weight)/mmol glucose.
Consumption of glucose or excretion of acetate or 2-oxo-glutarate (2-OGA) in mmol g −1 (dry cell weight) h−1.
mmol O2 h−1 g −1 (dry cell weight).
Flux analysis.
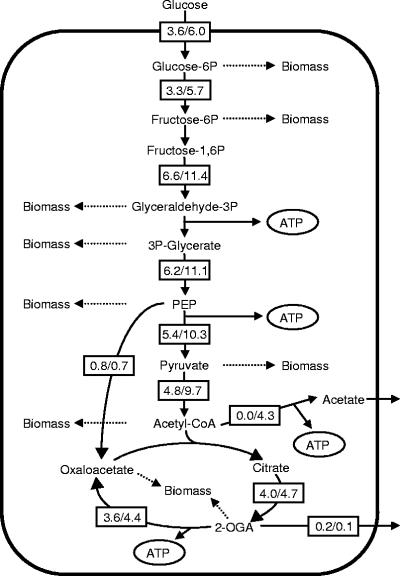
Based on the observed fermentation parameters (Table 1), the metabolic fluxes within the central metabolic pathways were estimated by a stoichiometric approach. As shown in Fig. 1, the glycolytic flux distribution in HBA-1 appeared to be approximately twice that in W1485. However, its distribution in the tricarboxylic acid (TCA) cycle was only 18% higher in the mutant than in the parent. This difference was due to the acetate excretion observed for HBA-1, whereby the increased flux within the glycolytic pathway overflows, thereby reducing the flow of acetyl-CoA entering the TCA cycle. Increased flow of carbon through the glycolytic pathway and in the TCA cycle and increased excretion of acetate were also observed in a different atp mutant growing in glucose minimal medium under batch culture conditions (21).
FIG. 1.
Flux analysis in the central metabolic pathways of the parent and mutant. The values in the boxes show the flux of the parent, W1485 (left), and the mutant, HBA-1 (right), in mmol g−1 (dry cell weight) h−1.
Transcriptome analysis.
To determine whether these alterations in glucose metabolism were accompanied by alterations in gene expression, a DNA array analysis was conducted to compare the genomic expression profiles of the parent and mutant. In general, the differences in the expression levels of most genes appeared to be relatively small (less than twofold) and not statistically significant. Moreover, when differences were detected, most of the genes showed decreased expression levels in the mutant. The similar tendency was observed in the DNA array analysis using cells cultured at a 50% higher dilution rate (D = 0.3 h−1) (data not shown). The results are summarized in Table 2, which lists only the genes of known function and relevance and shows the different expression levels for the two strains. Most striking is the absence of the genes involved in glycolysis, despite the enhanced glucose metabolism in the mutant. In contrast, the expression levels of several genes (gltA, icdA, sucA, sucB, sucD, and mdh) coding for enzymes in the TCA cycle were significantly decreased, to about 50% of the parental levels. Of these, the decreased expression of gltA (44% of the parental level), which codes for citrate synthase, appears to be important in the downregulation of the flux entering the TCA cycle. Also, aceE and aceF (126% of the parental level), which code for enzymes of the PDH complex, showed a tendency to upregulate. We consider these changes, in combination with reduced expression of gltA, to favor the redirection of the glycolytic flux into acetate formation in the mutant (Table 1 and Fig. 1). Moreover, the expression levels of the genes coding for the glyoxylate-shunt enzymes aceA and aceB were significantly decreased in the mutant (about 60% of the parental level). However, the most interesting finding involves the elevated expression levels of some genes coding for respiratory chain enzymes. Although the differences were not as prominent, the expression levels of ndh, coding for NDH-2, and cydA, coding for cytochrome bd oxidase subunit I, were elevated to about 130% of the parental levels. The cyoA expression also showed a tendency toward upregulation, but this was not statistically significant. These data suggest that increased respiration in the mutant may be accompanied by alterations in the composition of the respiratory enzymes, which are regulated at the transcription level. In addition to the genes involved in central metabolism, several genes coding for flagellar formation and cellular structures (ompF) appeared to be significantly repressed. The expression levels of flhC and flhD, which code for a flagellar transcriptional activator, were about 45% below those of the parent, and other flagellar genes (fliC, fliD, flgB, flgC, and flgL) were also repressed. Among the regulatory function genes, only hns, coding for the histone-like protein H-NS, was repressed in the mutant (∼50%). Given that H-NS is an abundant DNA-binding protein involved in numerous cellular processes, including the replication, recombination, and transcriptional regulation of a large number of genes, a decrease in H-NS protein may have profound effects on E. coli cell physiology (33). Besides the genes listed in Table 2, the following genes are worth referring to as significantly downregulated in the mutant (∼50%), with known function but with apparently less physiological relevance in response to bioenergetic stress: hupA and hupB (encoding the DNA-binding protein HU), topA (encoding DNA topoisomerase I), grpE (encoding heat shock protein GrpE), htpG (encoding heat shock protein HtpG), mopB (encoding the GroES protein), and cspD (encoding the cold shock-like protein CspD).
TABLE 2.
Summary of genes showing different expression levels between strains W1485 and HBA-1 grown in glucose-limited chemostat culture, as revealed by DNA array analysisa
| Gene | Gene product | Avg spot intensityb for:
|
SD for:
|
Ratio (HBA-1/W1485)c | ||
|---|---|---|---|---|---|---|
| W1485 | HBA-1 | W1485 | HBA-1 | |||
| aceE | Pyruvate dehydrogenase E1 component | 1.18E−04 | 1.48E−04 | 2.09E−05 | 3.27E−05 | 1.26 |
| aceF | E2 of pyruvate dehydrogenase | 1.52E−04 | 1.92E−04 | 2.79E−05 | 3.52E−05 | 1.26d |
| gltA | Citrate synthase | 4.05E−04 | 1.80E−04 | 7.80E−05 | 6.03E−05 | 0.45d |
| icdA | Isocitrate dehydrogenase | 2.95E−04 | 1.80E−04 | 4.53E−05 | 2.27E−05 | 0.61d |
| sucA | 2-Oxoglutalate dehydrogenase E1 component | 1.34E−04 | 7.63E−05 | 3.45E−05 | 2.25E−05 | 0.57d |
| sucB | Dihydrolipoamide succinyltransferase component (E2) | 1.29E−04 | 6.19E−05 | 2.94E−05 | 1.52E−05 | 0.48d |
| sucD | Succinyl-CoA synthetase α chain | 4.86E−04 | 1.72E−04 | 1.87E−04 | 8.94E−05 | 0.35d |
| mdh | Malate dehydrogenase | 3.18E−04 | 1.34E−04 | 1.14E−04 | 3.66E−05 | 0.42d |
| aceA | Isocitrate lyase | 2.37E−04 | 1.52E−04 | 5.90E−05 | 3.01E−04 | 0.64d |
| aceB | Malate synthase A | 3.82E−04 | 2.22E−04 | 7.72E−05 | 9.11E−05 | 0.58d |
| ndh | NADH dehydrogenase II | 8.25E−05 | 1.09E−04 | 1.18E−05 | 1.94E−05 | 1.32d |
| nuoA | NADH dehydrogenase I chain A | 5.78E−05 | 4.96E−05 | 1.27E−05 | 8.07E−06 | 0.86 |
| cydA | Cytochrome d oxidase subunit I | 3.00E−04 | 4.02E−04 | 1.61E−04 | 2.43E−04 | 1.34d |
| cyoA | Cytochrome o oxidase subunit II | 3.24E−04 | 4.19E−04 | 1.51E−04 | 2.49E−04 | 1.30 |
| flhC | Flagellar transcriptional activator | 1.99E−04 | 8.96E−05 | 4.14E−07 | 8.26E−06 | 0.45d |
| flhD | Flagellar transcriptional activator | 2.57E−04 | 1.12E−04 | 2.68E−07 | 2.28E−05 | 0.44d |
| fliC | Flagellin | 8.68E−04 | 3.64E−04 | 4.55E−05 | 1.00E−04 | 0.42d |
| fliD | Flagellar hook-associated protein 2 | 1.43E−04 | 8.07E−05 | 1.68E−05 | 2.70E−05 | 0.56d |
| flgB | Putative flagellar basal-body rod protein FlgB | 3.19E−04 | 1.85E−04 | 1.85E−05 | 6.18E−05 | 0.58d |
| flgC | Putative flagellar basal-body rod protein FlgC | 1.52E−04 | 8.88E−05 | 6.24E−06 | 2.77E−05 | 0.58d |
| flgL | Flagellar hook-associated protein 3 | 2.18E−04 | 1.20E−04 | 9.01E−06 | 1.79E−05 | 0.55d |
| ompF | Outer membrane protein F precursor | 2.58E−03 | 1.21E−03 | 5.83E−04 | 3.00E−04 | 0.47d |
| nmpC | Outer membrane porin protein NmpC precursor | 1.60E−04 | 1.10E−04 | 5.19E−05 | 1.84E−05 | 0.69d |
| hns | Histone-like protein H-NS | 1.20E−04 | 6.06E−04 | 3.18E−04 | 3.57E−04 | 0.50d |
The results are from eight independent DNA array analyses.
Expressed as the percentage of the total intensity of all the spots on the DNA array.
Expression ratio of HBA-1 to W1485.
Significant t test differences (P < 0.05).
To verify the results of the DNA array analysis (Table 2), the expression levels of four selected genes were monitored by either real-time PCR assay (ndh, cyoA, and cydA) or Northern blot analysis (hns). The results, summarized in Table 3, showed comparable tendencies between the DNA array experiments and the real-time PCR assay or Northern blot analysis. Although we did not monitor all the genes listed in Table 2, these results illustrate the reliability of the DNA array experiments in screening for and locating important changes in the mutant at the level of transcription.
TABLE 3.
Comparison of the expression ratios of several genes, as determined by DNA array and Northern blot analysis or real-time PCR analysis
| Gene | Expression ratio (HBA-1/W1485) determined by indicated analysis
|
||
|---|---|---|---|
| DNA arraya | Northern blotc | Real-time PCRc | |
| ndh | 1.32b | ND | 2.40 |
| cyoA | 1.30 | ND | 1.67 |
| cydA | 1.34b | ND | 1.38 |
| hns | 0.50b | 0.67 | ND |
Adapted from Table 1.
Significant t test differences (P < 0.05).
The assays were repeated two to three times. ND, not determined.
Enzyme activity in central carbon metabolism.
Based on the results of the fermentation analysis (Table 1 and Fig. 1) and the determination of the genomic expression profile (Table 2), several enzymes important to central carbon metabolism were measured to substantiate the observed metabolic changes, especially acetate production, in the mutant (Table 4). In the mutant, the activity of the PDH complex, a key enzyme complex in supplying acetyl-CoA, was twice that in the parent, whereas the activities of three TCA cycle enzymes (citrate synthase, succinyl-CoA synthetase, and malate dehydrogenase) in the mutant appeared to be about half those in the parent. These results agree well with the results obtained in the transcriptome analysis (Table 2), thereby providing validation at the level of enzyme activity. For further insight into the mechanism of acetate production in the mutant, we measured acetate kinase activity, although the transcriptome analysis (Table 2) detected no difference in this enzyme. However, as shown in Table 4, the acetate kinase activity in the mutant was 155% that in the parent. Thus, acetate production in the mutant appears to be triggered by increased acetyl-CoA accumulation attributable to the elevated activity of the PDH complex and the reduced activity of TCA cycle enzymes, especially citrate synthase. The accumulated acetyl-CoA is then readily metabolized into acetate through the acetate kinase-phosphotransacetylase pathway, in which acetate kinase activity is elevated.
TABLE 4.
Activities of several central carbon metabolism enzymes
| Enzyme | Sp acta (nmol/min/mg) for:
|
Ratio (HBA-1/W1485) | |
|---|---|---|---|
| W1485 | HBA-1 | ||
| PDH complex | 74 | 152 | 2.05 |
| Acetate kinase | 2,532 | 3,933 | 1.55 |
| Citrate synthase | 597 | 375 | 0.63 |
| Succinyl-CoA synthetase | 384 | 217 | 0.57 |
| Malate dehydrogenase | 13,064 | 6,039 | 0.46 |
Average values from at least two independent experiments.
Total activities of NDHs and terminal oxidases.
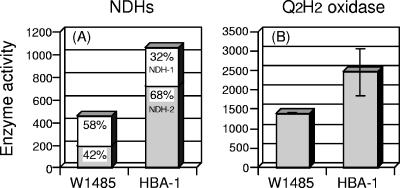
The enhanced rate of respiration (Table 1) as well as the transcriptional upregulation of ndh and cydA (Tables 2 and 3) observed in the mutant led us to measure the total activities of NDHs and terminal oxidases. Two types of NDHs are known in the E. coli respiratory chain, NDH-1 and NDH-2 (25). NDH-1 couples the oxidation of NADH to the creation of the electrochemical gradient of protons, whereas NDH-2 does not. Thus, NDH-1 is important primarily for energy recovery from NADH oxidation, and NDH-2 is thought to work as a bypass to modulate electron flow in response to the growth environment. As shown in Fig. 2A, the total activity of the NDHs (NDH-1 plus NDH-2) was increased 2.3-fold in the mutant compared with that in the parent. The mutant also showed a 1.8-fold increase in the total activity of the terminal oxidases (cytochrome bo3 oxidase plus cytochrome bd oxidase) (Fig. 2B). These changes in respiratory enzyme activity correspond to the increased respiration rate in the mutant (1.7-fold) (Table 1).
FIG. 2.
Analysis of respiratory chain components based on enzyme activity measurements, expressed in nmol min−1 mg protein−1. (A) NDH-1 activities: W1485, 267 ± 125 (n = 10); HBA-1, 338 ± 110 (n = 8). NDH-2 activities: W1485, 197 ± 87 (n = 10); HBA-1, 723 ± 112 (n = 8). (B) Q2H2 oxidase activities: W1485, 1,400 ± 14 (n = 2); HBA-1, 2,460 ± 594 (n = 2).
Analysis of the proportions of the NDHs.
The proportion of each NDH isozyme in the mutant (Fig. 2A) was particularly striking. In the parent, the total NDH activity was composed of about 60% NDH-1 and 40% NDH-2. However, the NDH-2 activity in the mutant was dramatically elevated to 3.7 times that in the parent, whereas only a slight increase (1.3-fold) was detected in the NDH-1 activity. Therefore, we concluded that the increased total activity of the NDHs was attained primarily through the preferential increase of NDH-2 activity in the mutant, which concurs with the observed upregulation of ndh transcription in the mutant (Tables 2 and 3). Consequently, NDH-2 activity is predominant in the mutant, composing up to about 70% of total NDH activity, in contrast to about 40% in the parent.
Analysis of the proportions of the terminal oxidases.
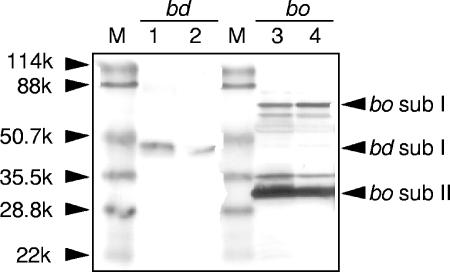
The increased total activity of the terminal oxidases in the mutant (Fig. 2B) prompted us to analyze in detail a possible alteration in the proportion of the terminal oxidases (cytochrome bo3 oxidase plus cytochrome bd oxidase). Thus, we have conducted an immunoblot analysis for the terminal oxidases to gain insight into the cytochrome components of the membrane (Fig. 3). The band corresponding to subunit I of the bd-type oxidase was more abundant in the membrane of the mutant (Fig. 3, lane 1) than of the parent (Fig. 3, lane 2), whereas the bands for subunits I and II of the bo-type oxidase in the membranes did not differ much between the mutant (Fig. 3, lane 3) and parent (Fig. 3, lane 4). These results correspond to the transcriptional upregulation observed for cydA in the mutant (Tables 2 and 3) and indicate an increase in the concentration of the bd-type oxidase relative to that of the bo-type oxidase in the mutant.
FIG. 3.
Immunoblot analysis of terminal oxidases. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the membrane preparations of the parent and mutant, the protein bands were transferred to polyvinylidene fluoride membranes and then probed with anti-cytochrome bo3 serum (bo) or anti-cytochrome bd serum (bd). Lanes 1 and 3, HBA-1; lanes 2 and 4, W1485; M, marker proteins; sub, subunit. The data shown are representative of at least three independent experiments that gave similar results.
DISCUSSION
This study illustrated the overall alterations in an E. coli K-12 cell that are associated with defective oxidative phosphorylation due to a mutation in the F1-ATPase. The results appear to explain how metabolic changes leading to enhanced glucose consumption are possible in response to energy shortages caused by the F1-ATPase defect.
The use of a glucose-limited chemostat for the culturing of the F1-ATPase-defective mutant and its parent enabled a precise characterization of both strains growing exponentially at the same rate (D = 0.2 h−1). The enhanced glucose metabolism in the mutant, as revealed by the fermentation parameters (Table 1), was further substantiated by flux analysis (Fig. 1). We calculated twice as much flux through the glycolytic pathway in the mutant. However, we obtained no evidence from the DNA array analysis to suggest that the transcriptional upregulation of genes is involved in glycolysis (Table 2). Thus, we suggest that the enhanced glycolytic flux was brought about by the allosteric activation of the key enzymes of this pathway, phosphofructokinase I (activation by ADP) (2) and pyruvate kinase II (activation by AMP) (23), under the reduced ATP/ADP ratio. Interestingly, in the mutant, the flux through the TCA cycle was only 18% higher than that in the parent, owing to a redirection of the flux into acetate (Fig. 1), which suggests a stringent metabolic regulation to prevent the flow of the glycolytic pathway from entering the TCA cycle. Results from the enzymatic activities (Table 4) revealed increased activities for the PDH complex and acetate kinase and decreased activities for several TCA cycle enzymes, including citrate synthase. The flux of the TCA cycle in E. coli has been shown to be controlled by citrate synthase through feedback inhibition by NADH as a negative effector (37). As the rate of NADH formation in the mutant with enhanced glucose metabolism would be higher than that in the parent, this inhibition, together with the observed alterations in the enzyme activities of the PDH complex, citrate synthase, and acetate kinase, might direct the glycolytic flux into acetate. From an energetic point of view, the number of ATPs generated by substrate-level phosphorylation from either the metabolism of acetyl-CoA through the TCA cycle (ATP generation at the succinyl-CoA synthetase reaction) or the acetate pathway (generation at the acetate kinase reaction) is the same (Fig. 1). Therefore, the physiological importance of the redirection of the glycolytic flux into acetate is not thought to be related to substrate-level phosphorylation in the acetate pathway, but rather to result from a different aspect. The best explanation is the suppression of additional NADH formation through the TCA cycle, because the F1-ATPase-defective mutant had already generated excess NADH by enhanced glycolysis. As shown in Table 2, the downregulated genes of the TCA cycle enzymes include three (icdA, sucA, and mdh) coding for dehydrogenases generating NADH. These changes, together with reduced citrate synthase activity, appear to reduce NADH formation by the reduced metabolism of acetyl-CoA through the TCA cycle. Furthermore, the downregulation of genes (aceA and aceB) coding for enzymes in the glyoxylate shunt was also demonstrated (Table 2), which could also reduce NADH formation through malate dehydrogenase, while saving acetyl-CoA for substrate-level phosphorylation coupled with acetate formation (acetate kinase) or the TCA cycle (succinyl-CoA synthetase).
In this study, we showed that both the reduction of flow in the TCA cycle and the alteration of respiratory chain components to increase the respiration rate are necessary for the F1-ATPase-defective mutant to achieve enhanced glucose metabolism. As shown in Fig. 2A and 3, preferential increases in NDH-2 activity and cytochrome bd oxidase content were discovered in the respiratory chain of the F1-ATPase-defective mutant. As a component of the respiratory chain of E. coli, each NDH and terminal oxidase isozyme exhibits a different efficiency in generating the electrochemical gradient of protons coupled with electron transfer (4, 15): NDH-1 (2H+/e−), NDH-2 (0H+/e−), bo-type oxidase (2H+/e−), and bd-type oxidase (1H+/e−). The increased components of the respiratory chain are bioenergetically less effective, and the net result is that the mutant can recycle the excess NADH formed in its enhanced central metabolism, thus avoiding the generation of excess proton-motive force. In fact, a 20% higher membrane potential has been measured in the atp deletion mutant (21). Therefore, this alteration in the respiratory components seems beneficial from a bioenergetics point of view for enabling the mutant to maintain homeostasis. The observed alterations in the respiratory chain components are a novel finding of an adaptive response in the F1-ATPase-defective mutant (probably common to all atp mutants), and this is in accord with the aforementioned metabolic redirection strategy that limits NADH formation in the TCA cycle. Another interesting aspect was the mechanism for the transcriptional upregulation of NDH-2 and bd-type oxidase in response to the atp mutation (Tables 2 and 3). Under anaerobic conditions, the expression of ndh, which codes for NDH-2, is subject to repression by Fnr, the fnr (fumarate nitrate reduction) gene product (18). Under aerobic conditions, Fis (the fis gene product; a factor for inversion stimulation) exhibits a growth phase-dependent modulation of transcription from the ndh promoter. In the early logarithmic growth phase, when Fis expression is maximal, ndh expression is repressed by Fis, thus ensuring that energetically efficient NDH-1 is used. This repression is relieved at the stationary phase, when Fis expression decreases. Thus, NDH-2 seems to be fully expressed when cellular energy is sufficient (17). In this context, the mechanism of the transcriptional upregulation of ndh in the F1-ATPase-defective mutant is difficult to interpret and needs to be clarified in future work. On the other hand, the cydAB operon, coding for the bd-type oxidase, has been shown to be regulated by the interplay of three global regulatory proteins, Fnr, ArcA (aerobic respiration control; the arcA gene product), and H-NS, in such a way that its expression is maximal under microaerobic conditions (11, 14, 16, 20). This is physiologically important because the bd-type oxidase has a high affinity for oxygen, thereby working effectively under microaerobic conditions. Under aerobic conditions, however, the expression of the cydAB operon is normally regulated at a low level because of repression by H-NS (16). In the F1-ATPase-defective mutant, bd-type oxidase content increased even under aerobic conditions (Fig. 3). Interestingly, in the mutant, the expression level of H-NS appeared to be repressed to half that of the parent (Tables 2 and 3). Thus, it seems reasonable to attribute the increase of bd-type oxidase content to the decrease of H-NS protein.
The transcription levels of seven genes involved in flagellar biogenesis and ompF coding for porin were found to be downregulated in the mutant (Table 2). The flhC and flhD genes constitute the master operon, the expression of which switches on the expression of all the other genes involved in flagellar biogenesis (10). As flhC and flhD are downregulated to less than half in the mutant (Table 2), the expression of the other genes (flgB, flgC, flgL, fliC, and fliD) seems to be affected accordingly. Once again, a decreased expression level for hns is implicated in these phenomena, because H-NS has been demonstrated to be the positive transcriptional regulator of the flhDC operon in vivo (35), and the hns mutation has been shown to cause a loss of motility due to the lack of flagella (3). The advantages of these responses are not clear. However, the reduced synthesis of such a large multicomponent apparatus (flagellum) and one of the most abundant proteins in E. coli in terms of mass (porin) may contribute to cost savings in biosynthesis (24, 28), especially with an atp mutant, in which the ATP supply is limited.
In this study, we clarified a series of physiological changes associated with an F1-ATPase-defective mutation in E. coli. The mutation produced not only alterations in central carbon metabolism but also changes in respiratory chain and cellular structure components. The overall results illustrate a novel, yet reasonable, strategy enabling E. coli to survive energetically difficult conditions brought about by impaired oxidative phosphorylation. Although experimental evidence is lacking, the observed qualitative changes in the mutant, especially the downregulation of TCA cycle enzymes and the upregulation of cytochrome bd oxidase, are associated with the operation of the global control network(s), such as the Arc two-component system. The possibility of the involvement of some global control network in the adaptive response of the atp mutant and the identification of the signal that is sensed by the network(s) remain to be elucidated.
Acknowledgments
We thank Hisao Ito and Akira Imaizumi at Ajinomoto Co., Inc. (Kawasaki, Japan), for their valuable advice in the DNA array experiments.
This study was supported in part by a grant-in-aid for Scientific Research (B) (10460033 to A.Y.), a grant-in-aid for Scientific Research (C) (13660072 to A.Y.) from the Japan Society for the Promotion of Science, and the Industrial Research Grant Program in 2004 (no. 04A07004 to M.W.) from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
REFERENCES
- 1.Aoki, R., M. Wada, N. Takesue, K. Tanaka, and A. Yokota. 2005. Enhanced glutamic acid production by a H+-ATPase-defective mutant of Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 69:1466-1472. [DOI] [PubMed] [Google Scholar]
- 2.Babul, J. 1978. Phosphofructokinases from Escherichia coli: purification and characterization of the nonallosteric isozyme. J. Biol. Chem. 253:4350-4355. [PubMed] [Google Scholar]
- 3.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogachev, A. V., R. A. Murtazina, and V. P. Skulachev. 1996. H+/e− stoichiometry for NADH dehydrogenase I and dimethyl sulfoxide reductase in anaerobically grown Escherichia coli cells. J. Bacteriol. 178:6233-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bragg, P. D., and C. Hou. 1977. Purification and characterization of the inactive Ca2+, Mg2+-activated adenosine triphosphatase of the unc A− mutant Escherichia coli AN120. Arch. Biochem. Biophys. 178:486-494. [DOI] [PubMed] [Google Scholar]
- 6.Bridger, W. A., R. F. Ramaley, and P. D. Boyer. 1969. Succinyl coenzyme A synthetase from Escherichia coli. Methods Enzymol. 13:70-75. [Google Scholar]
- 7.Butlin, J. D., G. B. Cox, and F. Gibson. 1971. Oxidative phosphorylation in Escherichia coli K12. Biochem. J. 124:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Causey, T. B., K. T. Shanmugam, L. P. Yomano, and L. O. Ingram. 2004. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc. Natl. Acad. Sci. USA 101:2235-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Causey, T. B., S. Zhou, K. T. Shanmugam, and L. O. Ingram. 2003. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: homoacetate production. Proc. Natl. Acad. Sci. USA 100:825-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. A., S. B. Melville, J. A. Albrecht, and R. P. Gunsalus. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25:605-615. [DOI] [PubMed] [Google Scholar]
- 12.Dulley, J. R., and P. A. Grieve. 1975. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal. Biochem. 64:136-141. [DOI] [PubMed] [Google Scholar]
- 13.Dunn, S. D. 1978. Identification of the altered subunit in the inactive F1ATPase of an Escherichia coli uncA mutant. Biochem. Biophys. Res. Commun. 82:596-602. [DOI] [PubMed] [Google Scholar]
- 14.Fu, H.-A., S. Iuchi, and E. C. C. Lin. 1991. The requirement of ArcA and Fnr for peak expression of the cyd operon in Escherichia coli under microaerobic conditions. Mol. Gen. Genet. 226:209-213. [DOI] [PubMed] [Google Scholar]
- 15.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 16.Govantes, F., A. V. Orjalo, and R. P. Gunsalus. 2000. Interplay between three global regulatory proteins mediates oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon. Mol. Microbiol. 38:1061-1073. [DOI] [PubMed] [Google Scholar]
- 17.Green, J., M. F. Anjum, and J. R. Guest. 1996. The ndh-binding protein (Nbp) regulates the ndh gene of Escherichia coli in response to growth phase and is identical to Fis. Mol. Microbiol. 20:1043-1055. [DOI] [PubMed] [Google Scholar]
- 18.Green, J., and J. R. Guest. 1994. Regulation of transcription at the ndh promoter of Escherichia coli by FNR and novel factors. Mol. Microbiol. 12:433-444. [DOI] [PubMed] [Google Scholar]
- 19.Holms, H. 1996. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19:85-116. [DOI] [PubMed] [Google Scholar]
- 20.Iuchi, S., V. Chepuri, H.-A. Fu, R. B. Gennis, and E. C. C. Lin. 1990. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J. Bacteriol. 172:6020-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, P. R., and O. Michelsen. 1992. Carbon and energy metabolism of atp mutants of Escherichia coli. J. Bacteriol. 174:7635-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koebmann, B. J., H. V. Westerhoff, J. L. Snoep, D. Nilsson, and P. R. Jensen. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184:3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotlarz, D., H. Garreau, and H. Buc. 1975. Regulation of the amount and of the activity of phosphofructokinases and pyruvate kinases in Escherichia coli. Biochim. Biophys. Acta 381:257-268. [DOI] [PubMed] [Google Scholar]
- 24.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 25.Matsushita, K., T. Ohnishi, and H. R. Kaback. 1987. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26:7732-7737. [DOI] [PubMed] [Google Scholar]
- 26.Murphey, W. H., C. Barnaby, F. J. Lin, and N. O. Kaplan. 1967. Malate dehydrogenases. II. Purification and properties of Bacillus subtilis, Bacillus stearothermophilus, and Escherichia coli malate dehydrogenases. J. Biol. Chem. 242:1548-1559. [PubMed] [Google Scholar]
- 27.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach, p. 133-173. Sinauer Associates, Inc., Sunderland, Mass.
- 28.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 29.Noumi, T., M. Futai, and H. Kanazawa. 1984. Replacement of serine 373 by phenylalanine in the α subunit of Escherichia coli F1-ATPase results in loss of steady-state catalysis by the enzyme. J. Biol. Chem. 259:10076-10079. [PubMed] [Google Scholar]
- 30.Oden, K. L., L. C. DeVeaux, C. R. T. Vibat, J. E. Cronan, Jr., and R. B. Gennis. 1990. Genomic replacement in Escherichia coli K-12 using covalently closed circular plasmid DNA. Gene 96:29-36. [DOI] [PubMed] [Google Scholar]
- 31.Riondet, C., R. Cachon, Y. Waché, G. Alcaraz, and C. Diviès. 2000. Extracellular oxidoreduction potential modifies carbon and electron flow in Escherichia coli. J. Bacteriol. 182:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santana, M., M. S. Ionescu, A. Vertes, R. Longin, F. Kunst, A. Danchin, and P. Glaser. 1994. Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176:6802-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schröder, O., and R. Wagner. 2002. The bacterial regulatory protein H-NS—a versatile modulator of nucleic acid structures. Biol. Chem. 383:945-960. [DOI] [PubMed] [Google Scholar]
- 34.Sekine, H., T. Shimada, C. Hayashi, A. Ishiguro, F. Tomita, and A. Yokota. 2001. H+-ATPase defect in Corynebacterium glutamicum abolishes glutamic acid production with enhancement of glucose consumption rate. Appl. Microbiol. Biotechnol. 57:534-540. [DOI] [PubMed] [Google Scholar]
- 35.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weitzman, P. D. J. 1969. Citrate synthase from Escherichia coli. Methods Enzymol. 13:22-26. [Google Scholar]
- 37.Weitzman, P. D. J. 1981. Unity and diversity in some bacterial citric acid-cycle enzymes. Adv. Microb. Physiol. 22:185-244. [DOI] [PubMed] [Google Scholar]
- 38.Yokota, A., S, Amachi, and F. Tomita. 1999. Pyruvate, production using defective ATPase activity, p. 2261-2268. In M. C. Flickinger and S. W. Drew (ed.), Encyclopedia of bioprocess technology: fermentation, biocatalysis, and bioseparation. John Wiley & Sons, Inc., New York, N.Y.
- 39.Yokota, A., H. Shimizu, Y. Terasawa, N. Takaoka, and F. Tomita. 1994. Pyruvic acid production by a lipoic acid auxotroph of Escherichia coli W1485. Appl. Microbiol. Biotechnol. 41:638-643. [DOI] [PubMed] [Google Scholar]
- 40.Yokota, A., Y. Terasawa, N. Takaoka, H. Shimizu, and F. Tomita. 1994. Pyruvic acid production by an F1-ATPase-defective mutant of Escherichia coli W1485lip2. Biosci. Biotechnol. Biochem. 58:2164-2167. [DOI] [PubMed] [Google Scholar]