Abstract
The phage HK022 Nun protein excludes phage λ by binding nascent λ pL and pR transcripts at nutL and nutR, respectively, and inducing transcription termination just downstream of these sites. Termination is more efficient at nutL than at nutR. One difference between nutL and nutR is the presence of RNase III processing sites (rIII) located immediately promoter distal to λ nutL. We found that deletion of rIII dramatically reduced Nun transcription arrest in vitro but had little effect on termination in vivo. However, consistent with the in vitro results, overexpression of a transcript carrying nutL and rIII efficiently titrated Nun, allowing λ to grow on a strain that expressed Nun, whereas a transcript carrying only nutL or nutL-rIII with nucleotides 97 to 141 deleted was ineffective. Rnc70, an RNase III mutant that binds but does not cleave rIII, also prevented Nun-mediated λ exclusion. We propose that rIII enhances the on-rate of Nun at nutL, stimulating Nun-mediated arrest in vitro. We have shown that a specific element in rIII, i.e., box C (G89GUGUGUG), strongly enhances arrest on rIII+ templates. Nun-rIII interactions do not stimulate Nun termination in vivo, presumably because formation of the Nun-nutL complex is normally not rate-limiting in the cell. In contrast to Nun, N is not occluded by Rnc70 and is not efficiently titrated by a nutL-rIII transcript.
The Nun protein of bacteriophage HK022 interacts with nascent phage λ RNA at nutL and nutR, inducing transcription termination at various points distal to these sites. Nun expressed from HK022 prophage thus excludes superinfecting λ (17). Host factors involved in Nun-dependent termination are identical to those for λ N-mediated antitermination (18). Both reactions are abrogated by certain mutations in nusA, nusB, nusE (S10), and nusG. In vitro, Nun inhibits transcription elongation but does not release the arrested transcription elongation complex (TEC). Nun activity in vitro requires a TEC that includes nascent nutL or nutR; Nus factors are not absolutely required but enhance the in vitro efficiency of Nun-mediated arrest (7).
The λ nutL and nutR sites are composed of three conserved motifs, including box A (8 nucleotides [nt]), box B (15 nt), and box C (8 nt) (Fig. 1). Box A RNA recruits NusB and NusE into a λ N antitermination complex that includes RNA polymerase (RNAP), NusA, and NusG (15, 16). Box B RNA forms a stem-loop that binds λ N or HK022 Nun and, subsequently, NusA (2, 6, 8, 27). Box B RNA alone binds λ N and Nun with similar affinities. This equivalent affinity for box B RNA does not reflect the inability of λ N to compete with Nun at nutL in vivo. A third conserved motif, box C (8 nt) (Fig. 1), lies downstream of nutL and nutR and does not appear to play a role in λ antitermination (5). The two nut sites differ in the spacer regions between box A, box B, and box C and by a single nt change in the box B loop and the sixth nt in box C.
FIG. 1.
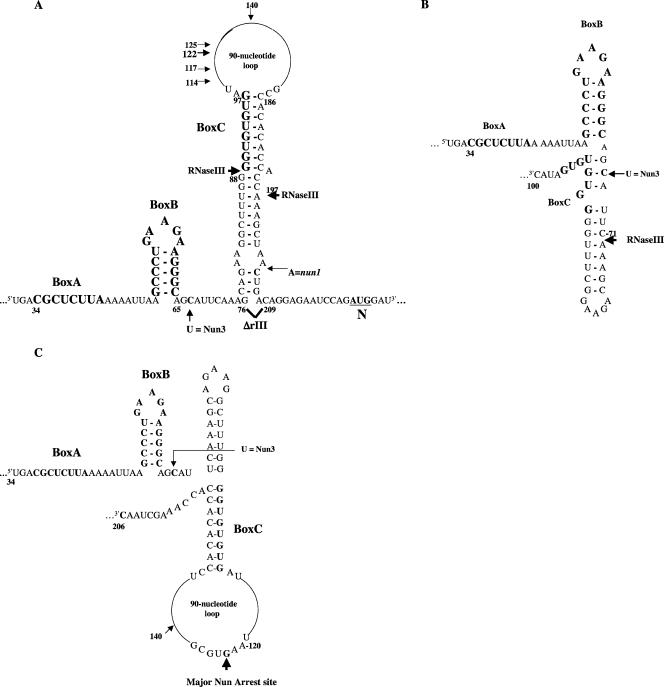
Structures of λ pL transcript. The structures show nutL, including box A (nt 34 to 41), the box B stem-loop (nt 50 to 64), box C (nt 89 to 96), and rIII (nt 76 to 208). The ΔrIII mutation is a precise deletion of rIII. The rIIIΔ76-140, rIIIΔ97-186, and rIIIΔ141-209 deletions remove nucleotides in the indicated ranges from the rIII site. Arrows indicate the two RNase III cleavage sites, at nt 88 and nt 197; the major Nun arrest site, at nt 122; and the minor arrest sites, at nt 114, nt 117, and nt 125. N denotes the translational start of the N gene. Also indicated are the sites of two point mutations, nun1 and nun3, that block translational repression of N by the termination-deficient Nun mutant Nun K106/107D. (A) rIII found in the mature pL transcript (23). (B and C) Proposed (computer-generated) transient RNase III sites formed during transcription, which are cleaved at nt 71.
Nun termination at nutL in vivo is both more efficient and less sensitive to nus mutations than termination at nutR (12, 18). nusG mutations have been isolated that block Nun termination only at nutR. Substitution of the box B sequence of nutL for that of nutR did not increase the sensitivity of the changed nutL transcript to the nusG mutant (1). Washburn et al. (26) proposed that the phenotypic difference between nutL and nutR might be explained by the relative distances of the two nut elements from their respective promoters. nutL is 34 nt from λ pL, whereas nutR lies 227 nt from its cognate promoter.
True termination of Nun-arrested TEC requires host Mfd protein, a DNA helicase that recognizes and dissociates stalled RNAP. Mfd appears to act at nutR but not at nutL, possibly because the short distance between pL and nutL precludes Mfd access. Thus, the off-rate of Nun-TEC complexes is lower at nutL than at nutR (26).
nutL also differs from nutR in that it lies immediately promoter proximal to RNase III cleavage sites (rIII) (Fig. 1). Nascent pL transcripts are cleaved at nt 71, 88, and 197, whereas the mature transcript is cleaved at nt 88 and 197 (13). This suggests that rIII forms transient RNase III substrate structures (Fig. 1B and C) prior to formation of a more stable structure (9) (Fig. 1A).
Cleavage at rIII prevents repression of λ N translation by N or Nun bound at nutL (4, 10). Isolation of rIII mutants resistant to Nun inhibition of N translation (nun3 and nun1) (Fig. 1) suggested that Nun might make contacts with rIII. We present evidence showing that a part of rIII does in fact interact with Nun and that this interaction enhances Nun activity in vitro but not in vivo.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used for this study are shown in Table 1. Standard bacteriological techniques used in strain construction, e.g., transformation, transduction, and medium preparation, were done as described by Silhavy et al. (21). Standard DNA cloning techniques were used as described by Sambrook and Russell (20). Plasmid pRSW101 was made by cloning the lacZ gene from pYW1 (26) into the EcoRI and EcoRV sites of pZero-2 (Invitrogen, Carlsbad, CA) and the lux gene from pYW1 into the XhoI and XbaI sites. Plasmids pRSW110, pRSW111, pRSW112, pRSW113, and pRSW114 were produced by cloning the nutL-rIII regions from λW336, λW335, pL-nutL-rIIIΔ97-106, pL-nutL-rIIIΔ76-140, and pL-nutL-rIIIΔ141-209, which were amplified by PCR using the DNA oligonucleotides 5′-CCGCTCGAGAGGTGACGCTCTTAAAAAT-3′ and 5′-CCGCTCGAGCCATCTGGATTCTCCTG-3′ and cloned into the XhoI site of pRSW101.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| N99 | Wild type | NIH collection |
| N7723 | N99 lacZA21 | Lab collection |
| N7726 | recA56 | Lab collection |
| N9478 | N99 rnc-14 lacZA21 [λcI857] pL-nutL-N-lacZ transcriptional fusion | 28 |
| N9479 | N99 rnc-14 lacZA21 [λcI857] pL-nutL-rIIIΔ76-209-N-lacZ transcriptional fusion | 28 |
| N9480 | N9478(HK022) | This study |
| N9481 | N9479(HK022) | This study |
| N9482 | N9478/pBad-NunK107A | This study |
| N9483 | N9479/pBad-NunK107A | This study |
| N9484 | N9480/pACS21 | This study |
| N9485 | N9480/pSDF701 | This study |
| N9486 | N9481/pACS21 | This study |
| N9487 | N9481/pSDF701 | This study |
| N9488 | N9478/pACS21 | This study |
| RSW149 | N7723/pBad-NunK107A | This study |
| RSW237 | RSW149/pRSW110 | This study |
| RSW238 | RSW149/pRSW111 | This study |
| RSW258 | N9478/pSDF701 | This study |
| RSW279 | N7726/pACS21 | This study |
| RSW280 | N7726/pSDF701 | This study |
| RSW353 | RSW149/pRSW112 | This study |
| RSW354 | RSW149/pRSW113 | This study |
| RSW355 | RSW149/pRSW114 | This study |
| Plasmids | ||
| pBad-NunK107A | pACYC1ori AmpraraC pBad-NunK107A | 26 |
| pACS21 | ColE1ori AmprPlacrnc+ | 25 |
| pSDF701 | ColE1ori AmprPlac rnc-70 | 4 |
| pRSW101 | ColE1ori KanrPlac lacZ-XhoI-lux | This study |
| pRSW110 | ColE1ori KanrPlac lacZ-nutL-rIIIΔ76-209-lux | This study |
| pRSW111 | ColE1ori KanrPlac lacZ-nutL-rIII-lux | This study |
| pRSW112 | ColE1ori KanrPlac lacZ-nutL-rIIIΔ97-186-lux | This study |
| pRSW113 | ColE1ori KanrPlac lacZ-nutL-rIIIΔ76-140-lux | This study |
| pRSW114 | ColE1ori KanrPlac lacZ-nutL-rIIIΔ141-209-lux | This study |
Proteins.
Escherichia coli RNAP and RNase III were purchased from Epicenter (Madison, WI). The Nun protein was a generous gift of Hyeong Kim.
β-Galactosidase assays.
Strains were grown in LB at 32°C with shaking to an optical density at 600 nm of 0.1 and then shifted to 42°C and grown to an optical density at 600 nm of 0.6. Where indicated, the appropriate antibiotic (100 μg/ml ampicillin or 50 μg/ml kanamycin) was present. Nun, when expressed from a plasmid under the control of the pBAD promoter, was induced by the addition of 0.05% arabinose. Cells were assayed for β-galactosidase activity as described by Miller (14).
Templates for in vitro transcription.
DNA templates were generated by PCR, using AmpliTaq DNA polymerase (Roche Diagnostics, Branchburg, NJ) and DNA oligonucleotides (5′-GGAATTCCATATGTCAGATCTCTCACCTACCAAAC-3′ and 5′-AGGGCGGTTAACTGGTTTTG-3′) to amplify a 500-bp fragment of phage λ including pL-nutL-rIII. A pL-nutL-ΔrIII fragment was prepared by using genomic DNA from λ W336 as the template. The remaining mutant templates were produced as follows: the 5′ ends of pL-nutL-rIIIΔ76-141 (5′-CCGTGATCACAATGTGCCAATCGC-3′) and pL-nutL-rIIIboxCmut (5′-CCGTGATCAGCAGAAGGCTTTGCCCACACACATACGAAACGAAGC) were amplified using the indicated DNA oligonucleotides paired with the oligonucleotide 5′-GGAATTCCATATGTCAGATCTCTCACCTACCAAAC-3′, digested with BclI, and ligated to the 3′ fragment of pL-nutL produced with the oligonucleotides 5′-AGGGCGGTTAACTGGTTTTG-3′ and 5′-CGGGATCCTTTGAATGCTGCCC-3′ and digested with BamHI. pL-nutL-rIIIΔ141-209 (5′-CCGTGATCAACAGGAGAATCCAGATG-3′) was amplified using the indicated DNA oligonucleotide paired with 5′-GGAATTCCATATGTCAGATCTCTCACCTACCAAAC-3′, digested with BclI, and ligated to the 3′ fragment of pL-nutL produced with the oligonucleotides 5′-AGGGCGGTTAACTGGTTTTG-3′ and 5′-CGGGATCCGCAGCTAATCCGGAATC-3′ and digested with BamHI. pL-nutL-rIIIΔ97-186 (5′-CCGTGATCACACACACCACCAAAG-3′) was amplified using the indicated DNA oligonucleotide paired with 5′-GGAATTCCATATGTCAGATCTCTCACCTACCAAAC-3′, digested with BclI, and ligated to the 3′ fragment of pL-nutL produced with the oligonucleotides 5′-AGGGCGGTTAACTGGTTTTG-3′ and 5′-CGGGATCCCACACACCCCAAAGC-3′ and digested with BamHI. pL-nutL-rIIIG114A, T117A, G122A, G125A was amplified using the DNA oligonucleotides 5′-CCGTGATCATACGAAACGAAGCATTGGCCG-3′ and 5′-GGAATTCCATATGTCAGATCTCTCACCTACCAAAC-3′, digested with BclI, and ligated to the 3′ fragment of pL-nutL produced with the oligonucleotides 5′-AGGGCGGTTAACTGGTTTTG-3′ and 5′-CGGGATCCGCAGCTAATCCGGAATTGCATTTACTGCTAATGCTTCG -3′ and digested with BamHI.
In vitro termination assay.
Open complexes were formed by preincubating 0.1 pmol template bound with 0.5 units RNAP (Epicenter) in 50 μl TB (20 mM Tris-acetate [pH 7.9], 60 mM potassium acetate, 4 mM magnesium acetate, 1 mm dithiothreitol, 0.25 mg/ml bovine serum albumin, and 5% glycerol) for 5 min at 32°C. The Nun protein, when included, was added at the indicated concentration (1.25, 2.5, or 5 pmol/reaction). Transcription was initiated by the addition of a 10 μM concentration of each nucleoside triphosphate plus 1 μCi [α-32P]ATP. After incubation at 32°C for 5 min, the reactions were terminated by the addition of 50 μl stop solution (375 mM sodium acetate [pH 5.2] and 62.5 mM EDTA). The reaction mixtures were extracted with an equal volume of phenol-chloroform-isoamylalcohol (Sigma) and ethanol precipitated with 3 volumes of 95% ethanol. Extracted RNAs were then resolved in a denaturing 12% polyacrylamide gel and analyzed by autoradiography.
Readthrough transcription was measured by excising the appropriate gel bands and measuring radioactivity in a liquid scintillation counter.
Efficiency of λ plating.
Bacteria (∼109 cells) of the appropriate strains were poured in top agar on LB or LB plus 50 μg/ml kanamycin plus 100 μg/ml ampicillin to produce lawns. IPTG (isopropyl-β-d-thiogalactopyranoside; 0.1 mM) was added to induce transcription of nutL or nutL-rIII from pRSW110-114. Efficiencies of plating were determined by spotting dilutions of λ phage on the bacterial lawns before incubating them overnight at 37°C.
RESULTS
Decreased Nun transcription arrest on rIII mutant templates in vitro.
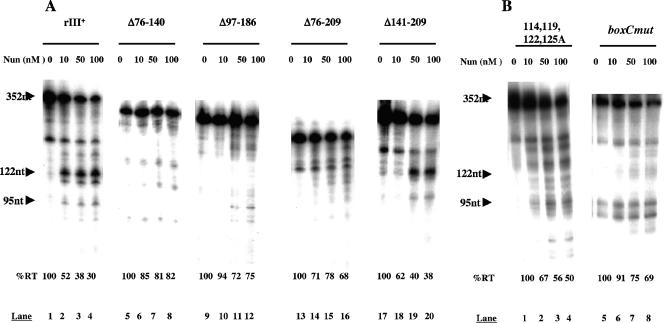
To determine the contribution of rIII to Nun-mediated transcription arrest, we measured Nun activity in vitro, comparing a wild-type λ pL-nutL-rIII template and the same template containing mutations within rIII (Fig. 1). The efficiency of arrest is indicated by the decrease in transcriptional readthrough induced by Nun. The presence of rIII clearly enhanced Nun activity (Fig. 2A). At 100 nM, Nun allowed only 30% readthrough on the rIII+ template, compared to 68% readthrough on the ΔrIII template (Fig. 2A, lanes 4 and 16). Reducing readthrough on the ΔrIII template to levels comparable to those with the wild-type template required 400 nM Nun (data not shown). Deleting the 5′ end of rIII (Δ76-141) was equivalent to deleting the entire rIII site (lanes 5 to 8), as was deleting the loop region (Δ97-186; lanes 9 to 12). Deleting sequences 3′ of nt 140 (Δ141-209) did not affect arrest (lanes 17 to 20).
FIG. 2.
In vitro transcription termination assay with pL-nutL-rIII+ and pL-nutL templates carrying rIII mutations. RT, readthrough transcription. Major Nun-dependent arrest sites are indicated by arrows. All experiments were performed at least three times. %RT is the percentage of readthrough transcription relative to reactions without Nun.
These results suggest that rIII contributes significantly to Nun-dependent arrest at nutL by increasing the affinity of Nun for the TEC. Note that the major Nun arrest site on the rIII+ template occurs at nt 122 (Fig. 2A) (7), after transcription of the proposed transient RNase III substrate structure shown in Fig. 1B but prior to transcription of the structures shown in Fig. 1A and C (9). Thus, Nun must recognize RNA structures or sequences deleted in the ΔrIII template that lie between nt 97 and 141.
We then examined the contributions made by the predominant Nun arrest sites, G114, T117, G122, and G125 (3). Figure 2B, lanes 1 to 4, shows Nun arrest on a nutL template with an A substitution at each site. The quadruple mutant template was less efficient for arrest than the rIII+ template (50% versus 30% readthrough) (Fig. 2A and B, lanes 1 to 4). The pattern of arrest was also changed: arrest was distributed over several sites 3′ of nt 122. These data confirm that rIII+ nt 114, 117, 122, and 125 contribute significantly to Nun arrest in vitro.
We then asked if box C (G89GUGUGUG), a conserved sequence on the ascending arm of rIII, enhanced the efficiency of Nun arrest (5). We constructed a template, boxCmut, which contains the complement of box C (C89CACACAC). Figure 2B, lanes 5 to 8, shows the results of a transcription assay with this template. Nun arrest was dramatically reduced compared to that with the rIII+ template (69% versus 30% readthrough) and was equivalent to the efficiency of Nun arrest on the rIIIΔ76-209 template (68%) (Fig. 2A, lanes 13 to 16). The boxCmut template has two strong Nun-independent spontaneous pauses introduced in the region of nt 89 to 96 (Fig. 2B, lane 5). Our interpretation of these results is that box C enhances arrest at G114, T117, G122, and G125 but is not sufficient for efficient arrest on templates with these sites deleted or mutated.
Titration of Nun by nutL-rIII transcript in vivo.
Our results suggest that rIII and Nun interact and that these interactions increase Nun arrest efficiency in vitro. We next looked for evidence that Nun and rIII interact in vivo. Accordingly, we compared the abilities of transcripts carrying nutL-rIII or nutL-rIII deletions (Δ76-209, Δ141-209, Δ76-140, and Δ97-186) to titrate Nun. The transcripts were expressed in a strain expressing a Nun mutant, NunK107A; Nun activity was monitored by λ plaque formation (Table 2). Although NunK107A arrests transcription with wild-type efficiency in vitro and in vivo (11), we found it to be titrated more easily than wild-type Nun. We previously showed that transcription of plasmid-borne nutL suppressed Nun exclusion of λ, partially in a wild-type host and fully in cells carrying an mfd mutation (26). We suggested that the nutL transcript sequestered and titrated Nun. In confirmation of this result, Nun inhibition of λ plating was partially reversed by transcription of nutL-rIIIΔ76-209 from the multicopy plasmid pRSW110 (efficiency of plating [EOP] of <10−5 [row 2 in Table 2] versus EOP of ∼10−2 [row 4]). In contrast, transcription of nutL-rIII from pRSW111 completely restored λ plating, even though the host was mfd+ (EOP = 1.0 [Table 2, row 3]). Transcription of nutL-rIIIΔ141-209 restored λ plating (EOP = 1.0 [Table 2, row 7]). However, λ plated with an EOP of ∼10−2 on cells expressing nutL-rIIIΔ76-140 or nutL-rIIIΔ97-186 transcripts (Table 2, rows 5 and 6). Thus, the same region of rIII identified as important for termination in vitro, nt 97 to 141, was found to titrate NunK107A in vivo. Alternatively, the sequences from nt 97 to 141 could be important for increasing the in vivo stability of the nutL RNA.
TABLE 2.
Transcription of nutL-rIII increases EOP of λ on a Nun+ straina
| Strain | Presence of nun | nut region | EOP |
|---|---|---|---|
| N7723 | − | 1.0 | |
| RSW149 | + | <10−5 | |
| RSW238 | + | nutL-rIII | 1.0 |
| RSW237 | + | nutL-rIIIΔ76-209 | ∼10−2* |
| RSW353 | + | nutL-rIIIΔ97-186 | ∼10−2* |
| RSW354 | + | nutL-rIIIΔ76-140 | ∼10−2* |
| RSW355 | + | nutL-rIIIΔ141-209 | 1.0 |
Nun was provided by plasmid pBad-NunK107A, where indicated. IPTG (1 mM) was added to induce transcription of nutL or nutL-rIII. Efficiencies of plating were determined at 37°C using λimm434. *, individual plaques were not visible.
Rnc70 suppresses Nun termination at nutL-rIII.
We then asked if RNase III, which also recognizes rIII, could compete with Nun and suppress termination in vivo. To test this notion, we expressed Rnc70, a noncatalytic mutant that binds but does not cleave rIII (4). Inhibition of Nun by Rnc70 was demonstrated by testing λ plating on HK022 lysogens (Table 3). Overexpression of Rnc+ had no effect on λ plating, whereas Rnc70 overexpression completely suppressed Nun exclusion (Table 3, rows 4 to 6). Rnc70 failed to protect λ ΔrIII from exclusion by Nun (data not shown).
TABLE 3.
Rnc70 restores λ plating on an HK022 lysogena
| Strain | Presence of nun | Rnc | EOP |
|---|---|---|---|
| N9478 | − | 1.0 | |
| N9488 | − | WT | 1.0 |
| N9487 | − | Rnc70 | 1.0 |
| N9480 | + | <10−4 | |
| N9484 | + | WT | <10−4 |
| N9485 | + | Rnc70 | 1.0 |
Nun, when present, was provided by an HK022 lysogen. The Rnc column indicates the presence of either a wild type (WT)- or mutant (Rnc70)-overproducing rnc plasmid. Efficiencies of plating relative to that of N9478 were determined using λimm434 at 37°C.
Termination in control HK022 lysogens in the absence of RNase III or in the presence of wild-type RNase III was highly efficient. Readthrough in either rnc+ or rnc mutant strains was <2% (Table 4, rows 2 and 3). Note that the absence of rIII made little difference in the efficiency of termination in this assay (Table 4, rows 6 and 7). We return to this point below (see Table 6).
TABLE 4.
Effects of rnc+ and rnc-70 mutation on Nun-dependent termination in vivoa
| Strain | Presence of sequence
|
β-Galactosidase activity (Miller units) | % Readthrough | ||
|---|---|---|---|---|---|
| rIII | nun | rnc | |||
| N9478 | + | − | − | 2,420 ± 480 | 100 |
| N9480 | + | + | − | 30 ± 2 | 1.2 |
| N9484 | + | + | WT | 18 ± 2 | 0.8 |
| N9485 | + | + | Rnc70 | 560 ± 38 | 23 |
| N9479 | − | − | − | 2,040 ± 200 | 100 |
| N9481 | − | + | − | 27 ± 14 | 1.4 |
| N9486 | − | + | WT | 15 ± 1 | 0.8 |
| N9487 | − | + | Rnc70 | 96 ± 5 | 4.8 |
All strains carry either pL-nutL-rIII-N-lacZ (rIII+) or pL-nutL-ΔrIII-N-lacZ (rIII mutant) under the control of the λ cI857 ts repressor. Nun, when present, was provided by an HK022 lysogen. The rnc indicates the presence of either a wild type (WT)- or mutant (Rnc70)-overproducing rnc plasmid. Strains were grown at 32°C to mid-log phase, shifted to 42°C for 1.5 h to induce pL, and assayed for β-galactosidase activity. Nun termination/arrest is indicated by the β-galactosidase level. Values are given in Miller units and represent the averages ± standard deviations for three or more experiments.
TABLE 6.
Rnc70 does not suppress N function at λ nutLa
| Strain | Presence of recA | Rnc | EOP
|
||
|---|---|---|---|---|---|
| λ | λ nutL400 | λ ΔrIII | |||
| N7723 | + | 1.0 | 1.0 | 1.0 | |
| RSW111 | − | 1.0 | <10−5 | 1.0 | |
| RSW279 | − | WT | 0.6 | <10−5 | 0.6 |
| RSW280 | − | Rnc70 | 0.3 | <10−5 | 0.2 |
recA-negative strains carry recA56. Efficiencies of plating of λ, λ nutL400, and λ ΔrIII were determined using strains W335, Y1167, and W336, respectively, at 37°C.
Overexpression of Rnc70, however, significantly reduced Nun-dependent transcription termination on the pL-nutL-rIII fusion (23% readthrough) (Table 4, row 4). Inhibition of Nun activity by Rnc70 was largely dependent on rIII (Table 4, row 8). This rules out a global effect of Rnc70 overexpression on Nun-dependent transcription termination accounting for the decrease in termination, although the partial suppression of termination in the rIIIΔ76-209 fusion (Table 4, row 8) might thus be explained. Our results support the ideas that Nun recognizes rIII and that Rnc70 competes with Nun for this interaction. The failure of wild-type RNase III to suppress Nun might be explained by a lower affinity for partially transcribed rIII.
Effect of rIII on Nun termination in vivo.
We then examined the contribution of rIII to Nun termination in strains N9482 and N9483 (Table 1). These strains carry the chromosomal N::lacZ transcription fusions λpL-nutL-rIII-N::lacZ and λ pL-nutL-N::lacZ, respectively. Plasmid-borne NunK107A was expressed from the pBAD promoter. Nun concentrations were increased over basal levels, where indicated, by induction of pBAD with 0.05% arabinose.
Nun efficiently terminated transcription on the rIII+ fusion (N9482). β-Galactosidase activity was reduced to 3% that of a control strain without the NunK107A plasmid. Induction of Nun expression with arabinose further reduced the β-galactosidase activity to 0.6% that of the control (Table 5, rows 1 to 3). Nun-mediated termination was almost as efficient in fusions lacking rIII. At basal levels of Nun, β-galactosidase activity was 6.5% that of the control. Nun overexpression reduced the β-galactosidase activity to 1.3% that of the control (Table 5, rows 4 to 6). These results indicate that deletion of rIII decreases the in vivo Nun termination efficiency at nutL twofold, at most. We concluded that Nun interactions with rIII contribute to but are not essential for termination in vivo.
TABLE 5.
In vivo termination activity in nutL-ΔrIII mutanta
| Strain | Presence of sequence
|
Addition of arabinose | β-Galactosidase activity (Miller units) | % Readthrough | |
|---|---|---|---|---|---|
| rIII | nun | ||||
| N9478 | + | − | − | 2,000 ± 440 | 100 |
| N9482 | + | + | − | 60 ± 30 | 3.0 |
| N9482 | + | + | + | 12 ± 0.6 | 0.6 |
| N9479 | − | − | − | 2,000 ± 220 | 100 |
| N9483 | − | + | − | 130 ± 24 | 6.5 |
| N9483 | − | + | + | 26 ± 5 | 1.3 |
All strains carry either pL-nutL-rIII-N-lacZ (rIII+) or pL-nutL-ΔrIII-N-lacZ (rIII mutant) under the control of the λ cI857 ts repressor. Nun was expressed from plasmid pBAD-NunK107A. Arabinose was added to 0.05%, where indicated, to induce Nun. Strains were grown at 32°C to mid-log phase, shifted to 42°C for 1.5 h to induce pL, and assayed for β-galactosidase activity. Values are given in Miller units and represent the averages ± standard deviations for three or more experiments.
Nus factors reduce the requirement for Nun and stimulate Nun arrest in vitro. We therefore asked if rIII might enhance the Nun reaction in HK022 lysogens unable to form the Nus ABEG complex. However, Nun termination in a strain deleted for nusB (D. L. Court, unpublished) was reduced only twofold in vivo and was equally efficient on rIII+ and rIII− templates. It is possible that an as yet unidentified host factor redundant with NusB might stimulate binding to the λ nascent transcript.
rIII and λ N antitermination in the pL operon.
We next asked if rIII affected the ability of λ N to antiterminate at nutL. The λ red and gam genes lie downstream of terminators in the pL operon (19). Although the products of these genes are not essential for λ growth in recA+ hosts, they are absolutely required in a recA-negative background. Table 6 confirms that antitermination is needed for λ plaque formation on a recA56 host. Thus, λ plates with an EOP of 1.0 on recA56, whereas λnutL400, which carries a mutation in box B that eliminates N antitermination (19), fails to grow (EOP, <10−5). In contrast, λ ΔrIII, which carries a precise deletion of rIII, plates with an EOP of 1.0. We concluded that deletion of rIII does not ablate N antitermination at nutL. Table 6 also shows that Rnc70 overexpression, which inhibits Nun termination, does not reduce λ plating on a recA56 host or the plating of a control phage, λ ΔrIII, which does not bind RNase III.
Finally, we found that overproduction of a nutL-rIII transcript did not titrate N. Thus, λ and λr32, a mutant that fails to grow when N is limiting, plated with an EOP of 1.0 on both RSW237, which overexpresses nutL, and RSW238, which overexpresses nutL-rIII (data not shown).
DISCUSSION
Transcription of the λ pL and pR operons is antiterminated by λ N or prematurely terminated by phage HK022 Nun. Both the N and Nun proteins recognize box B RNA sequences of nutL and nutR and act in complex with four host Nus proteins to modify the TEC. Phage λ fails to grow on an HK022 lysogen because Nun both competes with N to prevent antitermination and directly causes transcription termination distal to the nut sites (2, 7). The nut region is also subject to another form of regulation. Translation of N, the first gene in the pL operon, is partially depressed by the rIII secondary structure and strongly inhibited by the N antitermination or Nun termination complex at nutL (Fig. 1) (10, 27). The rIII structure and the N or Nun complex at nutL are thought to interfere with ribosomal attachment to the adjacent N ribosome-binding site (Fig. 1A).
In this work, we asked if rIII also plays a role in Nun termination. We provide evidence that Nun and rIII interact in vitro and in vivo. An rIII site on a template significantly enhanced Nun-dependent transcription arrest at nutL in vitro. Two elements in rIII contribute to the increased efficiency of Nun, namely, box C and four major arrest sites (nt 114, 117, 122, and 125). We propose that box C provides a recognition element for Nun that stimulates arrest at these sites.
In vivo titration assays confirmed that Nun binds rIII. Thus, overproduction of a transcript that included nutL-rIII permitted λ growth on a strain expressing NunK107A. A transcript containing only nutL or nutL-rIII with nt 97 to 141 deleted did not relieve λ exclusion. Finally, overexpression of Rnc70, a catalytically inactive RNase III mutant that binds rIII (4), inhibited Nun, presumably by occluding Nun binding to rIII.
However, experiments in vivo with transcriptional fusions linking pL-nutL-rIII or pL-nutL-ΔrIII and a lacZ reporter revealed, at most, a twofold difference in Nun termination between the two fusions. Our result is consistent with that of Sloan and Weisberg (22), who found that Nun termination was 90% efficient in a pL-nutL-ΔrIII fusion. We concluded that while rIII contributes to Nun activity at nutL, it does not play a major role under our in vivo assay conditions.
To explain the differences between these in vitro and in vivo results, we propose that the Nun reaction in vitro is rate-limited by the binding of Nun to the λ nascent transcript. Nun activity can be measured in a single-round transcription assay. Furthermore, in the absence of Nus factors, the in vitro reaction is suboptimal and requires a large excess of Nun relative to the TEC. In the minimal system described here, therefore, the Nun on-rate is limiting, and Nun binding to the TEC is stimulated by its interaction with rIII. We suggest that under certain, as yet unknown, in vivo conditions, the Nun on-rate might also be limiting, and the binding of Nun to rIII could improve the termination efficiency.
In contrast to deletion of rIII, overproduction of Rnc70, which binds but does not cleave rIII (4), did suppress Nun-dependent termination in a pL-nutL-rIII transcriptional fusion and also restored λ plating on an HK022 lysogen. It is plausible that Rnc70 bound to rIII occludes neighboring sequences that are important for Nun activity. Complete occlusion of nutL, however, is ruled out by the observation that N antitermination is resistant to Rnc70.
We found no evidence that rIII affects λ N antitermination. Unlike Nun, N is not inhibited by overproduction of the nutL-rIII transcript. Plating of λΔrIII on a recA56 host, which requires the pL operon genes red and gam, indicates that rIII is not required for N antitermination in the pL operon. Finally, overproduction of Rnc70 does not prevent λrIII+ propagation on a recA56 host.
These results also shed some light on the failure of N to compete efficiently with Nun at nutL (17). We propose that this is explained, in part, by assuming that Nun recognizes a larger motif than does N at nutL, enhancing its affinity for the pL transcript relative to N. Note that Nun carries 21 amino-terminal amino acids that are lacking in N (24). Whether these residues participate in RNA binding is under investigation. In vitro binding assays with box B RNA did not indicate a difference in affinity between Nun and N (2). It will be interesting to compare the binding of these factors to a larger transcript that includes box C.
Which sequences in the rIII deletion enhance Nun activity in the pL operon? Nun inhibition by Rnc70 indicates that an RNase III binding site is transcribed prior to Nun-mediated termination. Nun termination in vivo forms a series of transcripts within a region 100 nt promoter-distal of nutL (22). Consistent with this result, we (this work and reference 7) found that Nun arrest in vitro occurs predominantly at nt 122. Thus, the RNA sequences recognized by Nun must lie within the proximal 122 nt of the pL operon transcript. Figure 1 shows three possible rIII structures. The structures in Fig. 1B and C are formed transiently and are cleaved by RNase III at nt 71. Figure 1A represents a structure found in the mature pL transcript; this structure is cleaved at nt 88 and nt 197. Our data preclude the structures in Fig. 1A and C, whose endpoints lie beyond the major Nun termination site.
Several candidate sequences lie between nutL and nt 122. First, Nun3, which blocks translation repression by the termination-defective Nun K106/107D protein, is located at nt 67 (11). Two other sequences of potential interest lie in this region of the pL transcript. An inverted repeat, C71AAAGC and G82CUUUG, is found in phages λ, 21, and P22 (9). Box C (G89GUGUGUG) is likewise conserved among temperate phages (5). In this work, we found that box C is required in vitro for efficient arrest at nt 114, 117, 122, and 125. This is the first described role for box C in transcription elongation.
rIII promotes translational repression of N by Nun as well as Nun-mediated transcription termination at low concentrations of Nun. This does not explain, however, the higher efficiency of Nun termination at the pL operon than that at the pR operon (99% versus 87%) (12). Some of the difference is due to Mfd, which releases the Nun-arrested TEC at nutR but not nutL (26). Presumably, other factors also play a role in supporting Nun termination at nutL.
Acknowledgments
We thank Hyeong Kim and Helen Wilson for the generous gift of the Nun protein, for strains, and for helpful discussions.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by grant GM37219 from the National Institutes of Health.
REFERENCES
- 1.Burova, E., S. C. Hung, J. Chen, D. L. Court, J. G. Zhou, G. Mogilnitskiy, and M. E. Gottesman. 1999. Escherichia coli nusG mutations that block transcription termination by coliphage HK022 Nun protein. Mol. Microbiol. 31:1783-1793. [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyay, S., S. C. Hung, A. C. Stuart, A. G. Palmer III, J. Garcia-Mena, A. Das, and M. E. Gottesman. 1995. Interaction between the phage HK022 Nun protein and the nut RNA of phage lambda. Proc. Natl. Acad. Sci. USA 92:12131-12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels, D. L., J. L. Schroeder, W. Szybalski, F. Sanger, A. R. Coulson, G. F. Hong, D. F. Hill, G. B. Petersen, and F. R. Blattner. 1983. Complete annotated lambda sequence, p. 519-676. In R. Hendrix, J. Roberts, F. Stahl, and R. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Dasgupta, S., L. Fernandez, L. Kameyama, T. Inada, Y. Nakamura, A. Pappas, and D. L. Court. 1998. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III—the effect of dsRNA binding on gene expression. Mol. Microbiol. 28:629-640. [DOI] [PubMed] [Google Scholar]
- 5.Friedman, D. I., and M. E. Gottesman. 1983. The lytic mode of λ development, p. 22-51. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), The bacteriophage lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Horwitz, R. J., J. Li, and J. Greenblatt. 1987. An elongation and control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell 51:631-641. [DOI] [PubMed] [Google Scholar]
- 7.Hung, S. C., and M. E. Gottesman. 1995. Phage HK022 Nun protein arrests transcription on phage λ DNA in vitro and competes with the phage λ N antitermination protein. J. Mol. Biol. 247:428-442. [DOI] [PubMed] [Google Scholar]
- 8.Ishihama, A., A. Honda, H. Nagasawa-Fujimori, R. E. Glass, T. Maekawa, and F. Imamoto. 1987. Multivalent regulation of the nusA operon of Escherichia coli. Mol. Gen. Genet. 206:185-191. [DOI] [PubMed] [Google Scholar]
- 9.Kameyama, L., L. Fernandez, D. L. Court, and G. Guarneros. 1991. RNaseIII activation of bacteriophage lambda N synthesis. Mol. Microbiol. 5:2953-2963. [DOI] [PubMed] [Google Scholar]
- 10.Kim, H. C., J.-G. Zhou, H. R. Wilson, G. Mogilnitskiy, D. L. Court, and M. E. Gottesman. 2003. Phage HK022 Nun protein represses translation of phage λ N (transcription termination/translation repression). Proc. Natl. Acad. Sci. USA 100:5308-5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, H. C., and M. E. Gottesman. 2004. Transcription termination by phage HK022 Nun is facilitated by COOH-terminal lysine residues. J. Biol. Chem. 279:13412-13417. [DOI] [PubMed] [Google Scholar]
- 12.Kim, H. C., R. S. Washburn, and M. E. Gottesman. 2006. Role of E. coli NusA in phage HK022 Nun-mediated transcription termination. J. Mol. Biol. 359:10-21. [DOI] [PubMed] [Google Scholar]
- 13.Lozeron, L., J. E. Dahlberg, and W. Szybalski. 1976. Processing of the major leftward mRNA of coliphage lambda. Virology 71:262-277. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Nodwell, J., and J. Greenblatt. 1991. The nut site of bacteriophage λ is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Genes Dev. 5:2141-2151. [DOI] [PubMed] [Google Scholar]
- 16.Patterson, T. A., Z. Zhang, T. Baker, L. L. Johnson, D. I. Friedman, and D. L. Court. 1994. Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination. J. Mol. Biol. 236:217-228. [DOI] [PubMed] [Google Scholar]
- 17.Robert, J., S. B. Sloan, R. A. Weisberg, M. E. Gottesman, R. Robledo, and D. Harbrecht. 1987. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell 51:483-492. [DOI] [PubMed] [Google Scholar]
- 18.Robledo, R., B. L. Atkinson, and M. E. Gottesman. 1991. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J. Mol. Biol. 220:613-619. [DOI] [PubMed] [Google Scholar]
- 19.Salstrom, J. S., M. Fiandt, and W. Szybalski. 1979. N-independent leftward transcription in coliphage lambda: deletions, insertions and new promoters bypassing termination functions. Mol. Gen. Genet. 168:211-230. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Sloan, S. B., and R. A. Weisberg. 1993. Use of a gene encoding a suppressor tRNA as a reporter of transcription: analyzing the action of the Nun protein of bacteriophage HK022. Proc. Natl. Acad. Sci. USA 90:9842-9846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steege, D. A., K. C. Cone, C. Queen, and M. Rosenberg. 1987. Bacteriophage lambda N gene leader RNA. RNA processing and translational initiation signals. J. Biol. Chem. 262:17651-17658. [PubMed] [Google Scholar]
- 24.Stuart, A. C., M. E. Gottesman, and A. G. Palmer III. 2003. The N-terminus is unstructured, but not dynamically disordered, in the complex between HK022 Nun protein and lambda-phage box B RNA hairpin. FEBS Lett. 553:95-98. [DOI] [PubMed] [Google Scholar]
- 25.Takiff, H. E., S. M. Chen, and D. L. Court. 1989. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 174:1544-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Washburn, R. S., Y. Wang, and M. E. Gottesman. 2003. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 329:655-662. [DOI] [PubMed] [Google Scholar]
- 27.Watnick, R. S., and M. E. Gottesman. 1998. Escherichia coli NusA is required for efficient RNA binding by phage HK022 Nun protein. Proc. Natl. Acad. Sci. USA 95:1546-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, H. R., L. Kameyama, J.-G. Zhou, G. Guaneros, and D. L. Court. 1997. Transcriptional repression by a transcriptional elongation factor. Genes Dev. 11:2204-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]