Abstract
Despite its being a leading cause of nosocomal and community-acquired infections, surprisingly little is known about Staphylococcus aureus stress responses. In the current study, Affymetrix S. aureus GeneChips were used to define transcriptome changes in response to cold shock, heat shock, stringent, and SOS response-inducing conditions. Additionally, the RNA turnover properties of each response were measured. Each stress response induced distinct biological processes, subsets of virulence factors, and antibiotic determinants. The results were validated by real-time PCR and stress-mediated changes in antimicrobial agent susceptibility. Collectively, many S. aureus stress-responsive functions are conserved across bacteria, whereas others are unique to the organism. Sets of small stable RNA molecules with no open reading frames were also components of each response. Induction of the stringent, cold shock, and heat shock responses dramatically stabilized most mRNA species. Correlations between mRNA turnover properties and transcript titers suggest that S. aureus stress response-dependent alterations in transcript abundances can, in part, be attributed to alterations in RNA stability. This phenomenon was not observed within SOS-responsive cells.
Staphylococcus aureus is a leading cause of nosocomial and community-acquired infections. The organism owes its ability to cause disease, in part, to the production of a repertoire of virulence factors that modulate its ability to colonize host/inert surfaces, thwart host defenses, and disseminate to satellite sites (6, 39). Another facet of S. aureus pathogenesis is the organism's ability to maintain cellular homeostasis while enduring environmental challenges, such as changes in host cell core temperature or exposure to phagocyte-mediated reactive oxygen species (58). Although considerable progress has been made in defining S. aureus virulence factors and their regulatory networks, surprisingly little is known about the organism's ability to cope with environmental stresses.
Studies from Escherichia coli and, to a lesser extent, Bacillus subtilis indicate that bacteria have developed highly orchestrated responses to environmental stresses, which when elicited alter the organism's cellular physiology in a manner that enhances survival. For instance, DNA-damaging agents trigger induction (derepression) of the SOS response (reviewed in reference 16). Components of the SOS response increase the cell's capacity to inhibit cell division, repair DNA damage, and replicate noninstructive DNA lesions in an error-prone manner. Bacteria cope with conditions of nutrient limitation by eliciting the stringent response, which both reduces the cellular protein synthesis capacity and increases amino acid biosynthesis when substrates for protein synthesis are lacking (42, 52). Products of the cold shock response restore translation apparatus function, which is compromised at low temperatures, and resolve low temperature-mediated mRNA secondary structures that would otherwise impede the translation machinery (25, 26). At elevated temperatures, cells express heat shock factors that degrade/restructure heat-denatured proteins as well as factors that restore temperature-mediated alterations in chromosome topology (33, 61). It is currently unclear if S. aureus elicits similar responses and/or has developed novel strategies to cope with DNA damage, starvation, and temperature changes.
Recent studies indicate that bacterial stress responses are not merely controlled at the level of transcript synthesis. Rather, some responses modulate target mRNA stability to influence protein production (reviewed in reference 53). Perhaps the best example of this involves production of the major E. coli cold shock protein, CspA, an RNA structure-resolving protein that accounts for 13% of the total cellular protein at low temperatures (18). Under cold shock conditions, increased cspA mRNA stability, as opposed to changes in transcript synthesis, primarily accounts for the amount of CspA produced (14). Similarly, Klebsiella pneumoniae nitrogen fixation protein production corresponds to regulated mRNA turnover (10, 28, 29). Moreover, in Vibrio angustum, the cellular response to nutrient depravation is regulated by altering mRNA stability (53).
The focus of the current work is to define the members of S. aureus stress responses and their mechanisms of regulation. Doing so may provide a better understanding of the organism's ability to adapt to environmental challenges and provide novel strategies for the therapeutic intervention of bacterial infections. Accordingly, Affymetrix GeneChips were used to define the S. aureus SOS, stringent, cold shock, and heat shock responses and to measure the mRNA turnover properties of each response. The results indicate that each stress response influences the expression of distinct cellular processes, subsets of virulence factors, and antimicrobial resistance determinants. Many stress-responsive biological processes appear to be conserved across bacteria, whereas others are unique to S. aureus. Induction of the S. aureus cold shock, heat shock, and stringent responses caused dramatic global changes in mRNA turnover. This suggests that stress-mediated changes in mRNA abundances can, in part, be attributed to alterations in RNA stability as opposed to or in addition to changes in transcript synthesis. This phenomenon was not observed under SOS response-inducing conditions. Sets of S. aureus small stable RNA (SSR) molecules with no obvious open reading frames were also components of each stress response. Given the importance of SSR-like molecules in other organisms, it is likely that these stable RNA species influence stress-responsive functions (1, 51, 60).
MATERIALS AND METHODS
Bacterial strains.
S. aureus strain UAMS-1 is a well-characterized methicillin-susceptible clinical osteomyelitis isolate (2, 3, 5, 7, 46).
Growth conditions.
Overnight cultures of UAMS-1 cells were diluted 1:100 in 200 ml fresh brain heart infusion (BHI) medium and were incubated at 37°C at 225 rpm with a flask-to-medium-volume ratio of 5:1. Once cultures reached mid-log phase (optical density at 600 nm, 0.25), they were challenged with either mupirocin (60 μg ml−1; AppliChem, Cheshire, CT) or mitomycin C (1 μg ml−1; Sigma-Aldrich, St. Louis, MO) and were subsequently incubated at 37°C for 30 min with aeration for induction of the stringent or SOS response, respectively. For induction of the cold shock and heat shock responses, cultures of UAMS-1 were grown to mid-log phase (as described above) and were subsequently incubated for an additional 30 min with aeration at 10°C and 42°C, respectively. Following induction of each response, rifampin (200 μg ml−1; Sigma-Aldrich) was added to arrest transcription, and 21 ml of cells was removed at 0, 2.5, 5, 15, and 30 min post-rifampin treatment. Twenty milliliters of each aliquot was added to 20 ml ice-cold acetone-ethanol (1:1) and stored at −80°C overnight; 10−1 and 10−5 dilutions of the remaining 1 ml were plated on BHI-rifampin (200 μg ml−1) agar and BHI agar, respectively. Plates were incubated overnight at 37°C, and viable CFU ml−1 were calculated to ensure that cell proliferation was halted by the addition of rifampin. If rifampin-resistant colonies were detected, the experimental samples were discarded, and the experiment was repeated.
Antibiotic susceptibility assays.
Fifty milliliters of mid-log-phase UAMS-1 cells was either mock treated or challenged with mupirocin (60 μg ml−1) to induce the stringent response (as described above). Next, mock- and mupirocin-treated cultures at time zero (T0) were incubated in the absence or presence of either rifampin (200 μg ml−1) or ciprofloxacin (1.3 μg ml−1, which is the MIC) for an additional 4 h. Cell viability was monitored by determining the total CFU ml−1 at T0 and every hour thereafter. All susceptibility assays were repeated at least twice.
Microarray studies.
Total bacterial RNA was isolated from each sample, labeled, and hybridized to Affymetrix S. aureus GeneChips (Santa Clara, CA) as previously described (46). The S. aureus GeneChips used in this study are the most comprehensive commercially available Affymetrix arrays, representing genomic sequences from S. aureus strains NCTC 8325, COL, N315, and Mu50 as well as intergenic regions. The experiment for each response was repeated twice (biological replicates), and posttranscriptional arrest samples were prepared from each biological replicate. GeneChip signal intensity values for each qualifier at each time point (both pre- and posttranscriptional arrest) were then averaged and normalized to Affymetrix spike-in signals, using GeneSpring 6.2 software (Silicon Genetics, Redwood City, CA). The half-life of each transcript was calculated as the time point at which the T0 signal decreased by a factor of 2, as previously described (46, 49).
Real-time PCR.
Quantitative real-time PCR primers are shown in Table 1. Real time-PCRs were performed as previously described (46). Briefly, 25 ng of total bacterial RNA was reverse transcribed, amplified, and measured using a LightCycler RNA Master SYBR green I kit (Roche Applied Science, Indianapolis, IN) following the manufacturer's recommendations. As an internal control, 25 pg of RNA was used to quantitate rRNA. Transcript concentrations were calculated using LightCycler software, with a LightCycler control cytokine RNA (Roche Applied Science) titration kit as a standard, and were then normalized to the 16S rRNA abundance.
TABLE 1.
Sequences of oligonucleotides used for real-time PCR in this study
| Primer | Oligonucleotide sequence (5′→3′) |
|---|---|
| ilvB-F | ATCGAATATATCGGCAAAATTACAA |
| ilvB-R | AGCATACGACTGTTTTATCAGGATT |
| rpsL-F | ACCACAAAAACGTGGTGTATGTACT |
| rpsL-R | ACACCTGGTAAGTCTTTTACACGTC |
| recA-F | ATATGGAGAAATCTTTCGGTAAAGG |
| recA-R | CAGGACCATAAATTTCAATAATTCG |
| uvrB-F | AATATTCCCAGCCTCTAAAGAAGAA |
| uvrB-R | CTCATCTCGTAATTCTTTCAATCGT |
| 16SrRNA-F | TAACCTACCTATAAGACTGGGATAA |
| 16SrRNA-R | GCTTTCACATCAGACTTAAAAA |
| sarR-F | TTAGTCAACGCAACATTTCAAGTTA |
| sarR-R | GAACTCTGAGCACTTAGCAATCTCT |
| norA-F | AGTGATTTAGGGTTACTTGTTGCTG |
| norA-R | CAACTGCAAACATAAATTCTGACAC |
| clpC-F | AGTAGACGTACGAAAAACAATCCTG |
| clpC-R | GTTGGATTTCTTCCATAACCTTTTT |
| ctsR-F | ATTTGAAGAGTCGAATGAAGATGTC |
| ctsR-R | AATTTTAGTGATTCGGATGTAACCA |
| srtA-F | CTTATCCTAGTGGCAGCATATTTGT |
| srtA-R | GATTTATCTTTCGGAATTTGAGGTT |
| COLSA2731-F | AACGGTACAGTAAAATGGTTTAACG |
| COLSA2731-F | AGTTTGTACGTTAACTGCTTGGTCT |
RESULTS
Cold shock response.
To characterize the ability of S. aureus to adapt to changes in temperature, we first identified members of the organism's cold shock response. Accordingly, S. aureus strain UAMS-1 was grown at 37°C to the mid-log phase of growth, at which point cell cultures were incubated at 10°C for an additional 30 min to induce the cold shock response. Affymetrix S. aureus GeneChips were used to compare the transcript titers of cold-shocked and unshocked cells (grown at 37°C).
The cold shock condition studied did not appreciably affect cell viability (data not shown) but did increase the mRNA titers of 46 genes (Table 2). Transcription of the cold shock gene cspB was induced 9.3-fold at the lower temperature, confirming that the conditions used were appropriate for studying the S. aureus cold shock response. Transcription of the cold shock gene cspA was also upregulated 2.0-fold, but this was not considered significant by the t test (P ≤ 0.05). This correlates with E. coli cspA expression; cspA is strongly induced at 25°C but is marginally upregulated (at the transcriptional level) at a lower temperature (15°C) (57). The majority of cold shock-induced transcripts (36%) included hypothetical or conserved hypothetical genes; the latter are conserved within all publicly available sequenced S. aureus genomes. Two members of the cid regulon, lrgA and lrgB, which are believed to counteract the cell's programmed cell death machinery, were induced (23), as were four competence orthologs (SACOL0813, SACOL0814, SACOL1601, and SACOL1644). Several virulence determinants were induced during cold shock conditions, including two pathogenicity island genes (SACOL0901 and SACOL0902), an enterotoxin gene (seo), a lipase gene (lip), and a sortase gene (srtA). Two putative antimicrobial resistance determinants, mepA (27) and a beta-lactamase-like gene (yycJ), were induced by cold shock. The SOS repressor LexA (59) and a general stress-inducible protein (SACOL0958) that is predicted to bind mRNA were also upregulated at the low temperature. Real-time PCR confirmed that srtA and cspB transcripts were induced 2.2- and >1,000-fold, respectively, by cold shock conditions (data not shown). A total of 416 transcript titers decreased in response to low temperature (see Table S6 in the supplemental material).
TABLE 2.
S. aureus cold shock-induced transcripts
| Category and qualifiera | Fold inductionb | Common name | Locusc | Description |
|---|---|---|---|---|
| Amino acid metabolism | ||||
| sa_c4601s3932_a_at | 2.4* | rocF | SA2154 | Arginase |
| Carbohydrates | ||||
| sa_c8477s7437_at | 2.8 | SA0869 | Phosphoglycerate mutase | |
| Cell division and cell cycle | ||||
| sa_c1078s861_at | 3.3 | SA1191 | Conserved hypothetical protein | |
| Fatty acids and lipids | ||||
| sa_c6688s5833_a_at | 2.3* | lip | N315-SA2463 | Triacylglycerol lipase precursor |
| sa_c7744s6747_at | 2.5 | SA0621 | Substrate-CoA ligase, putative | |
| Hypothetical | ||||
| sa_c4550s9974_x_at | 2.2 | MSSA476-SAS070 | Conserved hypothetical protein | |
| sa_c7348s6387_a_at | 2.2* | N315-SA0413 | Conserved hypothetical protein | |
| sa_c8469s7429_a_at | 4.1 | N315-SA0751 | Conserved hypothetical protein | |
| sa_c9785s10434_at | 3.3* | N315-SA1186 | Conserved hypothetical protein | |
| sa_c6821s5955_a_at | 15.9* | N315-SA2496 | Conserved hypothetical protein | |
| sa_c3224s2774_a_at | 2.9 | SA0161 | Conserved hypothetical protein | |
| sa_c6917s6037_a_at | 3.4* | SA0299 | Hypothetical protein | |
| sa_c7132s6241_a_at | 2.9* | SA0436 | Conserved hypothetical protein | |
| sa_c7215s6283_a_at | 2.4* | SA0448 | Conserved hypothetical protein | |
| sa_c7352s6391_a_at | 2.8* | SA0497 | Conserved hypothetical protein | |
| sa_c7491s6511_at | 2.2 | SA0537 | Conserved hypothetical protein | |
| sa_c8897s7814_a_at | 2.1* | SA0692 | Conserved hypothetical protein | |
| sa_c485s314_at | 28.0* | SA1033 | Hypothetical protein | |
| sa_c10045s10498_at | 2.8* | SA1372 | Hypothetical protein | |
| sa_c1705s1441_a_at | 3.6 | SA1375 | Conserved hypothetical protein | |
| sa_c1960s1685_a_at | 2.4 | SA1438 | Conserved hypothetical protein | |
| sa_c2254s1952_a_at | 2.2 | SA1524 | Conserved hypothetical protein | |
| sa_c2266s1966_a_at | 2.1 | SA1526 | Conserved hypothetical protein | |
| sa_c2807s2375_a_at | 2.4* | SA1664 | Conserved hypothetical protein | |
| sa_c10133s10545_a_at | 2.3* | SA1777 | Conserved hypothetical | |
| sa_c4628s3951_a_at | 9.2 | SA2162 | Conserved hypothetical protein | |
| sa_c10213s10636_at | 2.1* | SA2218 | Conserved hypothetical protein | |
| sa_c6732s5872_a_at | 3.5 | SA2705 | Hypothetical protein | |
| Miscellaneous | ||||
| sa_c10345s9018_a_at | 3.0* | SA0162 | NAD-dependent formate dehydrogenase | |
| sa_c6230s5410_at | 10.4* | lrgA | SA0247 | Holin-like protein |
| sa_c6266s5446_a_at | 4.7* | lrgB | SA0248 | Holin-like protein |
| Regulation | ||||
| sa_c7020s6143_a_at | 4.4 | SA0404 | Transcriptional regulator, MarR family | |
| sa_c9575s8335_a_at | 5.0 | lexA | SA1374 | LexA repressor |
| Resistance | ||||
| sa_c9464s8272_a_at | 2.2 | yycJ | SA0023 | Metallo-beta-lactamase family |
| sa_c7024s6147_a_at | 2.7 | mepA | SA0405 | MATE efflux family protein |
| Stress response | ||||
| sa_c1956s1681_a_at | 9.3 | cspB | SA2731 | Cold shock protein |
| Transport | ||||
| sa_c5431s4700_a_at | 2.8 | SA0882 | ABC transporter | |
| sa_c8327s7303_a_at | 2.8* | SA0813 | Putative ComF protein 1 | |
| sa_c8331s7308_a_at | 3.9* | SA0814 | Competence protein F | |
| sa_c9678s8436_a_at | 3.5* | SA1601 | Putative competence protein ComGA | |
| sa_c2731s2305_a_at | 2.1* | SA1644 | Putative competence protein | |
| Virulence | ||||
| sa_c10522s10973_s_at | 2.6 | srtA | N315-SA2539 | Sortase |
| sa_c7169s10140_a_at | 7.6 | SA0901 | Pathogenicity island protein | |
| sa_c10151s10571_a_at | 2.4 | SA0902 | Pathogenicity island protein | |
| sa_c3571s9834_a_at | 2.2* | seo | SA1648 | Enterotoxin SeO |
Affymetrix S. aureus GeneChip descriptive representing indicated predicted open reading frame (ORF).
*, transcript was below the lower limits of sensitivity in unstressed cells and thus the amount of change represents an estimate.
S. aureus strain COL locus, unless otherwise indicated (strain preceeds locus identifier).
Heat shock response.
Next, we identified transcripts that are induced in heat-shocked UAMS-1 cells. To do so, mid-log-phase cells (grown at 37°C) were incubated for 30 min at 42°C to induce the S. aureus heat shock response. The transcript profile of heat-shocked cells was then compared to that of unstressed cells. Induction of the heat shock response did not affect cell viability (data not shown) but did induce the transcription of 98 genes.
As shown in Table 3, three well-characterized heat shock response genes, ctsR, clpB, and clpC, were upregulated 3.1-, 5.0-, and 2.3-fold, respectively, during growth at the elevated temperature, suggesting that the conditions tested are appropriate for studying aspects of the S. aureus heat shock response (12). Among the genes induced by heat shock were a number of putative S. aureus virulence factors, including (i) the alpha-hemolysin gene (hla), (ii) pathogenicity island genes (SACOL0900 and SACOL0901), (iii) an LPXTG motif-containing gene (SACOL2668), and (iv) members of the urease system (ureA-ureG), which are strongly upregulated in S. aureus biofilms (3). Thirty-six hypothetical or conserved hypothetical proteins were induced by heat shock conditions. Eleven cold shock genes were also induced within heat-shocked cells, indicating that they may constitute members of a general temperature-mediated stress response. Included among these were six conserved hypothetical genes, a pathogenicity island gene (SACOL0901), the MarR family regulator gene, and two competence orthologs (SACOL0813 and SACOL0814). Real-time PCR confirmed that clpC and ctsR were upregulated 65- and 95-fold, respectively, under heat shock conditions (data not shown). Forty-two transcripts decreased in response to heat shock conditions (see Table S6 in the supplemental material).
TABLE 3.
S. aureus heat shock-induced transcripts
| Category and qualifiera | Fold inductionb | Common name | Locusc | Description |
|---|---|---|---|---|
| Amino acids and derivatives | ||||
| sa_c7410s6434_a_at | 2.0* | gltB | SA0514 | Glutamate synthase large subunit |
| sa_c8813s7749_a_at | 2.2 | SA0569 | Guanido phosphotransferase family protein | |
| sa_c1659s1395_a_at | 2.4 | hom | SA1362 | Homoserine dehydrogenase |
| sa_c1922s1644_a_at | 2.4* | dapA | SA1430 | Dihydrodipicolinate synthase |
| sa_c1928s1652_a_at | 2.3* | dapD | SA1432 | 2,3,4,5-Tetrahydropyridine-2,6-dicarboxylate N-Succinyltransferase |
| sa_c5019s4319_a_at | 2.1* | SA2279 | Putative transporter | |
| sa_c5023s4322_at | 6.0 | ureA | SA2280 | Urease, gamma subunit |
| sa_c5029s4326_a_at | 5.2 | ureB | SA2281 | Urease, beta subunit |
| sa_c5031s4330_a_at | 3.8 | ureC | SA2282 | Urease, alpha subunit |
| sa_c5035s4334_at | 3.3 | ureE | SA2283 | Urease accessory protein |
| sa_c5039s4340_a_at | 3.2 | ureF | SA2284 | Urease accessory protein |
| sa_c5043s4344_a_at | 3.0 | ureG | SA2285 | Urease accessory protein |
| sa_c9293s8136_a_at | 3.4 | ureD | SA2286 | Urease accessory protein |
| sa_c9420s8234_a_at | 6.3 | betA | SA2627 | Choline dehydrogenase |
| Carbohydrates | ||||
| sa_c6411s5581_a_at | 3.8 | bglA | SA0251 | 6-Phospho-beta-glucosidase |
| sa_c2458s2040_a_at | 2.2* | malA | SA1551 | Alpha-glucosidase |
| sa_c4699s4019_a_at | 2.7* | lacG | SA2180 | 6-Phospho-beta-galactosidase |
| sa_c4703s4023_a_at | 7.0* | lacD | SA2183 | Tagatose 1,6-diphosphate aldolase |
| sa_c4709s4027_a_at | 3.6* | lacC | SA2184 | Tagatose-6-phosphate kinase |
| sa_c4711s4031_a_at | 4.5* | lacB | SA2185 | Galactose-6-phosphate isomerase |
| sa_c4715s4037_at | 5.9* | lacA | SA2186 | Galactose-6-phosphate isomerase |
| sa_c6417s5584_a_at | 6.4 | betB | SA2628 | Betaine aldehyde dehydrogenase |
| Cell wall and capsule | ||||
| sa_c6381s5549_a_at | 4.6 | SA0250 | PTS system, IIA component | |
| sa_c1330s1105_a_at | 3.5* | lytN | SA1264 | Cell wall hydrolase |
| sa_c9866s8605_at | 2.7 | SA1932 | Transglycosylase domain protein | |
| Cofactors | ||||
| sa_c3380s9339_a_at | 2.4 | ribH | SA1817 | Riboflavin synthase, beta subunit |
| sa_c3391s2919_a_at | 2.9 | ribE | SA1819 | Riboflavin synthase, alpha subunit |
| sa_c3395s2925_a_at | 2.4 | ribD | SA1820 | Riboflavin biosynthesis protein |
| DNA metabolism | ||||
| sa_c963s754_a_at | 2.3 | uvrC | SA1157 | Excinuclease C subunit |
| Hypothetical | ||||
| sa_c2938s2500_at | 5.0 | MW2-MW1600 | Conserved hypothetical protein | |
| sa_c4558s9982_x_at | 3.0 | MW2-MW2077 | Conserved hypothetical protein | |
| sa_c10133s10545_a_at | 2.0* | N315-SA1777 | Conserved hypothetical | |
| sa_c7157s10125_a_at | 9.8* | N315-SA1832 | Conserved hypothetical protein | |
| sa_c6011s10068_at | 2.3* | N315-SA2299 | Conserved hypothetical protein | |
| sa_c7036s9080_a_at | 2.8 | N315-SA0326 | Conserved hypothetical protein | |
| sa_c7819s6819_a_at | 4.5 | N315-SA0551 | Conserved mercuric reductase homologus | |
| sa_c7161s10131_at | 2.8* | N315-SA1829 | Conserved hypothetical protein | |
| sa_c3224s2774_a_at | 5.6 | SA0161 | Conserved hypothetical protein | |
| sa_c4120s3473_a_at | 3.7* | SA0191 | M23/M37 peptidase domain protein | |
| sa_c4917s4223_a_at | 4.3* | SA0215 | Putative propionate CoA-transferase | |
| sa_c6449s5616_a_at | 3.2 | SA0252 | Conserved hypothetical protein | |
| sa_c9259s8103_a_at | 2.2 | SA0255 | Putative membrane protein | |
| sa_c6700s5841_a_at | 2.8 | SA0259 | Hypothetical protein | |
| sa_c6967s6091_a_at | 2.5 | SA0314 | Conserved hypothetical protein | |
| sa_c7132s6241_a_at | 3.3* | SA0436 | Conserved hypothetical protein | |
| sa_c7571s6589_a_at | 2.6 | SA0568 | Conserved hypothetical protein | |
| sa_c7760s6763_at | 15.1 | SA0625 | Conserved hypothetical protein | |
| sa_c7821s6823_a_at | 6.3* | SA0641 | Conserved hypothetical protein | |
| sa_c8331s7308_a_at | 4.6* | SA0814 | Competence protein F | |
| sa_c485s314_at | 16.7* | SA1033 | Hypothetical protein | |
| sa_c582s408_a_at | 2.6 | SA1059 | Conserved hypothetical protein | |
| sa_c1071s9138_a_at | 4.2 | SA1189 | Putative acetyltransferase | |
| sa_c1618s1361_a_at | 3.7* | SA1349 | Conserved hypothetical protein | |
| sa_c1705s1441_a_at | 3.7 | SA1375 | Conserved hypothetical protein | |
| sa_c1960s1685_a_at | 5.0 | SA1438 | Conserved hypothetical protein | |
| sa_c2432s2017_a_at | 2.4 | SA1539 | Conserved hypothetical protein | |
| sa_c2942s2505_a_at | 8.7 | SA1705 | Hypothetical protein | |
| sa_c3692s3173_at | 2.5 | SA1927 | Conserved hypothetical protein | |
| sa_c9309s8152_at | 3.2 | SA2315 | Conserved hypothetical protein | |
| sa_c6395s5566_a_at | 3.2 | SA2401 | Conserved hypothetical protein | |
| sa_c5614s4868_a_at | 2.5* | SA2404 | Conserved hypothetical protein | |
| sa_c5616s4872_a_at | 3.1* | SA2405 | Conserved hypothetical protein | |
| sa_c9344s8176_a_at | 3.3 | SA2436 | Conserved hypothetical protein | |
| sa_c6112s5297_a_at | 2.0 | SA2551 | Conserved hypothetical protein | |
| sa_c6206s5387_a_at | 28.2* | SA2571 | Conserved hypothetical protein | |
| Miscellaneous | ||||
| sa_c4301s3654_a_at | 3.3 | kdpD | SA2070 | Sensor histidine kinase |
| sa_c6058s5252_a_at | 2.1 | SA2533 | Glyoxalase family protein | |
| sa_c9434s8247_a_at | 2.2 | SA2667 | Isochorismatase family protein | |
| Nucleosides and nucleotides | ||||
| sa_c1898s1619_a_at | 4.5* | deoD | SA0121 | Purine nucleoside phosphorylase |
| sa_c1167s950_a_at | 2.3 | pyrF | SA1216 | Orotidine 5-phosphate decarboxylase |
| Protein metabolism | ||||
| sa_c3341s2876_a_at | 2.0 | SA0164 | Gramicidin S synthetase 2-related protein | |
| sa_c256s9573_s_at | 2.3 | clpC | SA0570 | ATP-dependent protease |
| sa_c8966s7880_a_at | 2.6 | SA0815 | Ribosomal subunit interface protein | |
| sa_c278s123_a_at | 5.0 | clpB | SA0979 | ATP-dependent protease |
| Regulation | ||||
| sa_c9074s7960_a_at | 14.5* | N315-SA0882 | Similar to competence transcription factor | |
| sa_c6190s5368_a_at | 2.4 | N315-SA2340 | Putative transcriptional regulator, TetR family | |
| sa_c7020s6143_a_at | 2.6 | SA0404 | Transcriptional regulator, MarR family | |
| sa_c7405s6429_a_at | 3.2* | gltC | SA0513 | Transcriptional regulator |
| sa_c8807s7748_a_at | 3.1 | ctsR | SA0567 | Transcriptional regulator |
| sa_c4305s3658_at | 2.4* | kdpE | SA2071 | DNA-binding response regulator |
| sa_c9305s8148_a_at | 2.9 | SA2304 | Conserved regulatory domain protein | |
| sa_c5496s4759_a_at | 2.6 | SA2374 | Putative transcriptional regulator, TetR family | |
| Resistance | ||||
| sa_c1968s1693_a_at | 2.2 | tetR | SA0122 | Tetracycline resistance |
| Transport | ||||
| sa_c5355s4632_a_at | 2.8 | opuCA | N315-SA2237 | Glycine betaine/carnitine/choline ABC transporter |
| sa_c3997s3420_a_at | 2.4 | SA0184 | Peptide ABC transporter | |
| sa_c8327s7303_a_at | 3.6* | SA0813 | Putative ComF protein 1 | |
| sa_c3603s3083_a_at | 2.4 | SA1897 | Putative protein export protein | |
| sa_c9938s8633_a_at | 3.3* | lacE | SA2181 | PTS system, lactose-specific IIB components |
| sa_c9939s8637_a_at | 3.4* | lacF | SA2182 | PTS system, lactose-specific IIA component transporter CorA family |
| sa_c5500s4763_a_at | 2.2 | SA2375 | ||
| sa_c5773s5016_at | 2.0 | SA2450 | ABC transporter | |
| Virulence | ||||
| sa_c10620s11074_a_at | 4.5* | N315-SA0895 | Pathogenicity island protein | |
| sa_c10156s10581_at | 20.8* | N315-SA1833 | SaPI pathogenicity island protein | |
| sa_c7165s10136_a_at | 3.1* | SA0900 | Pathogenicity island protein | |
| sa_c7169s10140_a_at | 4.2 | SA0901 | Pathogenicity island protein | |
| sa_c1023s810_a_at | 3.4* | hla | SA1173 | Alpha-hemolysin precursor |
| sa_c1334s1109_a_at | 2.6* | eprH | SA1265 | Endopeptidase resistance |
| sa_c6575s5743_a_at | 2.8 | SA2668 | LPXTG cell wall surface anchor family protein |
Affymetrix S. aureus GeneChip descriptive representing indicated predicted ORF.
*, transcript was below the lower limits of sensitivity in unstressed cells, and thus the amount of change represents an estimate.
S. aureus strain COL locus, unless otherwise indicated (strain preceeds locus identifier).
Stringent response.
Mupirocin is an antimicrobial agent that induces the staphylococcal stringent response by inhibiting isoleucyl tRNA synthetase, thereby increasing the cellular concentration of uncharged tRNAIle molecules (8, 11). Using real-time PCR, we determined that 30 min of mupirocin treatment (60 μg ml−1) increased the transcript titers of the stringent response control factor relA and the ilv operon (3- and >1,000-fold, respectively), whereas rpsL transcript levels were decreased (5-fold); cell viability was marginally affected (data not shown). These results mimic the transcription profile for mupirocin-mediated E. coli stringent response induction and were the same conditions used by Crosse and colleagues to induce the stringent response in S. aureus strain 8325-4 (11, 47). Lower mupirocin concentrations did not appreciably alter the transcript titers of relA, ilvA, and rpsL, whereas higher concentrations increased toxicity.
Mupirocin treatment induced 248 open reading frame transcripts (Table 4). As expected, the stringent response element relA was upregulated (2.4-fold). Likewise, members of the ilv (ilvA-ilvD and ilvN), leu (leuA-leuD), and thr (thrB and thrC) operons were strongly upregulated by mupirocin challenge (30- to 157.1-fold); all have been shown to be responsive to isoleucyl tRNA limitation (47). Collectively, these results suggest that the conditions used were appropriate for studying components of the S. aureus stringent response. Eliciting the stringent response induced several classes of gene products, including (i) 37 transport proteins, (ii) 10 previously characterized virulence factors, (iii) 18 regulatory molecules, and (iv) 10 peptidases. The last class is a hallmark of the stringent response (22). Among the elevated transport proteins were three putative drug efflux pumps, i.e., NorA (10.1-fold), MepA (5-fold), and an EmrB/QacA-like protein encoded by the SACOL2413 gene (36, 64). Two loci, encoding a set of putative ABC transporter proteins (SACOL0504 to SACOL0506; average induction, 124-fold) and a set of oligopeptide transporter proteins (SACOL0991 to SACOL0995; average induction, 73-fold), were among the most dramatically upregulated transcripts in mupirocin-challenged cells. The virulence determinants of the stringent response included autolysin (alt), fibrinogen binding protein (fbp), sortase A (srtA), components of the intracellular adhesion locus (icaA and icaB), and extracellular proteases (sspA-sspC). Among the transcription factors that were upregulated were three well-characterized virulence factor regulators, i.e., sarR (3.2-fold), sarZ (8.7-fold), and a component of the agr locus (agrA; 3.1-fold). Real-time PCR demonstrated that the norA mRNA titer was induced 36.2-fold following induction of the stringent response (data not shown). A total of 814 transcripts decreased in abundance under stringent conditions (see Table S6 in the supplemental material).
TABLE 4.
S. aureus stringent response-induced transcripts
| Category and qualifiera | Fold inductionb | Common name | Locusc | Description |
|---|---|---|---|---|
| Amino acid metabolism | ||||
| sa_c2576s2153_a_at | 2.4 | aroK | SA1596 | Shikimate kinase |
| sa_c1918s1640_a_at | 75.1* | asd | SA1429 | Aspartate-semialdehyde dehydrogenase |
| sa_c1922s1644_a_at | 87.1* | dapA | SA1430 | Dihydrodipicolinate synthase |
| sa_c1924s1648_a_at | 57.6* | dapB | SA1431 | Dihydrodipicolinate reductase |
| sa_c1928s1652_a_at | 32.7* | dapD | SA1432 | 2,3,4,5-Tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase |
| sa_c2572s2149_a_at | 2.5 | gcvT | SA1595 | Glycine cleavage system T protein |
| sa_c7410s6434_a_at | 29.6* | gltB | SA0514 | Glutamate synthase large subunit |
| sa_c7412s6438_a_at | 15.6 | gltD | SA0515 | Glutamate synthase small subunit |
| sa_c8240s7220_a_at | 7.1 | hisC | SA0784 | Histidinol-phosphate aminotransferase |
| sa_c6724s5865_a_at | 4.1* | hisG | SA2703 | ATP phosphoribosyltransferase |
| sa_c1659s1395_a_at | 28.9 | hom | SA1362 | Homoserine dehydrogenase |
| sa_c4243s3594_a_at | 48.2* | ilvA | SA2050 | Threonine dehydratase |
| sa_c4213s3565_a_at | 142.1* | ilvB | SA2043 | Acetolactate synthase large subunit |
| sa_c9931s8627_a_at | 156.7* | ilvC | SA2045 | Ketol-acid reductoisomerase |
| sa_c4209s3561_a_at | 131.9* | ilvD | SA2042 | Dihydroxy-acid dehydratase |
| sa_c4217s3569_at | 153.6* | ilvN | SA2044 | Acetolactate synthase small subunit |
| sa_c4223s3575_a_at | 157.1* | leuA | SA2046 | 2-Isopropylmalate synthase |
| sa_c4225s3576_a_at | 100.8* | leuB | SA2047 | 3-Isopropylmalate dehydrogenase |
| sa_c4229s3580_a_at | 80.0* | leuC | SA2048 | 3-Isopropylmalate dehydratase large subunit |
| sa_c4239s3588_a_at | 71.1* | leuD | SA2049 | 3-Isopropylmalate dehydratase small subunit |
| sa_c1940s1663_at | 3.5 | lysA | SA1435 | Diaminopimelate decarboxylase |
| sa_c1912s1635_a_at | 67.6* | lysC | SA1428 | Aspartokinase alpha and beta subunits |
| sa_c10721s11169cv_s_at | 2.8 | metB | N315-SA0419 | Cystathionine gamma-synthase |
| sa_c9702s8459_a_at | 3.6 | proC | SA1546 | Pyrroline-5-carboxylate reductase |
| sa_c3376s2908_a_at | 3.9* | putA | SA1816 | Proline dehydrogenase |
| sa_c3548s3054_a_at | 3.9* | rocD | SA0170 | Ornithine aminotransferase |
| sa_c3204s2753_a_at | 19.4* | serA | SA1773 | d-3-Phosphoglycerate dehydrogenase |
| sa_c1994s9145_a_at | 2.4 | sucA | SA1449 | 2-Oxoglutarate dehydrogenase E1 component |
| sa_c1669s1406_a_at | 36.4 | thrB | SA1364 | Homoserine kinase |
| sa_c1665s1401_a_at | 30.0 | thrC | SA1363 | Threonine synthase |
| sa_c1810s1538_a_at | 8.5* | tyrA | SA1401 | Prephenate dehydrogenase |
| sa_c47s43_a_at | 7.4* | SA0012 | Putative homoserine O-acetyltransferase | |
| sa_c7106s6219_a_at | 6.7* | SA0430 | trans-Sulfuration enzyme family protein | |
| sa_c7112s6224_a_at | 5.8* | SA0431 | trans-Sulfuration enzyme family protein | |
| sa_c7368s9209_a_at | 2.1 | SA0502 | Cysteine synthase/cystathionine beta-synthase family protein | |
| sa_c7372s10188cs_s_at | 3.0 | SA0503 | trans-Sulfuration enzyme family protein | |
| sa_c7649s6662_a_at | 4.4 | SA0595 | Peptidase, M20/M25/M40 family | |
| sa_c574s400_a_at | 9.5 | SA1058 | Aminotransferase class I | |
| sa_c9581s8342_a_at | 20.6* | SA1360 | Aspartate kinase | |
| sa_c1934s1655_a_at | 8.0 | SA1433 | Peptidase, M20/M25/M40 family | |
| sa_c1936s1659_a_at | 8.3 | SA1434 | Alanine racemase family protein | |
| sa_c2570s2145_a_at | 2.6 | SA1594 | Glycine cleavage system P protein | |
| sa_c3262s2810_a_at | 6.4 | SA1787 | Chorismate mutase/phospho-2-dehydro-3-Deoxyheptonate aldolase | |
| sa_c3945s3412_a_at | 2.4 | SA2000 | Putative aminotransferase | |
| sa_c6224s5400_a_at | 7.4 | SA2575 | Aminotransferase | |
| sa_c6718s5857_a_at | 2.3* | SA2701 | Putative histidinol-phosphate aminotransferase | |
| Carbohydrates | ||||
| sa_c9528s8308_a_at | 2.4 | SA0111 | Oxidoreductase, short-chain-dehydrogenase/reductase family | |
| sa_c8477s7437_at | 11.1 | SA0869 | Phosphoglycerate mutase family protein | |
| sa_c629s448_at | 3.5 | SA1071 | Chitinase-related protein | |
| sa_c2791s2362_a_at | 3.3 | SA1661 | Putative acetyl-CoA carboxylase | |
| sa_c5142s4440_at | 4.0 | SA2313 | Hydrolase haloacid dehalogenase-like family | |
| sa_c6220s5396_a_at | 3.7 | SA2574 | 2-Hydroxyacid dehydrogenase family protein | |
| Cell wall and capsule | ||||
| sa_c6371s5542_a_at | 2.3 | budA | SA2617 | Alpha-acetolactate decarboxylase |
| sa_c1124s904_a_at | 2.2 | SA1205 | Putative cell division initiation protein | |
| sa_c4589s3921_a_at | 4.8* | fmtB | N315-SA1964 | FmtB protein |
| sa_c3228s2778_a_at | 3.6 | SA1779 | Transglycosylase domain protein | |
| sa_c9866s8605_at | 5.7 | SA1932 | Transglycosylase domain protein | |
| sa_c4494s3841_a_at | 3.6 | SA2125 | Peptidase, M20/M25/M40 family | |
| sa_c5176s4476_a_at | 26.3* | SA2322 | Peptidase, M20/M25/M40 family | |
| DNA metabolism | ||||
| sa_c8705s9226_a_at | 2.0 | rexB | SA0970 | Exonuclease |
| sa_c2436s2020_a_at | 2.9 | xerD | SA1540 | Site-specific recombinase |
| sa_c1804s1534_a_at | 3.6* | SA1400 | ImpB/MucB/SamB family protein | |
| sa_c2977s2534_a_at | 6.4 | SA1711 | DNA-3-methyladenine glycosylase | |
| Hypothetical | ||||
| sa_c2287s9677_a_at | 2.4* | N315-SA0141 | Conserved hypothetical protein | |
| sa_c7819s6819_a_at | 3.5 | N315-SA0551 | Mercuric reductase homologue | |
| sa_c8469s7429_a_at | 16.1 | N315-SA0751 | Conserved hypothetical protein | |
| sa_c4084s9951_at | 2.6* | N315-SA1802 | Conserved hypothetical protein | |
| sa_c7157s10125_a_at | 6.9* | N315-SA1832 | Conserved hypothetical protein | |
| sa_c6385s5555_a_at | 127.9* | N315-SA2397 | Conserved hypothetical protein | |
| sa_c8467s10245_s_at | 4.6 | N315-SAS018 | Conserved hypothetical protein | |
| sa_c41s39_a_at | 31.0* | SA0011 | Conserved hypothetical protein | |
| sa_c648s458_a_at | 2.2 | SA0076 | Hypothetical protein | |
| sa_c2222s1923_a_at | 3.3* | SA0129 | Conserved hypothetical protein | |
| sa_c3008s2564_a_at | 12.5 | SA0157 | Conserved hypothetical protein | |
| sa_c3158s2704_a_at | 16.5 | SA0160 | Conserved hypothetical protein | |
| sa_c3224s2774_a_at | 9.2 | SA0161 | Conserved hypothetical protein | |
| sa_c3976s9867_at | 2.9* | SA0181 | Conserved domain protein | |
| sa_c4729s4046_at | 6.2* | SA0208 | Hypothetical protein | |
| sa_c6917s6037_a_at | 5.1* | SA0299 | Hypothetical protein | |
| sa_c6967s6091_a_at | 4.7 | SA0314 | Conserved hypothetical protein | |
| sa_c7055s6165_a_at | 9.0* | SA0414 | Putative lipoprotein | |
| sa_c7061s6172_a_at | 2.8* | SA0415 | Hypothetical protein | |
| sa_c7088s6200_a_at | 3.7 | SA0425 | Hypothetical protein | |
| sa_c7094s6206_a_at | 3.6 | SA0427 | Conserved hypothetical protein | |
| sa_c7132s6241_a_at | 4.9* | SA0436 | Conserved hypothetical protein | |
| sa_c7203s6269_a_at | 5.3* | SA0445 | Conserved hypothetical protein | |
| sa_c7313s9398_a_at | 16.5 | SA0480 | Hypothetical protein | |
| sa_c7760s6763_at | 2.7 | SA0625 | Conserved hypothetical protein | |
| sa_c7813s6815_at | 4.1 | SA0639 | Conserved hypothetical protein | |
| sa_c7821s6823_a_at | 3.7* | SA0641 | Conserved hypothetical protein | |
| sa_c8897s7814_a_at | 5.0* | SA0692 | Conserved hypothetical protein | |
| sa_c7985s6972_at | 2.2 | SA0703 | Conserved hypothetical protein | |
| sa_c8069s7049_a_at | 2.6 | SA0730 | Conserved hypothetical protein | |
| sa_c8928s7841_a_at | 3.1 | SA0755 | Conserved hypothetical protein | |
| sa_c8196s7173_a_at | 3.5 | SA0768 | Conserved hypothetical protein | |
| sa_c8244s7224_a_at | 2.7 | SA0785 | Conserved hypothetical protein | |
| sa_c8348s7321_a_at | 5.3 | SA0821 | HD domain protein | |
| sa_c10288s8968_a_at | 3.3* | SA0822 | Conserved hypothetical protein | |
| sa_c6027s5223_a_at | 7.5* | SA0870 | LysE/YggA family protein | |
| sa_c10616s11070_s_at | 15.1 | SA0871 | Putative acetyltransferase | |
| sa_c8548s7508_a_at | 4.5* | SA0920 | Hypothetical protein | |
| sa_c485s314_at | 103.2* | SA1033 | Hypothetical protein | |
| sa_c525s350_at | 2.3 | SA1044 | Conserved hypothetical protein | |
| sa_c750s552_a_at | 2.4 | SA1101 | Conserved hypothetical protein | |
| sa_c827s629_a_at | 2.9 | SA1117 | Conserved hypothetical protein | |
| sa_c1078s861_at | 3.1 | SA1191 | Conserved hypothetical protein | |
| sa_c1112s893_a_at | 2.1 | SA1200 | Conserved hypothetical protein | |
| sa_c1705s1441_a_at | 33.0 | SA1375 | Conserved hypothetical protein | |
| sa_c9771s8515_a_at | 2.5 | SA1418 | Conserved hypothetical protein | |
| sa_c2481s2059_at | 4.3 | SA1556 | Hypothetical protein | |
| sa_c2779s2349_a_at | 18.1 | SA1658 | Hypothetical protein | |
| sa_c2783s2353_a_at | 4.9* | SA1659 | Conserved hypothetical protein | |
| sa_c2807s2375_a_at | 5.8* | SA1664 | Conserved hypothetical protein | |
| sa_c2819s2389_at | 2.8 | SA1670 | Conserved hypothetical protein | |
| sa_c3298s2841_at | 2.7 | SA1796 | Conserved hypothetical protein | |
| sa_c3306s2847_a_at | 2.5 | SA1798 | Conserved hypothetical protein | |
| sa_c3357s2894_a_at | 2.3 | SA1810 | Conserved hypothetical protein | |
| sa_c9141s8010_at | 3.4* | SA1896 | Conserved hypothetical protein | |
| sa_c3786s3258_a_at | 2.8 | SA1956 | Conserved hypothetical protein | |
| sa_c3842s3311_at | 2.3 | SA1972 | Conserved hypothetical protein | |
| sa_c3900s3368_at | 5.8 | SA1986 | Conserved hypothetical protein | |
| sa_c4134s3487_a_at | 12.3 | SA2019 | Putative SdrH protein | |
| sa_c4171s3522_a_at | 7.3 | SA2033 | Conserved hypothetical protein | |
| sa_c4173s3526_a_at | 4.1 | SA2034 | Conserved hypothetical protein | |
| sa_c9799s8540_a_at | 2.3 | SA2123 | Conserved hypothetical protein | |
| sa_c4492s3837_a_at | 4.3 | SA2124 | Conserved hypothetical protein | |
| sa_c4628s3951_a_at | 2.8 | SA2162 | Conserved hypothetical protein | |
| sa_c5081s4376_a_at | 2.4 | SA2294 | Conserved hypothetical protein | |
| sa_c9305s8148_a_at | 5.7 | SA2304 | Conserved domain protein | |
| sa_c5156s4454_a_at | 2.3 | SA2318 | Conserved hypothetical protein | |
| sa_c5174s4471_a_at | 4.8 | SA2321 | Oxidoreductase, short-chain-dehydrogenase/reductase family | |
| sa_c10593s9072_a_at | 2.9 | SA2338 | Hypothetical protein | |
| sa_c5299s4579_a_at | 4.9 | SA2354 | Putative membrane protein | |
| sa_c5458s4723_a_at | 3.8 | SA2365 | Conserved hypothetical protein | |
| sa_c5516s4772_a_at | 2.0 | SA2379 | Conserved hypothetical protein | |
| sa_c5614s4868_a_at | 11.9* | SA2404 | Conserved hypothetical protein | |
| sa_c5616s4872_a_at | 19.1* | SA2405 | Conserved hypothetical protein | |
| sa_c5624s4878_a_at | 3.2 | SA2408 | Conserved hypothetical protein | |
| sa_c9344s8176_a_at | 3.4 | SA2436 | Conserved hypothetical protein | |
| sa_c5788s5026_a_at | 2.2 | SA2456 | Conserved hypothetical protein | |
| sa_c5815s5055_a_at | 6.3 | SA2467 | Putative lipoprotein | |
| sa_c10598s11052_s_at | 2.6 | SA2526 | Putative membrane protein | |
| sa_c6270s5451_a_at | 2.2 | SA2587 | Conserved hypothetical protein | |
| sa_c6274s5452_at | 4.4* | SA2588 | Hypothetical protein | |
| sa_c6278s5456_a_at | 13.6* | SA2589 | Conserved hypothetical protein | |
| sa_c6728s5871_a_at | 8.8* | SA2704 | Conserved hypothetical protein | |
| sa_c6750s5891_a_at | 16.1* | SA2709 | Conserved hypothetical protein | |
| sa_c6752s5895_a_at | 26.3* | SA2710 | Conserved hypothetical protein | |
| Miscellaneous | ||||
| sa_c6461s5629_a_at | 2.2 | cysJ | SA2639 | Sulfite reductase |
| sa_c1739s1473_a_at | 3.2 | mscL | SA1383 | Mechanosensitive channel protein |
| sa_c10345s9018_a_at | 4.6* | SA0162 | NAD-dependent formate dehydrogenase | |
| sa_c8485s7447_a_at | 2.3 | SA0872 | OsmC/Ohr family protein | |
| sa_c10627s11083cv_s_at | 2.6 | SA0941 | Putative NADH dehydrogenase | |
| sa_c594s417_at | 5.2 | SA1063 | Acetyltransferase GNAT family | |
| sa_c1671s1408_a_at | 4.1 | SA1365 | Hydrolase, haloacid dehalogenase-like family | |
| sa_c2466s2048_a_at | 2.1 | SA1553 | Glyoxalase family protein | |
| sa_c8677s7626_a_at | 3.6* | SA0962 | Putative glycerophosphoryl diester phosphodiesterase | |
| sa_c3202s2750_a_at | 25.5* | SA1772 | Aminotransferase class V | |
| sa_c3210s2757_a_at | 15.3 | SA1774 | Hydrolase, haloacid dehalogenase-like family | |
| sa_c6764s5905_a_at | 6.2 | SA2713 | Rhodanese-like domain protein | |
| sa_c5009s4311_a_at | 4.7 | SA2276 | Inosine-uridine-preferring nucleoside hydrolase | |
| Protein metabolism | ||||
| sa_c1128s910_a_at | 2.5 | ileS | SA1206 | Isoleucyl-tRNA synthetase |
| sa_c1800s1530_a_at | 2.2 | msrA | SA1397 | Peptide methionine sulfoxide reductase |
| sa_c6770s5910_a_at | 3.3* | pcp | SA2714 | Pyrrolidone-carboxylate peptidase |
| sa_c7051s6163_a_at | 15.1* | SA0413 | Putative ribosomal protein-serine acetyltransferase | |
| sa_c8966s7880_a_at | 10.7 | SA0815 | Ribosomal subunit interface protein | |
| sa_c2024s1734_a_at | 2.8 | SA1455 | Carboxyl-terminal protease | |
| sa_c5620s4877_a_at | 5.7 | SA2407 | Putative lipoprotein | |
| Regulation | ||||
| sa_c4149s3503_a_at | 3.1 | agrA | SA2026 | Accessory gene regulator protein A |
| sa_c4530s3875_a_at | 11.7 | czrA | SA2137 | Transcriptional regulator |
| sa_c9575s8335_a_at | 4.7 | lexA | SA1374 | LexA repressor |
| sa_c2879s2445_a_at | 2.4 | relA | SA1689 | GTP pyrophosphokinase |
| sa_c5047s4347_at | 3.2 | sarR | SA2287 | Staphylococcal accessory regulator R |
| sa_c5529s4783_a_at | 8.7 | sarZ | SA2384 | Staphylococcal accessory protein Z |
| sa_c10319s10704cv_s_at | 3.4 | N315-SA0142 | Putative transcription factor | |
| sa_c9074s7960_a_at | 7.2* | N315-SA0882 | Putative competence transcription factor | |
| sa_c7020s6143_a_at | 14.0 | SA0404 | Transcriptional regulator, MarR family | |
| sa_c9104s7978_a_at | 2.5 | SA1003 | Negative regulator of competence | |
| sa_c9058s7951_a_at | 10.8 | SA1060 | Transcriptional regulator, MarR family | |
| sa_c9706s8463_a_at | 3.2 | SA1541 | Transcriptional regulator, Fur family | |
| sa_c9202s8059_a_at | 3.1 | SA1906 | Putative sensor histidine kinase | |
| sa_c3661s3140_a_at | 2.8 | SA1917 | PTS system IIC component | |
| sa_c3663s3145_a_at | 4.2 | SA1919 | Transcriptional regulator, Fur family | |
| sa_c9297s8140_at | 2.2 | SA2302 | Putative transcriptional regulator | |
| sa_c6050s5246_a_at | 2.7 | SA2531 | Transcriptional regulator, MarR family | |
| sa_c6262s5443_a_at | 78.4* | SA2585 | Putative regulatory protein | |
| Resistance | ||||
| sa_c7024s6147_a_at | 5.5 | mepA | SA0405 | MATE efflux family protein |
| sa_c8155s7139_a_at | 10.1 | norA | SA0754 | Multidrug resistance protein |
| sa_c346s186_a_at | 2.0 | SA2413 | Drug resistance transporter, EmrB/QacA subfamily | |
| sa_c5721s4964_a_at | 6.7 | bcr | SA2437 | Bicyclomycin resistance protein |
| Stress response | ||||
| sa_c1956s1681_a_at | 2.4 | cspB | SA2731 | Cold shock protein |
| sa_c3134s2687_a_at | 3.6 | SA1753 | Universal stress protein family | |
| sa_c8744s7687_a_at | 13.0 | SA1759 | Universal stress protein family | |
| sa_c9791s8532_a_at | 4.6 | SA2131 | Dps family protein | |
| sa_c5530s4787_a_at | 6.0* | SA2385 | Heat shock protein, Hsp20 family | |
| Transport | ||||
| sa_c3567s9159_a_at | 10.0 | brnQ | SA0171 | Branched-chain amino acid transport |
| sa_c324s166_a_at | 100.8* | oppC | SA0992 | Oligopeptide ABC transporter |
| sa_c328s170_a_at | 64.2* | oppD | SA0993 | Oligopeptide ABC transporter |
| sa_c332s172_a_at | 12.3* | oppF | SA0994 | Oligopeptide ABC transporter |
| sa_c37s34_a_at | 17.4* | SA0010 | AzlC family protein | |
| sa_c9538s8314_a_at | 2.9* | SA0128 | Phosphonate ABC transporter | |
| sa_c3043s2596_a_at | 35.4 | SA0158 | ABC transporter | |
| sa_c3118s2672_a_at | 24.0 | SA0159 | ABC transporter | |
| sa_c3997s3420_a_at | 29.2 | SA0184 | Peptide ABC transporter | |
| sa_c7236s6302_a_at | 5.3 | SA0454 | Sodium:dicarboxylate symporter family protein | |
| sa_c5418s4689_a_at | 133.0* | SA0504 | ABC transporter | |
| sa_c7374s6406_a_at | 124.9* | SA0505 | ABC transporter | |
| sa_c7378s6412_a_at | 114.6* | SA0506 | ABC transporter | |
| sa_c8252s7229_a_at | 2.1 | SA0788 | Oligopeptide transporter family protein | |
| sa_c5431s4700_a_at | 27.7 | SA0882 | ABC transporter | |
| sa_c8512s7471_a_at | 25.9 | SA0883 | ABC transporter | |
| sa_c8518s7475_a_at | 28.5 | SA0884 | ABC transporter | |
| sa_c10571s9056_a_at | 124.5* | SA0991 | Oligopeptide ABC transporter | |
| sa_c350s191_a_at | 63.5* | SA0995 | Oligopeptide ABC transporter | |
| sa_c1675s1413_a_at | 2.5 | SA1367 | Amino acid permease | |
| sa_c3810s3279_a_at | 4.9 | SA1963 | Proline permease | |
| sa_c4110s3463_a_at | 3.5* | SA2011 | Sodium transport family protein | |
| sa_c4536s3879_a_at | 17.0 | SA2138 | Cation efflux family protein | |
| sa_c5416s4682_a_at | 2.4 | SA2211 | ABC transporter | |
| sa_c5126s4423_a_at | 2.4 | SA2309 | Amino acid permease | |
| sa_c5148s4444_a_at | 17.0 | SA2314 | Sodium/bile acid symporter family protein | |
| sa_c5160s4458_a_at | 3.1 | SA2319 | Na+/H+ antiporter family protein | |
| sa_c5632s4887_a_at | 3.0* | SA2411 | Amino acid ABC transporter | |
| sa_c9340s8173_a_at | 2.5* | SA2416 | Cation efflux family protein | |
| sa_c5769s5011_a_at | 3.0 | SA2449 | Putative drug transporter | |
| sa_c5795s5035_a_at | 4.1 | SA2458 | Amino acid permease | |
| sa_c5875s5111_a_at | 31.9 | SA2483 | Putative transporter | |
| sa_c6017s5213_a_at | 3.2 | SA2521 | Putative transporter | |
| sa_c5345s4618_a_at | 2.2 | SA2525 | ABC transporter | |
| sa_c6186s5364_a_at | 3.5 | SA2566 | Putative MmpL efflux pump | |
| sa_c6378s5547_a_at | 17.2 | SA2619 | Amino acid permease | |
| Virulence | ||||
| sa_c592s9345_a_at | 2.4 | atl | SA1062 | Bifunctional autolysin |
| sa_c1007s793_a_at | 7.9 | fbp | SA1168 | Fibrinogen-binding protein |
| sa_c5630s4882_a_at | 4.4 | fmhA | SA2409 | FmhA protein |
| sa_c6975s6099_a_at | 3.1 | geh | N315-SA0309 | Glycerol ester hydrolase |
| sa_c3951s9849_a_at | 12.5* | hlb | SA2003 | Phospholipase C |
| sa_c9442s8255_a_at | 9.2* | icaA | SA2689 | Intercellular adhesion protein A |
| sa_c6681s9106_a_at | 3.1* | icaB | SA2691 | Intercellular adhesion protein B |
| sa_c6688s5833_a_at | 17.5* | lip | N315-SA2463 | Triacylglycerol lipase precursor |
| sa_c9390s8214_a_at | 7.4 | srtA | SA2539 | Sortase |
| sa_c570s397_a_at | 4.0* | sspA | N315-SA0901 | Serine protease |
| sa_c568s393_a_at | 5.1 | sspB | SA1056 | Cysteine protease |
| sa_c564s391_a_at | 5.2* | sspC | SA1055 | Protease |
| sa_c10149s10564_at | 2.2* | N315-SA1819 | Toxic shock syndrome toxin 1 | |
| sa_c10156s10581_at | 11.9* | N315-SA1833 | SaPI pathogenicity island | |
| sa_c6859s5993_a_at | 3.8 | SA0270 | Putative staphyloxanthin biosynthesis protein | |
| sa_c7169s10140_a_at | 2.5 | SA0901 | Pathogenicity island protein | |
| sa_c1181s961_a_at | 3.8 | SA1220 | Fibronectin/fibrinogen binding-related protein |
Affymetrix S. aureus GeneChip descriptive representing indicated predicted ORF.
*, transcript was below the lower limits of sensitivity in unstressed cells, and thus the amount of change represents an estimate.
S. aureus strain COL locus, unless otherwise indicated (strain preceeds locus identifier).
SOS response.
Mitomycin C is an antimicrobial and anticancer agent that causes DNA intra- and interstrand cross-linking as well as monofunctional alkyl lesions and is a potent inducer of the bacterial SOS regulon (41). To define the optimal conditions for induction of the SOS regulon, log-phase UAMS-1 cells were treated with 0.5, 1.0, 2.5, or 5 μg ml−1 mitomycin C for 30 min. Real-time PCR was used to compare recA and uvrB transcript titers at each drug concentration to those in mock-treated cells; recA and uvrB are well-characterized components of the SOS response (24, 32, 48). The cell viability of each mitomycin C-challenged sample was also compared to that of mock-treated cells. It was determined that 1 μg ml−1 mitomycin C induced both recA (2.0-fold) and uvrB (15-fold) transcription and simultaneously produced the least amount of toxicity (data not shown), suggesting that these conditions were appropriate for studying mitomycin C-mediated induction of the SOS response. Accordingly, log-phase UAMS-1 cells were treated with 1 μg ml−1 mitomycin C for 30 min, and transcript titers were compared to those for untreated cells.
A total of 73 genes were induced by mitomycin C challenge (Table 5). Among these were the genes for the SOS repressor protein LexA (4.6-fold), components of the nucleotide excision repair machinery, namely, UvrA (2.4-fold) and UvrB (4.1-fold), the single-stranded binding protein (ssb; 44.5-fold), and the recombination repair proteins SbcC (4.9-fold) and SbcD (4.3-fold), all of which are known members of the bacterial SOS response (13). Additionally, a umuC-like gene (SACOL1400) was dramatically upregulated (36.2-fold) by mitomycin C challenge; UmuC is a component of the E. coli SOS response that promotes replicative lesion bypass of noninstructive DNA lesions (44). Collectively, these results suggest that the conditions used were appropriate for studying the SOS system.
TABLE 5.
S. aureus SOS response-induced transcripts
| Category and qualifiera | Fold inductionb | Common name | Locusc | Description |
|---|---|---|---|---|
| DNA metabolism | ||||
| sa_c10340s10724_s_at | 4.9* | sbcC | SA1382 | Exonuclease SbcC |
| sa_c1727s1464_a_at | 4.3 | sbcD | SA1381 | Exonuclease SbcD |
| sa_c4052s9933_a_at | 44.5 | ssb | N315-SA1792 | Single-stranded DNA-binding protein |
| sa_c8354s7326_a_at | 2.4 | uvrA | SA0824 | Excinuclease ABC, A subunit |
| sa_c10546s11006_s_at | 4.1 | uvrB | SA0823 | Excinuclease ABC, B subunit |
| sa_c10309s10698_a_at | 14.5* | N315-SA1196 | ImpB/MucB/SamB family protein | |
| sa_i875ur_x_at | 36.2* | SA1400 | ImpB/MucB/SamB family protein | |
| Hypothetical | ||||
| sa_c2404s9766cs_s_at | 46.8 | MSSA476-SAS064 | Conserved hypothetical protein | |
| sa_c4550s9974_at | 2.2 | MSSA476-SAS070 | Conserved hypothetical protein | |
| sa_c4556s9980_x_at | 2.1 | MSSA476-SAS072 | Conserved hypothetical protein | |
| sa_c4049s9931_a_at | 42.1* | MSSA476-SAS1903 | Putative phage regulatory protein | |
| sa_c4031s9915_at | 8.0* | Mu50-SAV0881 | Conserved hypothetical protein | |
| sa_c10677s11128_at | 2.3 | Mu50-SAV2001 | Putative lipoprotein | |
| sa_c4046s9927_at | 17.4* | MW2-MW1918 | Conserved hypothetical protein | |
| sa_c4075s9947_at | 86.9* | MW2-MW1930 | Conserved hypothetical protein | |
| sa_c8469s7429_a_at | 2.2 | N315-SA0751 | Conserved hypothetical protein | |
| sa_c4058s9936_a_at | 40.9 | N315-SA1795 | Conserved hypothetical protein | |
| sa_c4070s9942_a_at | 83.0 | N315-SA1799 | Conserved hypothetical protein | |
| sa_c10141s10553_s_at | 35.1 | N315-SA1803 | Conserved hypothetical protein | |
| sa_c7177s10148_a_at | 7.6* | N315-SA1821 | Conserved hypothetical protein | |
| sa_c6248s5424_a_at | 2.7* | N315-SA2352 | Conserved hypothetical protein | |
| sa_c7132s6241_a_at | 15.5* | SA0436 | Conserved hypothetical protein | |
| sa_c1071s9138_a_at | 2.0 | SA1189 | Acetyltransferase | |
| sa_c1705s1441_a_at | 30.8 | SA1375 | Conserved hypothetical protein | |
| sa_c1960s1685_a_at | 2.8 | SA1438 | Conserved hypothetical protein | |
| sa_c3900s3368_at | 33.3 | SA1986 | Conserved hypothetical protein | |
| sa_c3910s3377_a_at | 8.0 | SA1988 | Conserved hypothetical protein | |
| sa_c10125s8848_a_at | 20.6 | SA1999 | Conserved hypothetical protein | |
| sa_c4628s3951_a_at | 6.1 | SA2162 | Conserved hypothetical protein | |
| Miscellaneous | ||||
| sa_c8264s7244_at | 2.3 | nrdI | SA0791 | NrdI protein |
| sa_c1810s1538_a_at | 6.9* | tyrA | SA1401 | Prephenate dehydrogenase |
| sa_c6244s5421_a_at | 2.1 | SA2579 | Phytoene dehydrogenase | |
| Prophage | ||||
| sa_c10417s10840_s_at | 32.2 | dut | SA0357 | Deoxyuridine 5-triphosphate nucleotidohydrolase |
| sa_c9918s10462_a_at | 3.2 | int | N315-SA1810 | Integrase |
| sa_c10465s10903_s_at | 33.5 | BA000017///BA000018 | Phage antirepressor protein | |
| sa_c10466s10905_a_at | 42.5 | BA000017///BA000018 | Phage antirepressor protein | |
| sa_c4046s9927_a_at | 67.8* | BA000017///BA000018 | Endodeoxyribonuclease RusA | |
| sa_i10903u_x_at | 34.9 | BA000017///BA000018 | Phage antirepressor protein | |
| sa_c4040s9922cs_s_at | 26.2 | BA000017///BA000018 | Conserved hypothetical protein | |
| sa_c10668s11120_a_at | 7.4 | MRSA252-SAR2051 | Hypothetical protein | |
| sa_c2375s1984_a_at | 2.7* | Mu50-SAV0874 | phi PVL ORF 51 homolog | |
| sa_c3991s9882_a_at | 4.3 | Mu50-SAV1954 | phi PVL ORF 18-19-like protein | |
| sa_c3992s9884_a_at | 5.8 | Mu50-SAV1955 | phi PVL ORF 15 and 16 homolog | |
| sa_c4039s9920_at | 6.9* | Mu50-SAV1979 | phi PVL ORF 50 homolog | |
| sa_c3994s9885_s_at | 4.6 | MW2-MW1895 | Conserved hypothetical protein | |
| sa_c4057s9934_a_at | 38.7 | MW2-MW1923 | Conserved hypothetical protein | |
| sa_c3995s9886_a_at | 6.8 | N315-SA1767 | Conserved hypothetical protein | |
| sa_c4005s9893_a_at | 4.0 | N315-SA1769 | Conserved hypothetical protein | |
| sa_c4009s9897_a_at | 4.0 | N315-SA1770 | Conserved hypothetical protein | |
| sa_c4010s9899_at | 4.9 | N315-SA1771 | Conserved hypothetical protein | |
| sa_c4014s9903_a_at | 4.2 | N315-SA1773 | Conserved hypothetical protein | |
| sa_c10670s11122_a_at | 5.2 | N315-SA1774 | Conserved hypothetical protein | |
| sa_c10672s11124_a_at | 6.0 | N315-SA1775 | Conserved putative Clp protease | |
| sa_c4020s9905_at | 11.5 | N315-SA1776 | Conserved hypothetical protein | |
| sa_c10134s10547_a_at | 15.6 | N315-SA1777 | Conserved hypothetical protein | |
| sa_c4022s9907_a_at | 23.9 | N315-SA1778 | Conserved hypothetical protein | |
| sa_c4024s9909_a_at | 42.7 | N315-SA1779 | Conserved hypothetical protein | |
| sa_c4026s9911_at | 41.9 | N315-SA1780 | Conserved hypothetical protein | |
| sa_c4048s9929_a_at | 43.3 | N315-SA1790 | Conserved hypothetical protein | |
| sa_c9913s10456_a_at | 75.3* | N315-SA1793 | Conserved hypothetical protein | |
| sa_c4060s9937_a_at | 41.0* | N315-SA1797 | Conserved hypothetical protein | |
| sa_c7182s10152cs_s_at | 8.6 | SA2014 | Phage terminase family protein | |
| Protein metabolism | ||||
| sa_c1812s1540_a_at | 9.0 | SA1402 | Putative glutamyl aminopeptidase | |
| Regulation | ||||
| sa_c9575s8335_a_at | 4.6 | lexA | SA1374 | LexA repressor |
| sa_c10143s10555cv_s_at | 28.4 | N315-SA1804 | Putative transcriptional regulator | |
| sa_c1737s1469_a_at | 4.8* | mscL | N315-SA1182 | Large-conductance mechanosensitive channel |
| Virulence | ||||
| sa_c4094s3450_a_at | 3.4 | hlb | SA2003 | Phospholipase C |
| sa_c10522s10973_s_at | 2.3 | srtA | SA2539 | Sortase |
| sa_c7169s10140_a_at | 24.9 | SA0901 | Pathogenicity island protein | |
| sa_c10151s10571_a_at | 9.9 | SA0902 | Pathogenicity island protein | |
| sa_c10150s10567_a_at | 7.4 | SA0903 | Pathogenicity island protein | |
| sa_c7173s10144_at | 8.1 | SA0904 | Pathogenicity island protein |
Affymetrix S. aureus GeneChip descriptive representing indicated predicted ORF.
*, transcript was below the lower limits of sensitivity in unstressed cells, and thus the amount of change represents an estimate.
S. aureus strain COL locus, unless otherwise indicated (strain preceeds locus identifier).
Among the SOS-induced transcripts were several bacteriophage replication/packaging genes, including those for N315 SA1791 (42.1-fold), which has homology to the replication initiation protein DnaB, a dUTPase (COLSA0357; 34.3-fold), putative phage tail components (SAV1954 [4.3-fold] and SAV1955 [5.8-fold]), and a small terminase protein (SACOL0906; 8.6-fold). Induction of these phage transcripts correlated with the dramatic SOS-mediated upregulation of a putative phage antirepressor gene, SAV1994 (42.5-fold). Two loci, each harboring a set of genes, were among the highest SOS-induced transcripts. They included members of a bovine pathogenicity island (SACOL0901 to SACOL0904; average induction, 12.5-fold) and a set of genes encoding conserved hypothetical proteins (N315 SA1767 to SA1780; average induction, 12.9-fold). A putative Holliday junction resolvase gene (rusA) was also dramatically upregulated (67.8-fold). These results were validated, in part, by real-time PCR, which demonstrated that uvrB was upregulated in SOS-induced cells (41.5-fold) (data not shown). RecA transcript titers saturated the microarray, and thus we could not accurately determine what, if any, effect mitomycin C challenge had on recA expression. Nonetheless, real-time PCR did indeed indicate that recA was upregulated (2.4-fold) in mitomycin C-challenged cells, as opposed to the case in untreated cells. As shown in Table S6 in the supplemental material, 453 transcripts decreased in response to SOS-inducing conditions.
Global effects of stress responses on RNA half-lives.
As stated above, studies have linked stress response-mediated changes in protein production to alterations in target transcript mRNA stability, suggesting that modulating mRNA turnover plays a role in bacterial adaptability to environmental challenges. Admittedly, most of those studies have been limited to a few transcripts. Nonetheless, we set out to determine whether induction of the S. aureus cold shock, heat shock, stringent, or SOS response globally influences mRNA turnover. To do so, either log-phase UAMS-1 cells were mock treated or the corresponding stress response was induced (conditions described above). Rifampin was then added to inhibit de novo transcript synthesis, as previously described (46). Aliquots were removed at 0, 2.5, 5.0, 15, and 30 min post-transcriptional arrest, and cell viability and rifampin resistance were measured (see Materials and Methods). Total bacterial RNA was isolated, and the mRNA half-lives of transcripts produced in mock-treated, cold-shocked, heat-shocked, stringent response-induced, and SOS response-induced cells were determined using Affymetrix S. aureus GeneChips as previously described (46, 49).
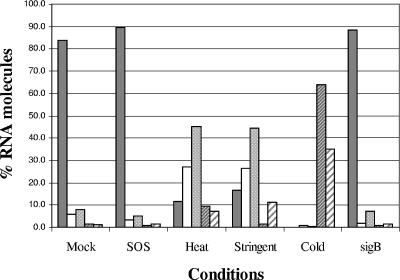
The results (Fig. 1) indicate that log-phase transcripts are degraded rapidly within mock-treated cells; 89.7% of all transcripts had half-lives of ≤5 min, 206 transcripts (9.2%) had intermediate half-lives (>5 min but ≤30 min), and 25 (1.1%) RNA species were stable (half-lives of >30 min). These results are in agreement with previous studies using custom-made S. aureus GeneChips (Saur2a), which found that the half-lives of 89.6% of all UAMS-1 log-phase transcripts were <5 min, those of 9.5% of transcripts were intermediate, and those of 0.7% of transcripts were stable (>60 min) (46). Induction of the SOS response did not appreciably affect global RNA turnover properties, whereas induction of the heat shock, cold shock, and stringent responses appeared to dramatically stabilize RNA species (Fig. 1). Within heat-shocked cells, 38.5% of log-phase transcripts had half-lives of ≤5 min, 54.5% demonstrated intermediate rates of mRNA turnover, and 7.1% were stable. Similarly, the half-lives of transcripts in stringent response-induced cells were as follows: 42.9% were ≤5 min, 45.7% were intermediate, and 11.4% were stable. Cold-shocked cells had a unique RNA turnover profile, as only 0.7% of transcripts had half-lives of ≤5 min, while the majority (64.1%) had half-lives of between 5 and 30 min or were stable (35.1%).
FIG. 1.
Global RNA turnover properties of S. aureus log-phase transcripts within untreated (mock) cells and under SOS response-, heat shock-, stringent response-, and cold shock-inducing conditions. RNA degradation properties of sigB-deficient cells are also plotted. Percentages of total transcripts with RNA half-lives of <2.5 min (gray bars), 2.5 to 5 min (white bars), 5 to 15 min (dotted bars), 15 to 30 min (hatched bars), and >30 min (widely hatched bars) are shown.
One possible explanation for the observed stress-mediated increases in mRNA stability could be that rifampin is not active within heat-shocked, cold-shocked, and stringent response-induced cells. Indeed, most of these stress conditions induce transport functions and/or drug efflux pumps (stringent response; NorA and MepA). However, several lines of evidence suggest that the efflux of rifampin does not contribute to this phenomenon and that the antibiotic arrests de novo transcript synthesis under each stress condition. First, Williams and Piddock have shown that efflux inhibitors do not influence rifampin accumulation within S. aureus cells (63). Second, a large portion of the transcriptome is rapidly degraded (half-lives of ≤5 min) under both heat shock and stringent response conditions, suggesting that de novo transcript synthesis is arrested. Third, rifampin challenge has similar effects on unstressed and stressed cell proliferation at 2.5, 5.0, 15, and 30 min post-rifampin treatment (data not shown). Moreover, the results in Fig. 2 demonstrate that induction of the stringent response confers resistance to the fluoroquinolone ciprofloxacin, presumably via norA and/or mepA upregulation, but does not reduce rifampin susceptibility. More specifically, UAMS-1 viability was decreased 2,600-fold during prolonged exposure (3 h) to ciprofloxacin (Fig. 2A). In contrast, induction of the stringent response decreased UAMS-1 susceptibility to ciprofloxacin, resulting in a threefold reduction in cell viability (Fig. 2B). In contrast, induction of the stringent response had no measurable effect on S. aureus rifampin susceptibility; cells challenged with rifampin demonstrated dramatic reductions in cell viability, in both stringent response-induced and noninduced cells (compare Fig. 2A and B). Finally, these results fit directly with studies of other organisms (53). Collectively, these results suggest that the stringent, cold shock, and heat shock responses influence molecular components that influence mRNA turnover in S. aureus cells.
FIG. 2.
Stringent response-inducing conditions decrease S. aureus susceptibility to ciprofloxacin but not rifampin. The graphs show cell viabilities of unstressed (A) and mupirocin-treated (B) log-phase S. aureus UAMS-1 cells (0 h) in the absence (diamonds) or presence of either rifampin (triangles) or ciprofloxacin (squares). Cell viability was monitored for 4 h and then plotted.
The global increases in mRNA stability in stringent response-induced, cold-shocked, and heat-shocked cells could be explained by two other scenarios, as follows: (i) these stress conditions induce/activate cellular RNA-stabilizing capacities or (ii) the conditions repress RNase production/function. Interestingly, we found that homologues of putative B. subtilis RNase genes (15 genes) are positively regulated by the alternative sigma factor σB (P. M. Dunman, P. D. Olson, and K. L. Anderson, unpublished data). However, as shown in Fig. 1, no global differences in mRNA turnover were observed between UAMS-1 sigB+ and UAMS-1 sigB mutant cells. Thus, it is likely that stress-induced σB-dependent alterations in RNase expression do not account for differences in mRNA stability.
Correlation between stress responses and target transcript stability.
Table S6 in the supplemental material lists all loci represented on the S. aureus GeneChip that were up- or downregulated by each stress response and the RNA half-lives of these transcripts under each stress condition as well as in mock-treated cells. A comparison of stress-mediated changes in transcript titers and their corresponding RNA turnover properties indicated that stress response-dependent alterations in transcript abundances can be attributed, in part, to alterations in RNA stability. In other words, induction of a stress response appears to alter both transcript synthesis and stability, suggesting that modulating RNA turnover may be an important component of the ability of S. aureus to cope with environmental challenges.
More specifically, as shown in Table S6 in the supplemental material, 164 of the 277 stringent response-induced transcripts are also expressed in mock-treated cells. A comparison of their RNA half-lives indicated that 147 (89.6%) of these transcripts are more stable when the stringent response is elicited than in mock-treated cells. Similarly, 65 heat shock-induced transcripts were also detected within mock-treated cells. Sixty-three (96.9%) of these transcripts were more stable in heat-shocked cells than in mock-treated cells. Most (56%) cold shock-induced transcripts were not detected in mock-treated cells, and thus their RNA half-lives could not be compared. The remaining 28 transcripts were more stable under cold shock conditions than in mock-treated cells. Sixty-two SOS-induced transcripts were detected within mock-treated cells. A comparison of their half-lives found that five (8%) SOS-induced transcripts were more stable in mitomycin C-stressed cells than in mock-treated cells. Real-time PCR was used to validate these results in part. As shown in Table 6, GeneChip-based RNA turnover measurements correlated with real-time PCR-determined RNA half-lives for each mRNA species analyzed.
TABLE 6.
Comparison of real-time PCR- and GeneChip-based mRNA half-life determinations
| Assay | mRNA half-life (min)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Cold shock
|
Heat shock
|
Stringent response
|
SOS response
|
|||||
| srtA | cspB | clpC | ctsR | sarR | norA | recA | uvrB | |
| GeneChip microarray | 15-30 | ≥30 | ≥30 | ≥30 | 15-30 | ≥30 | ≤2.5 | ≤2.5 |
| Real-time PCR | 30 | 30 | 15 | 15-30 | 15 | 15-30 | 2.5 | 2.5 |
We also investigated whether stress response-mediated decreases in transcript titers correlated with increased rates of mRNA turnover. Although induction of the stringent response globally increased RNA stability (Fig. 1), 20% of the stringent response-downregulated transcripts were degraded by the first posttranscriptional arrest sampling time (half-lives of ≤2.5 min) or were more rapidly degraded under stringent response conditions than under mock treatment conditions (see Table S6 in the supplemental material). Forty-three genes were downregulated within heat-shocked cells. Of these, 8 transcripts were not detected under heat shock conditions, and a comparison of mRNA half-lives of the remaining 35 transcripts indicated that 3 (8%) were less stable within heat-shocked cells or had half-lives of ≤2.5 min. No cold shock-repressed transcript demonstrated more rapid turnover under cold shock conditions. RNA turnover was more rapid under SOS-induced conditions than in mock-treated cells for 15% of the SOS-repressed transcripts.
Stable RNA species.
We have previously shown that log-phase UAMS-1 cells produce a set of SSR (half-lives of >60 min) molecules that are not expected to code for protein products (46). Given the importance of the S. aureus agr-encoded RNAIII molecule and small noncoding RNAs within other pathogens, it is likely that many of these molecules play an important role(s) in S. aureus biological processes. As shown in Table S8 in the supplemental material, 126 stable transcripts (half-lives of >30 min) that map to short S. aureus intergenic regions but have no defined function were identified to be produced under mock and/or stressed conditions.
Most SSRs were found to be stress-responsive; 12 small stable RNAs were produced in untreated cells, whereas 90% of SSRs were produced in response to stress. More specifically, two RNA species were stable under all conditions examined. Six transcripts were stable under four of the five conditions. Five transcripts were stable under three conditions, with the majority of these (4) being stable under stringent response, cold shock, and heat shock conditions but having half-lives of between 2.5 and 15 min under mock treatment and SOS-induced conditions. Eleven RNA species were stable under two conditions studied, and 102 transcripts were stable under one condition. Based on the surrounding genomic content and directionality of each SSR, it is likely that some stable RNA molecules are cotranscribed as part of an operon, whereas others are more likely to behave as antisense RNA molecules.
DISCUSSION
Infection is a dynamic process, during which the invading organism is subjected to an array of environmental challenges. Bacteria have developed highly orchestrated processes to respond to environmental stresses, which when elicited alter the cellular physiology in a manner that enhances the organism's survival and its ability to cause disease. Much effort has been devoted toward defining the members of bacterial stress responses by identifying genes whose transcript synthesis is controlled in a stress-specific manner. Only recently has it become recognized that modulation of mRNA degradation is a highly regulated process that also plays an essential role in many stress responses.
Despite S. aureus being a leading cause of nosocomial and community-acquired infections, surprisingly little is known about S. aureus stress responses. Here we have used Affymetrix S. aureus GeneChips to define transcript species that are altered under cold shock, heat shock, stringent, and SOS response-inducing conditions. In addition, we have defined the mRNA turnover characteristics of each response and identified a set of small stable RNA molecules with no obvious open reading frames that are produced as a component of each stress response.
Collectively, our results suggest that S. aureus stress response-dependent alterations in transcript abundances can be attributed, in part, to alterations in RNA stability. This was especially true for conditions of heat shock, cold shock, and stringent response induction, where most (89 to 100%) stress-induced transcripts had increased stability compared to those in untreated cells. Admittedly, it is not yet clear what, if any, effect the observed alterations in transcript stability have on protein production. However, studies of other organisms suggested that modulation of RNA turnover directly influences protein abundance. Thus, it seems likely that the ability of S. aureus to modulate mRNA turnover in a stress-responsive manner correlates with changes at the protein level. Nonetheless, currently it would be premature to interpret the effects of stress-mediated mRNA stabilization on the cellular physiology of S. aureus cells, simply because we do not know whether increases in transcript stability increase or decrease protein production (of all or subsets of mRNA species). However, our expression data do provide many insights about how S. aureus copes with various types of stress.
In general, our results suggest that, like the case for other bacteria, S. aureus stress responses are distinct, but response members do overlap. Moreover, there is a high degree of similarity between the ways that different bacteria cope with environmental stresses. For instance, induction of the S. aureus cold shock response profoundly stabilized most RNA species, increased the transcript titer of the SOS repressor LexA, and decreased expression of the stringent response control factor relA. This suggests that cold shock conditions repress both SOS and stringent responses. Studies have demonstrated that cold-shocked E. coli cells behave similarly, as low temperatures increase RNA stability and reduce RelA activity (61). Although low temperature appears to repress the stringent response, 27 S. aureus cold shock response genes were also components of the stringent response (see Table S6 in the supplemental material), indicating that they may represent members of a generalized stress response. Included among these 27 genes was the cold shock factor cspB. In E. coli, CspB and the major cold shock protein, CspA, are believed to act as RNA chaperones, although their RNA binding specificities differ (25, 43). Transcription of CspA was only marginally induced in cold-shocked cells, yet cspA mRNA was profoundly stabilized during low-temperature growth (half-life of <2.5 min at 37°C versus >30 min at 10°C), which based on E. coli cspA studies, suggests that CspA production was increased. Indeed, preliminary proteomics studies indicated that CspA levels are dramatically increased within cold-shocked UAMS-1 cells (S. Slater and P. M. Dunman, unpublished). Interestingly, cspA mRNA was also significantly stabilized in stringent response-induced cells (see Table S6 in the supplemental material), with an intermediate half-life. Given that cspB expression and cspA mRNA stability correlate with decreased mRNA turnover within both cold-shocked and stringent response-induced cells, it is conceivable that CspB and/or CspA may directly modulate transcript stability.
Induction of the cold shock response primarily increased the transcription of genes with no previously determined function. These gene products may play a role in rescuing stalled ribosomes, which is a requirement for cellular survival at low temperatures. Cold shock conditions also induced the expression of a protein with an S1 RNA binding domain, which is thought to mediate single-stranded RNA and RNA-pseudoknot binding (45) and may contribute to RNA stability at low temperatures. As shown in Fig. 3A, the predominant effect of cold shock conditions was the general decrease in mRNA titers involved in most cellular functions, despite globally increasing mRNA stability. This suggests that low temperatures promote basal S. aureus gene expression, but because RNA species are stable, templates for translation are available for the cell to efficiently respond to changes in growth conditions without having to expend energy for de novo gene expression.
FIG. 3.
Biological processes that are regulated in response to cold shock (A), heat shock (B), stringent response-inducing (C), and SOS response-inducing (D) conditions.
Heat shock conditions induced clpB and clpC transcription. ClpC is proposed to play a role in targeting heat-denatured proteins for degradation by ClpP (15). ClpB is believed to interact with DnaK and other heat shock proteins to mediate solubilization of protein aggregates (19, 37, 38). Although neither clpP nor dnaK transcripts were induced by the heat shock conditions studied, their transcripts were stabilized at the higher temperature (data not shown). The upregulation of clpB and clpC suggests that, like the case for other bacteria, coping with aberrant proteins is a vital component of S. aureus's ability to endure elevated temperatures. The urease operon (ureA-ureG), whose gene products convert urea to ammonia and CO2, was also upregulated at the high temperature. The thermodependent protein denaturation properties of urea have been well documented. Thus, it is conceivable that heat-shocked cells may guard against urea-based protein denaturation by metabolizing endogenous urea. As shown in Fig. 3B, amino acid biosynthetic genes are in the largest class of genes that are upregulated at elevated temperatures, suggesting that replenishing substrates for protein synthesis is also a priority under heat shock conditions. Paradoxically, the heat shock repressor protein CtsR was also upregulated under heat shock conditions. Frees and colleagues observed this same phenomenon and suggested that CtsR may need a cofactor for repressor function (15). No obvious contributor to the observed increase in heat shock-mediated mRNA stability was found. It is possible that elevated temperatures cause the denaturation of mRNA secondary structures that are recognized by the S. aureus RNA degradation machinery.
Induction of the SOS response increased the expression of nucleotide excision repair pathway and recombinational repair components, suggesting that, like the case for other organisms, repairing DNA damage is an important aspect of S. aureus SOS-induced cells. In a related study, the S. aureus transcriptional response to the oxidizing agent hydrogen peroxide, which also induces the bacterial SOS response, was determined for strain NCTC 8325 (9). The current work differs significantly from that study in several respects. First, H2O2 damages DNA, lipids, and proteins (50). Thus, in addition to inducing the SOS response, H2O2 also induces other bacterial oxidative stress responses (50), whereas the SOS-inducing agent used here, mitomycin C, primarily causes DNA damage (54, 56). Second, the former study was based on S. aureus strain NCTC 8325, which is functionally deficient for production of the primary stress response sigma factor, SigB (17). SigB has been shown to modulate the expression of a number of genes, including many that contribute to S. aureus pathogenesis (4). Moreover, Giachino and colleagues have shown that by virtue of their SigB deficiency, NCTC 8325 cells are less tolerant to DNA-damaging agents, suggesting that the strain may also be deficient in SOS functions (17). Comparison of our results to the transcriptional changes within H2O2-challenged NCTC 8325 cells suggested that SigB does not play a major role in DNA damage-mediated repair functions or the replication of DNA lesions; both studies found that repair components and a homologue to the replication protein UmuC (incorrectly annotated as UvrA in the preceding study) are induced (30). The current work also suggests that SOS induction activates the expression of numerous bacteriophage (Fig. 3D) and pathogenicity island genes. The concerted functions of these factors may account, in part, for the observation that SOS induction results in phage-mediated pathogenicity island dissemination among staphylococci (34, 55).
Striking features were observed within S. aureus stringent response-induced cells. As shown in Fig. 3C, amino acid biosynthetic processes were dramatically upregulated, and members of the translation machinery were downregulated (25 genes), mimicking the effects in mupirocin-treated E. coli cells (47). This implies that S. aureus copes with nutrient-limiting conditions by slowing bulk protein synthesis while increasing the cellular concentration of free amino acids. It was also observed that secreted proteases (i.e., SspA, SspB, and SspC) and transport processes are upregulated (Table 4). These functions may constitute another mechanism by which S. aureus increases its cellular amino acid concentration; the organism may increase digestion of extracellular proteins within its milieu and transport degradation products into the cell to bolster the abundance of amino acids. Another aspect of induction of the stringent response was the profound increase in mRNA stability of most transcripts, but as stated above, we do not yet know the consequences of this phenotype. Interestingly, we found that 60 μg ml−1 mupirocin is optimal for stringent response induction within UAMS-1 cells, which produce high levels of SigB. Crosse and colleagues independently found that the same experimental conditions effectively induce the stringent response within 8325-4 cells, which are SigB deficient (11). This suggests that SigB is not likely to play a major role in stringent response induction.
Each stress condition studied caused alterations in virulence factor expression. In general, S. aureus virulence determinants are expressed in a cell density-dependent manner. Cell surface components that are involved in attachment to host tissue are expressed within exponential-phase cultures, whereas extracellular virulence enzymes/toxins are preferentially produced during the postexponential growth phase (40). Within cold-shocked log-phase cells, extracellular virulence factor regulators (mgrA, sarA, and saeRS) were significantly downregulated in comparison to those in cells grown at 37°C, presumably ensuring low-level production of extracellular virulence determinants. The expression of cell surface virulence factors, including clfA, clfB, and fnbA, did not change at the low temperature. The best-characterized upregulated virulence determinant was sortase (srtA), which is an enzyme that tethers cell surface components (such as host attachment components) to the cell wall (35). Taken together, these results suggest that cold-shocked cells are poised to express cell surface factors on their exterior. Within heat-shocked cells, clpC and the urease operon were upregulated, and as stated above, it is possible that these factors influence the protein turnover properties of the cell. A number of uncharacterized pathogenicity island genes and the alpha-hemolysin gene (hla) were also upregulated at elevated temperature. No known virulence determinants were downregulated under heat shock conditions. Further characterization of the heat-dependent increase in overall mRNA stability may provide a better understanding of pathogenic features of heat-shocked cells. Induction of the S. aureus stringent response upregulated the production of various extracellular virulence determinants, including several proteases. This suggests that stringent response cells are poised to degrade host tissues.
The importance of short noncoding RNAs (microRNAs) in regulating eukaryotic cell development, cell death, and chromosome silencing is well documented. Recent studies have demonstrated that microRNAs also play essential roles in prokaryotic processes, including the regulation of bacterial stress responses and pathogenesis (reviewed in reference 20). For instance, noncoding RNAs are required for Vibrio sp. quorum sensing and Pseudomonas aeruginosa iron homeostasis (31, 62). Within E. coli, over 60 microRNAs have been identified, most of which are thought to act by binding to a protein and modifying its activity or by base pairing with mRNAs and affecting target transcript stability and translation (reviewed in reference 21). We recently identified eight putative S. aureus noncoding small stable RNA molecules (46), which taken together with the results of the current study, suggests that S. aureus produces an array of SSRs both during log-phase growth and in a stress-responsive manner. Based on the genomic context of SSRs, it is likely that many behave as antisense molecules, whereas others are components of operons (see Table S8 in the supplemental material). Studies are currently under way to better characterize these molecules and determine what influence, if any, they have on S. aureus biological processes and stress responses.
Supplementary Material
Acknowledgments
We thank Gail Crowell for technical assistance.
This work was partially supported by a University of Nebraska Medical Center Assistantship to K.L.A.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altuvia, S., D. Weinstein-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43-53. [DOI] [PubMed] [Google Scholar]
- 2.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blevins, J. S., M. O. Elasri, S. D. Allmendinger, K. E. Beenken, R. A. Skinner, J. R. Thomas, and M. S. Smeltzer. 2003. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 71:516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 7.Cassat, J. E., P. M. Dunman, F. McAleese, E. Murphy, S. J. Projan, and M. S. Smeltzer. 2004. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J. Bacteriol. 187:576-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassels, R., B. Oliva, and D. Knowles. 1995. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 177:5161-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, W., D. A. Small, F. Toghrol, and W. E. Bentley. 2006. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188:1648-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, J. J., G. P. Roberts, and W. J. Brill. 1986. Posttranscriptional control of Klebsiella pneumoniae nif mRNA stability by the nifL product. J. Bacteriol. 168:173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosse, A. M., D. L. Greenway, and R. R. England. 2000. Accumulation of ppGpp and ppGp in Staphylococcus aureus 8325-4 following nutrient starvation. Lett. Appl. Microbiol. 31:332-337. [DOI] [PubMed] [Google Scholar]
- 12.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 13.Eisen, J. A., and P. C. Hanawalt. 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435:171-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang, L., W. Jiang, W. Bae, and M. Inouye. 1997. Promoter-independent cold-shock induction of cspA and its derepression at 37 degrees C by mRNA stabilization. Mol. Microbiol. 23:355-364. [DOI] [PubMed] [Google Scholar]
- 15.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 16.Fry, R. C., T. J. Begley, and L. D. Samson. 2005. Genome-wide responses to DNA-damaging agents. Annu. Rev. Microbiol. 59:357-377. [DOI] [PubMed] [Google Scholar]
- 17.Giachino, P., S. Engelmann, and M. Bischoff. 2001. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goloubinoff, P., A. Mogk, A. P. Zvi, T. Tomoyasu, and B. Bukau. 1999. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 96:13732-13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399-404. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 22.Gottesman, S., and M. R. Maurizi. 2001. Cell biology. Surviving starvation. Science 293:614-615. [DOI] [PubMed] [Google Scholar]
- 23.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudas, L. J., and D. W. Mount. 1977. Identification of the recA (tif) gene product of Escherichia coli. Proc. Natl. Acad. Sci. USA 74:5280-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 26.Jones, P. G., M. Mitta, Y. Kim, W. Jiang, and M. Inouye. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:76-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaatz, G. W., F. McAleese, and S. M. Seo. 2005. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49:1857-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn, D., M. Hawkins, and R. R. Eady. 1982. Metabolic control of Klebsiella pneumoniae mRNA degradation by the availability of fixed nitrogen. J. Gen. Microbiol. 128:3011-3018. [DOI] [PubMed] [Google Scholar]
- 29.Kaluza, K., and H. Hennecke. 1981. Regulation of nitrogenase messenger RNA synthesis and stability in Klebsiella pneumoniae. Arch. Microbiol. 130:38-43. [DOI] [PubMed] [Google Scholar]
- 30.Koch, W. H., A. R. Fernandez de Henestrosa, and R. Woodgate. 2000. Identification of mucAB-like homologs on two IncT plasmids, R394 and Rts-1. Mutat. Res. 457:1-13. [DOI] [PubMed] [Google Scholar]
- 31.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 32.Little, J. W., S. H. Edmiston, L. Z. Pacelli, and D. W. Mount. 1980. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc. Natl. Acad. Sci. USA 77:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Garcia, P., and P. Forterre. 2000. DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. Bioessays 22:738-746. [DOI] [PubMed] [Google Scholar]
- 34.Maiques, E., C. Ubeda, S. Campoy, N. Salvador, I. Lasa, R. P. Novick, J. Barbe, and J. R. Penades. 2006. β-Lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 188:2726-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 36.McAleese, F., P. Petersen, A. Ruzin, P. M. Dunman, E. Murphy, S. J. Projan, and P. A. Bradford. 2005. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother. 49:1865-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mogk, A., E. Deuerling, S. Vorderwulbecke, E. Vierling, and B. Bukau. 2003. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50:585-595. [DOI] [PubMed] [Google Scholar]
- 38.Mogk, A., T. Tomoyasu, P. Goloubinoff, S. Rudiger, D. Roder, H. Langen, and B. Bukau. 1999. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18:6934-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 40.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 41.Palom, Y., G. Suresh Kumar, L. Q. Tang, M. M. Paz, S. M. Musser, S. Rockwell, and M. Tomasz. 2002. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbamoyl mitomycin C and mitomycin C. Chem. Res. Toxicol. 15:1398-1406. [DOI] [PubMed] [Google Scholar]
- 42.Pao, C. C., P. P. Dennis, and J. A. Gallant. 1980. Regulation of ribosomal and transfer RNA synthesis by guanosine 5′-diphosphate-3′-monophosphate. J. Biol. Chem. 255:1830-1833. [PubMed] [Google Scholar]
- 43.Phadtare, S., and M. Inouye. 1999. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 33:1004-1014. [DOI] [PubMed] [Google Scholar]
- 44.Pham, P., S. Rangarajan, R. Woodgate, and M. F. Goodman. 2001. Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:8350-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ringquist, S., T. Jones, E. E. Snyder, T. Gibson, I. Boni, and L. Gold. 1995. High-affinity RNA ligands to Escherichia coli ribosomes and ribosomal protein S1: comparison of natural and unnatural binding sites. Biochemistry 34:3640-3648. [DOI] [PubMed] [Google Scholar]
- 46.Roberts, C., K. L. Anderson, E. Murphy, S. J. Projan, W. Mounts, B. Hurlburt, M. Smeltzer, R. Overbeek, T. Disz, and P. M. Dunman. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 188:2593-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabina, J., N. Dover, L. J. Templeton, D. R. Smulski, D. Soll, and R. A. LaRossa. 2003. Interfering with different steps of protein synthesis explored by transcriptional profiling of Escherichia coli K-12. J. Bacteriol. 185:6158-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schendel, P. F., M. Fogliano, and L. D. Strausbaugh. 1982. Regulation of the Escherichia coli K-12 uvrB operon. J. Bacteriol. 150:676-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selinger, D. W., R. M. Saxena, K. J. Cheung, G. M. Church, and C. Rosenow. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semchyshyn, H., T. Bagnyukova, K. Storey, and V. Lushchak. 2005. Hydrogen peroxide increases the activities of soxRS regulon enzymes and the levels of oxidized proteins and lipids in Escherichia coli. Cell Biol. Int. 29:898-902. [DOI] [PubMed] [Google Scholar]
- 51.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 52.Stephens, J. C., S. W. Artz, and B. N. Ames. 1975. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino-acid deficiency. Proc. Natl. Acad. Sci. USA 72:4389-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takayama, K., and S. Kjelleberg. 2000. The role of RNA stability during bacterial stress responses and starvation. Environ. Microbiol. 2:355-365. [DOI] [PubMed] [Google Scholar]
- 54.Tomasz, M. 1995. Mitomycin C: small, fast and deadly (but very selective). Chem. Biol. 2:575-579. [DOI] [PubMed] [Google Scholar]
- 55.Ubeda, C., E. Maiques, E. Knecht, I. Lasa, R. P. Novick, and J. R. Penades. 2005. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 56:836-844. [DOI] [PubMed] [Google Scholar]
- 56.Utzat, C. D., C. C. Clement, L. A. Ramos, A. Das, M. Tomasz, and A. K. Basu. 2005. DNA adduct of the mitomycin C metabolite 2,7-diaminomitosene is a nontoxic and nonmutagenic DNA lesion in vitro and in vivo. Chem. Res. Toxicol. 18:213-223. [DOI] [PubMed] [Google Scholar]
- 57.Vasina, J. A., and F. Baneyx. 1996. Recombinant protein expression at low temperatures under the transcriptional control of the major Escherichia coli cold shock promoter cspA. Appl. Environ. Microbiol. 62:1444-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voyich, J. M., K. R. Braughton, D. E. Sturdevant, A. R. Whitney, B. Said-Salim, S. F. Porcella, R. D. Long, D. W. Dorward, D. J. Gardner, B. N. Kreiswirth, J. M. Musser, and F. R. DeLeo. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907-3919. [DOI] [PubMed] [Google Scholar]
- 59.Walker, G. C. 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C.
- 60.Wassarman, K. M. 2002. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109:141-144. [DOI] [PubMed] [Google Scholar]
- 61.Wick, L. M., and T. Egli. 2004. Molecular components of physiological stress responses in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89:1-45. [DOI] [PubMed] [Google Scholar]
- 62.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams, K. J., and L. J. Piddock. 1998. Accumulation of rifampicin by Escherichia coli and Staphylococcus aureus. J. Antimicrob. Chemother. 42:597-603. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 172:6942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.